Abstract
Activation of caspases is a hallmark of apoptosis. Several methods, therefore, were developed to identify and count the frequency of apoptotic cells based on the detection of caspases activation. The method described in this chapter is based on the use of fluorochrome-labeled inhibitors of caspases (FLICA) applicable to fluorescence microscopy, and flow- and image-cytometry. Cell-permeant FLICA reagents tagged with carboxyfluorescein or sulforhodamine when applied to live cells in vitro or in vivo, exclusively label cells that are undergoing apoptosis. The FLICA labeling methodology is simple, rapid, robust, and can be combined with other markers of cell death for multiplexed analysis. Examples are presented on FLICA use in combination with a vital stain (propidium iodide), detection of the loss of mitochondrial electrochemical potential, and exposure of phosphatidylserine on the outer surface of plasma cell membrane using Annexin V fluorochrome conjugates. Following cell fixation and stoichiometric staining of cellular DNA, FLICA binding can be correlated with DNA ploidy, cell cycle phase, DNA fragmentation, and other apoptotic events whose detection requires cell permeabilization. The “time window” for the detection of apoptosis with FLICA is wider compared to that with the Annexin V binding, making FLICA a preferable marker for the detection of early phase apoptosis and more accurate for quantification of apoptotic cells.
Keywords: FLICA, Caspases, Apoptosis, Flow cytometry, Laser scanning cytometry, Cell death, Mitochondrial potential, Annexin V binding
1. Introduction
Caspases are cysteine-aspartic acid-specific proteases that are activated in response to different cell death-inducing stimuli (1). Their activation initiates specific cleavage of the respective target proteins and, therefore, is considered to be a marker of the irreversible steps leading to cell demise. Although there are exceptions (2), caspase activation is considered a characteristic event of apoptosis, and identification of apoptotic cells often relies on detection of their activity (3, 4). Caspases specifically recognize a four-amino acid sequence on their substrate proteins; the carboxyl end of aspartic acid within this sequence is the target for cleavage. Several approaches have been developed to detect the process of caspases activation. Because the activation involves the transcatalytic cleavage of the zymogen pro-caspases (1), the cleavage products having lower molecular weight than the zymogen can be revealed electrophoretically and identified in Western blots using caspase-specific antibodies. Another approach utilizes the fluorogenic (or chromogenic) substrates of caspases. Peptide substrates were developed, which are colorless or non-fluorescent, but upon cleavage, generate colored or fluorescing products (5, 6). These two approaches, however, are primarily used to detect caspases activation in cell extracts, thereby providing no information on their activation in individual cells that otherwise is needed to reveal heterogeneity of cell populations or to correlate caspases activation with other cell attributes such as cell cycle position.
Among the approaches that can be applied to study activation of caspases in situ are the methods based on immunocytochemical detection of the epitope that is characteristic of the caspases’ active form. Antibodies that react only with the activated caspases are now available and have been used in cytometric assays (7, 8). Activation of caspases also can be detected indirectly, by immunocytochemical identification of the specific cleavage products, e.g., the p89 fragment of poly(ADP-ribose) polymerase, and this method has been adapted to cytometry as well (9).
The method described here relies on the use of the fluorochrome-labeled inhibitors of caspases (FLICA; (10, 11)). The principle of this methodology was introduced long ago in the studies of the esterases and proteases utilizing radio-labeled specific inhibitors that bound to the active centers of these enzymes and were detected by autoradiography (12). In the case of caspases, the ligands that bind to their active centers are carboxyfluorescein (FAM)- or fluorescein (FITC)-tagged peptide-fluoromethyl ketone (FMK) inhibitors. These ketone reagents penetrate the plasma membrane of live cells and are relatively nontoxic to the cells, at least in short-term incubations (13). Actually, in some cell systems by virtue of caspase inhibition, FLICA slow down the process of apoptosis and prevent total cell disintegration, making it possible to quantify the frequency of apoptotic cells more accurately (apoptotic index; AI) (14). The recognition peptide moieties of these reagents were expected to provide some level of specificity of their binding to particular caspases. Thus the FLICA containing VDVAD, DEVD, VEID, YVAD, LETD, LEHD, and AEVD peptide moieties were expected to preferentially bind to activated caspases-2, -3, -6, -1, -8, -9, and -10, respectively. On the contrary, the FAM-VAD-FMK containing the valylalanyl aspartic acid sequence was designed to be pan-caspase reactive, binding to activated caspases-1, -3 -4, -5, -7, -8, and -9 (15). It was later observed, however, that because of the FMK group reactivity, likely with thiol groups of intracellular proteins that become available upon cleavage by caspases, binding of the sequence is not as caspase specific as initially thought (7, 16). Despite the apparent lack of specificity in labeling caspases, the FLICA probes have consistently shown themselves to be highly reliable reporters of caspases activation and convenient markers of apoptotic cells.
