Abstract
The measles virus entry system, constituted of attachment (hemagglutinin, H) and fusion proteins, operates based on a variety of natural and targeted receptors. However, the mechanism triggering fusion of the viral envelope with the plasma membrane is not understood. Here we tested a model considering that the two heads of an H-dimer, which are covalently linked at their base, after binding two receptor molecules, move relative to each other to transmit the fusion-triggering signal. Indeed, stabilizing the H-dimer interface by additional inter-molecular disulfide bonds prevented membrane fusion, an effect reversed by a reducing agent. Moreover, a membrane-anchored designated receptor efficiently triggered fusion, provided it engaged the H-dimer at locations proximal to where the natural receptors bind, and distal to the H-dimer interface. We discuss how receptors may force H-heads to switch partners and transmit the fusion-triggering signal.
INTRODUCTION
Enveloped viruses have evolved proteins to fuse viral and cellular membranes. Many use trimeric fusion proteins that share structural characteristics with cellular proteins of similar function 1. Since refolding of most viral fusion proteins is irreversible, triggering must be strictly regulated. While many enveloped viruses, including influenza-, rhabdo-, alpha- and flaviviruses, take advantage of low pH in the endosomal compartment to trigger fusion 2, the filovirus Ebola uses proteases 3. Other enveloped viruses, including measles virus (MV) and HIV fuse directly with the plasma membrane at neutral pH 1,4.
MV, while being targeted for eradication, still causes more than 150,000 deaths yearly 5,6. MV is a member of the Paramyxoviridae 4,7, a family including deadly emerging viruses like Hendra and Nipah, and prevalent human viruses like mumps, parainfluenza, and respiratory syncytial viruses. Moreover, the MV entry system has become a paradigm for targeting oncolysis and therapeutic gene delivery 8–10 because it can be triggered by cell surface proteins differing in size, trans-membrane organization, and quaternary structure 11. Since the mechanism triggering MV cell entry is not understood, we set out to characterize it.
Cell entry of Paramyxoviridae requires the concerted action of two envelope glycoproteins. The attachment protein mediates receptor binding and triggers a refolding event of the metastable fusion (F) protein that results in membrane fusion 4,7. The attachment proteins are named hemagglutinin (H) for MV