Exposure of live cells to FLICA results in the rapid uptake of these reagents followed by their covalent binding to apoptotic cells containing activated caspase enzymes. Unbound FLICA are removed from the nonapoptotic cells by rinsing with wash-buffer. The protocols given below describe labeling cells that contain activated caspases using FAM-VAD-FMK. The same protocol can be applied to other FLICAs. Concurrent exposure of cells to propidium iodide (PI) strongly labels all cells with a compromised plasma membrane integrity that cannot exclude this cationic dye; the loss of membrane integrity is a key feature of mid-late apoptotic cells or cells undergoing necrosis. On the contrary, simultaneous staining with the mitochondrial electrochemical potential probe chloromethyl-X-rosamine (CMXRos; MitoTracker Red) (17) and FLICA allows one to discriminate between two sequential events of apoptosis: dissipation of the inner mitochondrial membrane potential and activation of the caspase enzyme cascade, respectively. Likewise, simultaneous staining with FLICA and Annexin V conjugated to red fluorescing dyes such as Cy5 reveals the relationship between caspases activation and exposure of phosphatidylserine (PS) residues on the external surface of plasma membrane during apoptosis (7, 18). Cells labeled with FLICA and PI, CMXRos, or Annexin V–Cy5 can be examined by fluorescence microscopy or subjected to quantitative analysis by flow or image cytometry such as laser scanning cytometry (LSC). By virtue of the ability to measure large cell populations rapidly to analyze cell images, LSC is particularly useful in studies of apoptosis (19).
2. Materials
Cells to be analyzed: can be grown on slides (see Subheading 3.1) or in suspension.
Microscope slides, coverslips: see Subheading 3.1
Instrumentation: Fluorescence microscope, or multiparameter Laser Scanning Cytometer (LSC; iCys® Research Imaging Cytometer), available from CompuCyte, Inc. (Westwood, MA). Flow cytometers of different types, offered by several manufacturers, can be used to measure cell fluorescence following staining according to the procedures described below. The manufacturers of the most common flow cytometers are Beckman /Coulter Corporation (Miami, FL), BD Biosciences (formerly Becton Dickinson Immunocytometry Systems; San Jose, CA), iCyt (Urbana-Champain, IL), and PARTEC GmbH (Zurich, Switzerland). The software to deconvolute the DNA content frequency histograms, to analyze the cell cycle distributions, is available from Phoenix Flow Systems (San Diego, CA) or Verity Software House (Topham, MA).
Phosphate-buffered saline (PBS).
Dimethylsulfoxide (DMSO; Sigma Chemical Co., (St. Louis, MO)).
Stock solution of PI: dissolve 1 mg of PI (Invitrogen/Molecular Probes, Eugene, OR) in 1 mL of distilled water. This solution can be stored at 4°C in the dark for several months.
Stock FLICA solution: dissolve lyophilized FLICA (e.g., FAM-VAD-FMK; available as a component of the FLICA™ Apoptosis Detection (“Green FLICA”) kit from Immunochemistry Technologies LLC, Bloomington, MN) in dimethylsulfoxide (DMSO) as specified in the kit to obtain 150×concentrated (stock) solution of this inhibitor. Also available from this vendor are caspase-2 (VDVAD), caspase-3 (DEVD), caspase-6 (VEID), caspase-1 (YVAD), caspase-8 (LETD), caspase-9 (LEHD), and caspase-10 (AEVD) FLICA. Aliquots of FLICA solution may be stored at −20°C in the dark for 6 months.
Intermediate (30×concentrated) FLICA solution: prepare a 30× concentrated solution of FAM-VAD-FMK by diluting the stock solution 1:5 in PBS. Mix the vial until the contents become transparent and homogenous. This solution should be made freshly. Protect all FLICA solutions from light.
FLICA staining solution: just prior to the use, add 3 µL of 30× concentrated FAM-VAD-FMK solution into 100 µL of culture medium.
Rinsing solution: 1% (w/v) BSA in PBS.
Staining solution of PI: add 10 µL of stock solution of PI to 1 mL of the rinsing solution.
MitoTracker Red (Invitrogen/Molecular Probes).
Annexin V–Cy5 conjugate and the binding buffer (Abcam, Cambridge, MA, or Alexis Biochemicals, San Diego, CA).
3. Methods
3.1. Attachment of Cells to Slides (Cells to be Analyzed by Microscopy and/or LSC)
The procedure requires incubation of live (unfixed, not permeabilized) cells with solutions of FLICA. A variety of adherent cells are available for growth in cell culture flasks. Such cells can be attached to microscope slides by culturing them on slides or coverslips. Culture vessels having microscope slides at the bottom of the chamber are commercially available (e.g., “Chamberslide,” Nunc, Inc., Naperville, IL, or single- or multi-chambered Falcon CultureSlides, BD Biosciences). The cells growing in these chambers spread and attach to the slide surface after incubation at 37°C for several hours. Alternatively, the cells can be grown on coverslips, e.g., placed on the bottom of Petri dishes. The cover-slips are then inverted over shallow (<1 mm) wells on the microscope slides. The wells can be prepared by constructing the well walls (~2 × 1 cm size) either with a pen that deposits a hydrophobic barrier (“Isolator,” Shandon Scientific), nail polish, or melted paraffin. The wells also may be made by preparing a strip of Parafilm “M” (American National Can, Greenwich, CT) of the size of the slide, cutting a hole ~2 × 1 cm in the middle of this strip, placing the strip on the microscope slide, and heating the slide on a warm plate until the Parafilm starts to melt. It should be stressed, however, that because the cells detach during late stages of apoptosis, these cells may be selectively lost if the analysis is limited to attached cells.
Cells that normally grow in suspension can be attached to glass slides by electrostatic forces. This is due to the fact that sialic acid residues that cover the cell surface have a net negative charge in contrast to the glass surface which is positively charged. Incubation of cells on microscope slides in the absence of any serum or serum proteins (which otherwise neutralize the charge), thus, leads to their attachment. The cells taken from culture should be rinsed in PBS in order to remove serum proteins contained in the cell culture media and then resuspended in PBS at a concentration of 2 × 105 cells/mL. An aliquot (50–100 µL) of this suspension should be deposited within a shallow well (prepared as described above) on the horizontally placed microscope slide. To prevent drying, a small piece (~2 × 2 cm) of a thin polyethylene foil or Parafilm may be placed atop the cell suspension drop. A short (15–20 min) incubation of such cell suspension at room temperature in a closed box containing wet tissue or filter paper that provides 100% humidity is adequate to ensure that most cells firmly attach to the slide surface. Cells attached in this manner remain viable for several hours and can be subjected to surface immunophenotyping, viability tests, or intracellular enzyme kinetics assays. Such preparations can then be fixed (e.g., in formaldehyde) without significant loss of cells from the slide. However, as in the case of cell growth on glass, late apoptotic cells have a tendency to detach even after the initial attachment.
It should be stressed that the microscope slide to which the cells are going to be attached electrostatically should be extra clean. Fingerprints leave oils on the slide that interfere with cell attachment. To remove possible contamination of the glass surface that may interfere with cell attachment, soak the microscope slides in a household detergent, then rinse in water and 100% ethanol, respectively. The slides should be allowed to air dry and used the same day they were cleaned.
3.2. Cell Staining and Analysis by Microscopy or LSC(iCys® Research Imaging Cytometer)
Attach the cells to the microscope slide as described in Subheading 3.1. Keep the cells immersed in the culture medium by adding 100 µL of the medium (with 10% serum) into the well on the microscope slide to cover the area with the cells. In the case of cells growing on microscope slide chambers, move directly to step 2.
Remove the medium and replace it with 100 µL of 1 × FLICA (e.g., FAM-VAD-FMK) staining solution.
Place ~2 × 4 cm strip of Parafilm atop the staining solution to prevent drying. Incubate the slides horizontally for 1 h at 37°C in a closed box with wet tissue or filter paper to ensure 100% humidity, in the dark.
Remove the staining solution with Pasteur pipette. Rinse twice with the rinsing solution, each time adding a new aliquot, gently mixing, and after 2 min replacing with the next rinse.
Apply 100 µL of the PI staining solution atop the cells deposited on the slide. Cover with a coverslip and seal the edges to prevent drying (see Notes 1–4).
During the subsequent 30 min after the addition of PI solution, observe the cells under fluorescence microscope (blue light illumination) or measure cell fluorescence with LSC/iCys. Use the argon ion laser (488 nm) of LSC/iCys to excite fluorescence, contour on light scatter, and measure green fluorescence of FLICA at 530 ± 20 nm and red fluorescence of PI at >600 nm (Fig. 1) (See Note 5).
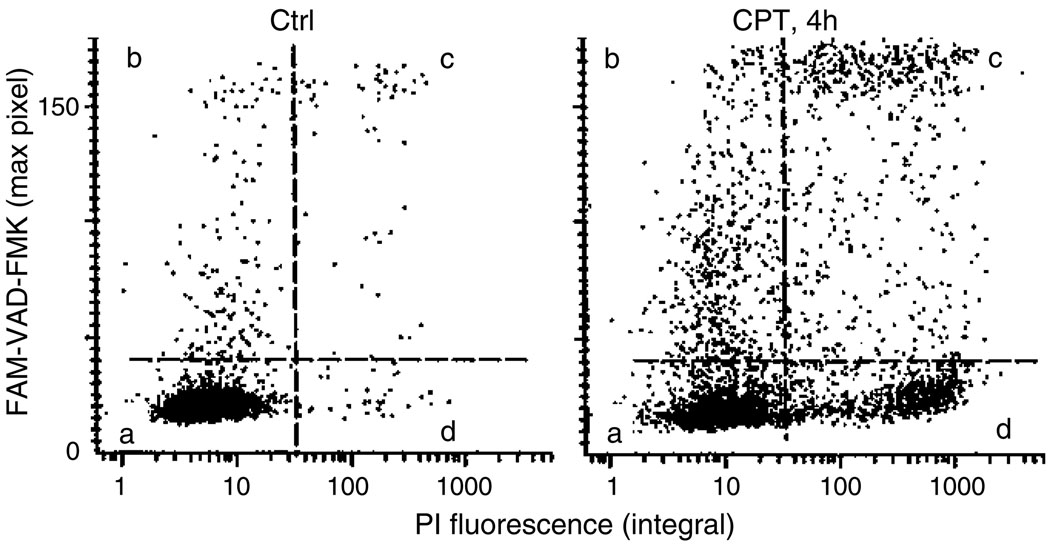
Fig. 1.
The bivariate distributions (scatterplots) of FAM-VAD-FMK (FLICA; green maximal pixel) vs. PI (red integral) fluorescence of the control (Ctrl) and CPT-treated HL-60 cells. To induce apoptosis, the cells were treated for 4 h with DNA topoisomerase I inhibitor camptothecin (0.15 µM; CPT). The cells were then stained according to the protocol (Subheading 3.2) and their fluorescence was measured by iCys® Research Imaging Cytometer. The live non-apoptotic cells, which are predominant in Ctrl, are unlabeled (quadrant a). Early apoptotic cells have increased FLICA fluorescence but minimal fluorescence of PI (quadrant b). The cells that are more advanced in apoptosis show variable degrees of both, FLICA and PI, fluorescence (quadrant c). At very late stages of apoptosis (“necrotic stage”), caspases either leak out of cells or are not reactive with FLICA, and the cells become FLICA negative/PI positive (quadrant d) (see Notes 6–11).
3.3. Cell Staining and Analysis by Flow Cytometry
Suspend 5 × 105–106 cells in 0.3 mL of complete culture medium (with 10% serum) in a centrifuge tube.
Add 10 µL of the 30× concentrated (“intermediate”) FLICA solution to this cell suspension. Mix the cell suspension by flicking the tube.
Incubate for 60 min at 37°C in atmosphere of air with 5% CO2, at 100% humidity, in the dark.
To the cell suspension with FLICA, add 5 mL of the rinsing solution (PBS with BSA) or 1× “wash buffer” provided with the FLICA kit and gently mix the cell suspension.
Centrifuge at 300 × g for 5 min at room temperature and remove supernatant by aspiration.
Resuspend cell pellet in 2 mL of the rinsing solution or in 1× “wash buffer.”
Centrifuge at 300 × g for 5 min and aspirate the supernatant (See Note 5).
Resuspend cells in 1 mL of the PI staining solution. Place the tube on ice (see Notes 2–4).
- Measure cell fluorescence by flow cytometry:
- Excite cell fluorescence with 488-nm laser line.
- Measure green fluorescence of FLICA at 530 ± 20 nm.
- Measure red fluorescence of PI at >600 nm.
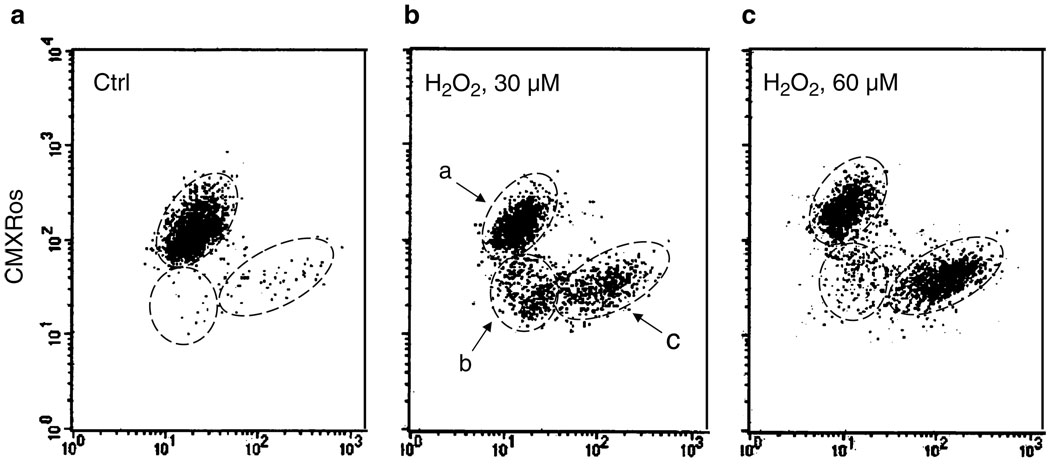
Fig. 2.
Concurrent detection of caspases activation by FLICA (FAM-VAD-FMK) and the loss of mitochondrial electrochemical potential (Δψm) during apoptosis. To induce apoptosis, human leukemic Jurkat cells were treated with 30 or 60 µM H2O2 for 7 h (7). The cells were then subjected to staining with FLICA and CMXRos (MitoTracker Red), and their fluorescence was measured by flow cytometry as described in the protocol (Subheading 3.3 and Note 3). Based on differential binding of FLICA and CMXRos, one can distinguish three cell subpopulations: (a) live, non-apoptotic cells; (b) cells having decreased mitochondrial potential (CMXRos binding), prior to caspases activation, and (c) cells showing both decreased mitochondrial potential and increased caspases activation (c). The data illustrate that the reduction of mitochondrial potential precedes FLICA binding. Treatment with 60 µM H2 O2 accelerates the process of apoptosis as evidenced by an increase in frequency of FLICA-positive cells. (For more details, see ref. 7) (see Note 9).
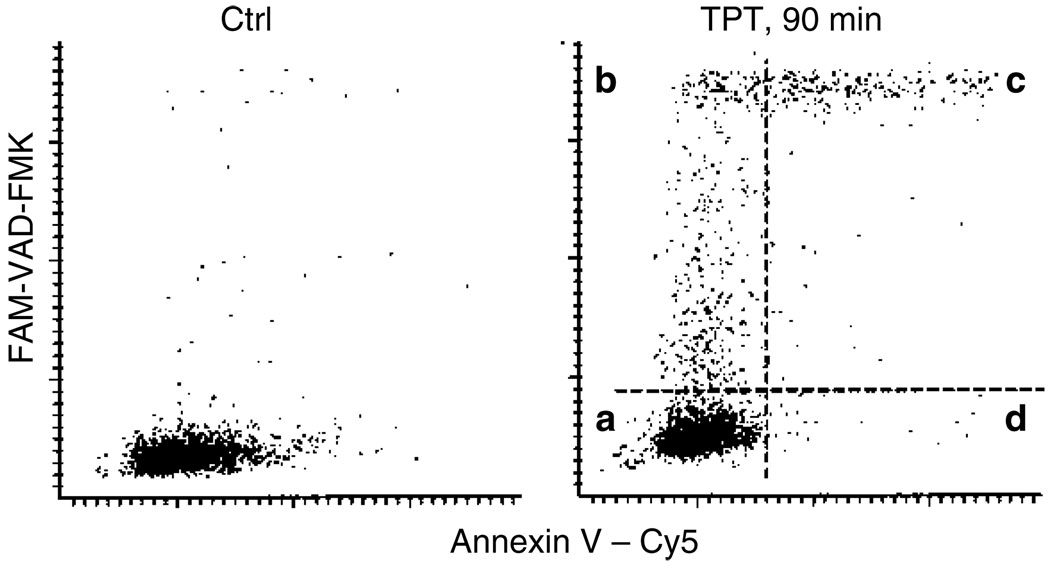
Fig. 3.
Relationship between caspases activation as detected by FLICA binding and externalization of phosphatidylserine revealed by Annexin V binding during apoptosis. Apoptosis of HL-60 cells was induced by treatment with the DNA topoisomerase I inhibitor topotecan (TPT, 0.15 µM, 90 min). The cells were then labeled with FLICA (FAM-VAD-FMK), subsequently with Annexin V–Cy5 conjugate, and their fluorescence was measured by iCys® Research Imaging Cytometer as described in the protocol (see Subheading 3.2 and Note 4). Four quadrants shown in the right panel identify cells that are (a) non-apoptotic, (b) early apoptotic cells that bind FLICA but do not bind Annexin V–Cy5, (c) apoptotic cells that bind both FLICA and Annexin V–Cy5, and (d) very late apoptotic or necrotic cells that are FLICA negative. The data show that the “time window” to detect apoptosis is wider for the FLICA assay as the early apoptotic cells are FLICA positive (b) and not detectable by the Annexin V–Cy5 assay (see Notes 6–11).
Acknowledgment
This study was supported by NCI grant RO1 28704.
Footnotes
Protect cells from light throughout the procedure.
Staining with PI is optional. It allows us to identify the cells that have compromised cell membrane integrity features. Cells bearing compromised cell membrane integrity structure (necrotic and mid-late apoptotic cells, cells with mechanically damaged membranes, isolated cell nuclei will stain red as a result of their inability to exclude PI (Fig. 1).
If concurrently with FLICA cells are stained with CMXRos (MitoTracker Red), instead of adding PI solution as described above, add 100 µL of PBS containing 0.2 µM CMXRos. Alternatively, the CMXRos potentiometric dye can be replaced by a tetramethylrhodamine derivative dye such as tetramethylrhodamine methyl ester perchlorate (TMRM, available from Sigma, Invitrogen/Molecular Probes, or in kit form from Immunochemistry Technologies). Cells can be stained with 0.4 µM of TMRM for 15 min at 37°C. After addition of either the CMXRos or TMRM potentiometric dyes, rinse the cells with rinsing solution, suspend in rinsing solution, and measure their green (FLICA) and red (CMXRos or TMRM) fluorescence the same way as in the case of cells stained with FLICA and PI (Fig. 2).
If concurrently with FLICA cells are stained with Annexin V–Cy5 conjugate, instead of adding PI solution, add 100 µL of the 1× binding buffer containing Annexin V–Cy5 at the concentration suggested by the vendor. Incubate for 15 min, rinse once with the binding buffer, and suspend in rinsing buffer. The calcium containing binding buffer must be used to maintain the affinity interaction between the Annexin and PS residues on the apoptotic cell membrane surface. Excite cell fluorescence with both 488 nm and red diode (647 nm) lasers and measure the emission of Cy5 at far red (>650 nm) wavelength (Fig. 3). This procedure, however, can be combined with staining with PI. High intensity of PI fluorescence at red wavelength (590–620 nm, excited at 488 nm) allows one to identify and gate out the cells with a damaged plasma membrane (PI positive) and analyze the PI negative cells with respect to their Annexin V–Cy5 and FLICA fluorescence.
After step 4 (Subheading 3.2) or step 7 (Subheading 3.3), the cells may be fixed in 1% formaldehyde (10 min on ice) followed by 70% ethanol and then subjected to staining with PI in the presence of RNase A or stained with 7-aminoactinomycin D. In this format, PI is used to stain the DNA of all cells to facilitate the assessment of DNA ploidy parameters. Analysis of the FLICA vs. PI fluorescence by LSC or flow cytometry allows for the correlation of caspases activation with cellular DNA content, i.e., the cell cycle position or DNA ploidy. Details of this procedure are provided in reference (7). Alternatively, when two-laser excitation is available and one of the lasers produces UV light the cellular DNA may be counterstained with Hoechst 33342 or with 4′, 6-diamidino-2-phenylindole (DAPI), both available from Invitrogen/Molecular Probes.
One has to keep in mind that FLICA are not passive reagents that mark the activated caspases, but react directly with the caspase by covalent interaction with the active site of the enzyme. This inhibits caspase activity, suppressing the process of apoptosis. Thus, the rate of apoptosis progression and all the events related to caspases activity are suppressed by FLICA (14).
Another problem that should be taken into an account when using FLICA to mark the activated caspases in live cells pertains to fragility of apoptotic cells. The flow cytometric assay requires incubation of live cells with these reagents followed by repeated rinsing to remove unbound FLICA from the non-apoptotic cells. Apoptotic cells, particularly at late stages of apoptosis, are fragile and may be preferentially lost during the centrifugations. A certain degree of stability is derived from the presence of serum (up to 20% v/v) or BSA (up to 2% w/v) in the rinsing buffers. Also, the cells should be sedimented with minimal g force and short centrifugation time. Because of a possibility of a selective loss of apoptotic cells, one has to be careful when drawing conclusions about the absolute frequency of apoptosis based on the percentage of FLICA positive cells in the samples assayed by flow cytometry. In the case of cell analysis by LSC/iCys® Research Imaging Cytometer, the propensity of apoptotic cells to detach in cultures and thus escape from analysis should also be taken into account, as it may favor a downward bias in the estimation of the apoptotic index as well. The above technical difficulties in the analysis of frequency of apoptosis pertain to any cytometric methodology, not only by FLICA, and are discussed in extent elsewhere (20).
It is difficult to assess the specificity of in situ bound individual FLICA sequences designed to be markers for their respective caspases in the light of the evidence of a nonspecific component in FLICA binding (7, 16). While activation of caspases is definitely associated with FLICA binding (7), it is likely that products of cleavage of other proteins by caspases have exposed thiol groups reactive with FLICA as well (16).
Also available are FLICA red fluorescing reagents containing sulforhodamine (SR) instead of FAM. Their availability extends multiparameter FLICA applications in combination with other markers.
A combination of FLICA with SYTO dyes offers an attractive marker of apoptosis concurrently revealing activation of caspases and condensation of nucleic acids (21).
It should be stressed that FLICA reagents at concentrations used to label apoptotic cells do not show toxicity to the non- apoptotic cells, at least in short-term (up to 48 h) in vitro experiments (13). Therefore, they are ideally suited to serve as in vivo markers of apoptotic cells. Defined as FLIVO™ in vivo apoptosis detection and imaging reagents (Immunochemistry Technologies, LLC), these reagents were successfully used to image and mark apoptotic cells in rats and mice following induction of apoptosis by different reagents (22–24). Increased levels of tumor cell apoptosis resulting from suppression of gp78 protein in sarcoma cells were assessed using in vivo injections of the SR-VAD-FMK (Red FLIVO) probe (24). These FLIVO probes were also used successfully in vivo, to measure increased levels of apoptosis in rat liver ischemia studies (25, 26).
Contributor Information
Zbigniew Darzynkiewicz, Email: darzynk@nymc.edu, Brander Cancer Research Institute, New York Medical College, Valhalla, NY, USA.
Piotr Pozarowski, Brander Cancer Research Institute, New York Medical College, Valhalla, NY, USA, Dept. of Clinical Immunology, Medical University, Lublin, Poland.
Brian W. Lee, Immunochemistry Technologies, 55431, Bloomington, MN, USA
Gary L. Johnson, Immunochemistry Technologies, 55431, Bloomington, MN, USA
References
- 1.Salvesen GS, Riedl SJ. Caspase mechanisms. Adv. Exp. Med. Biol. 2008;615:13–23. doi: 10.1007/978-1-4020-6554-5_2. [DOI] [PubMed] [Google Scholar]
- 2.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nunez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovski B, Melino G. Classification of cell death: recommendations of the nomenclature committee on cell death 2009. Cell Death Differ. 2008;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson DW. Caspase structure, proteolytic substrates and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 5.Komoriya A, Packard BZ, Brown MJ, Wu ML, Henkart PA. Assessment of caspase activities in intact apoptotic thymocytes using cell-permeable fluorogenic caspase substrates. J. Exp. Med. 2000;191:1819–1828. doi: 10.1084/jem.191.11.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee BW, Johnson GL, Hed SA, Darzynkiewicz Z, Talhouk JW, Mehrota S. DEVDase detection in intact apoptotic cells using the cell permeant fluorogenic substrate, (z-DEVD)2-cresyl violet. BioTechniques. 2003;35:1080–1085. doi: 10.2144/03355pf01. [DOI] [PubMed] [Google Scholar]
- 7.Pozarowski P, Huang X, Halicka DH, Lee B, Johnson G, Darzynkiewicz Z. Interactions of fluorochrome-labeled caspase inhibitors with apoptotic cells. A caution in data interpretation. Cytometry A. 2003;55A:50–60. doi: 10.1002/cyto.a.10074. [DOI] [PubMed] [Google Scholar]
- 8.Huang X, Okafuji M, Traganos F, Luther E, Holden E, Darzynkiewicz Z. Assessment of histone H2AX phosphorylation induced by DNA topoisomerase I and II inhibitors topotecan and mitoxantrone and by DNA crosslinking agent cisplatin. Cytometry A. 2004;58A:99–110. doi: 10.1002/cyto.a.20018. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Darzynkiewicz Z. Cleavage of poly(ADP-ribose) polymerase measured in situ in individual cells: relationship to DNA fragmentation and cell cycle position during apoptosis. Exp. Cell Res. 2000;255:125–132. doi: 10.1006/excr.1999.4796. [DOI] [PubMed] [Google Scholar]
- 10.Bedner E, Smolewski P, Amstad P, Darzynkiewicz Z. Activation of caspases measured in situ by binding of fluorochrome-labeled inhibitors of caspases (FLICA): correlation with DNA fragmentation. Exp. Cell Res. 2000;260:308–313. doi: 10.1006/excr.2000.4955. [DOI] [PubMed] [Google Scholar]
- 11.Smolewski P, Bedner E, Du L, Hsieh T-C, Wu JM, Phelps DJ, Darzynkiewicz Z. Detection of caspases activation by fluorochrome-labeled inhibitors: multiparameter analysis by laser scanning cytometry. Cytometry. 2001;44:73–82. doi: 10.1002/1097-0320(20010501)44:1<73::aid-cyto1084>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 12.Darzynkiewicz Z, Barnard EA. Specific proteases of mast cells. Nature. 1966;212:1198–1203. [Google Scholar]
- 13.Smolewski P, Grabarek J, Lee BW, Johnson GL, Darzynkiewicz Z. Kinetics of HL-60 cell entry to apoptosis during treatment with TNF-α or camptothecin assayed by the stathmo-apoptosis method. Cytometry. 2002;47:143–149. doi: 10.1002/cyto.10062. [DOI] [PubMed] [Google Scholar]
- 14.Smolewski P, Grabarek J, Phelps DJ, Darzynkiewicz Z. Stathmo-apoptosis: arresting apoptosis by fluorochrome-labeled inhibitor of caspases. Int. J. Oncol. 2001;19:657–663. doi: 10.3892/ijo.19.4.657. [DOI] [PubMed] [Google Scholar]
- 15.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Valliancourt JP, Chapman KT, Nicholson DW. A combinatorial approach defines specificities of members of the caspase family and granzyme B. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 16.Darzynkiewicz Z, Pozarowski P. All that glitters is not gold: all that binds FLICA is not caspase. A caution in data interpretation - and new opportunities. Cytometry A. 2007;71A:536–537. doi: 10.1002/cyto.a.20425. [DOI] [PubMed] [Google Scholar]
- 17.Pendergrass W, Wolf, Poot M. Efficacy of MitoTracker Green and CMXRosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry. 2004;61:162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- 18.Koopman G, Reutelingsperger CPM, Kuijten GAM, Kechnen RMJ, Pals ST, van Oers MHH. Annexin V for flow cytometric detection of phosphatidylderine expression of B cells undergoing apoptosis. Blood. 1994;84:1415–1420. [PubMed] [Google Scholar]
- 19.Pozarowski P, Holden E, Darzynkiewicz Z. Laser scanning cytometry. Principles and applications. Methods Mol. Biol. 2005;319:165–192. doi: 10.1007/978-1-59259-993-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darzynkiewicz Z, Bedner E, Traganos F. Difficulties and pitfalls in analysis of apoptosis. Methods Cell Biol. 2001;63:527–559. doi: 10.1016/s0091-679x(01)63028-0. [DOI] [PubMed] [Google Scholar]
- 21.Wlodkowic D, Skommer J, Hillier C, Darzynkiewicz Z. Multiparameter detection of apoptosis using red-excitable SYTO probes. Cytometry A. 2008;73A:563–569. doi: 10.1002/cyto.a.20564. [DOI] [PubMed] [Google Scholar]
- 22.Griffin RJ, Williams BW, Bischof JC, Olin M, Johnson GL, Lee BW. Use of fluorescently labeled poly-caspase inhibitor for in vivo detection of apoptosis related to vascular-targeting agent arsenic trioxide for cancer therapy. Technol. Cancer Res. Treat. 2007;6:651–654. doi: 10.1177/153303460700600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee BW, Olin MR, Johnson GL, Griffin RJ. In vitro and in vivo apoptosis detection using membrane permeant fluorescent-labeled inhibitors of caspases. Methods Mol. Biol. 2008;414:109–135. doi: 10.1007/978-1-59745-339-4_10. [DOI] [PubMed] [Google Scholar]
- 24.Tsai YC, Mendoza A, Mariano JM, Zhou M, Kostova Z, Chen B, Veenstra T, Hewitt SM, Helman LJ, Khanna C, Weissman AM. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat. Med. 2007;13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
- 25.Cursio R, Colosetti P, Auberger P, Gugenheim J. Liver apoptosis following normothermic ischemia-reperfusion: in vivo evaluation of caspase activity by FLIVO assay in rats. Transplant. Proc. 2008;40:2038–2041. doi: 10.1016/j.transproceed.2008.05.039. [DOI] [PubMed] [Google Scholar]
- 26.Cursio R, Miele C, Filippa N, Colosetti P, Auberger P, Van Obberghen E, Gugenheim J. Tyrosine phosphorylation of insulin receptor substrates during ischemia/reperfusion-induced apoptosis in rat liver. Langenbecks Arch. Surg. 2009;394:123–131. doi: 10.1007/s00423-008-0394-3. [DOI] [PubMed] [Google Scholar]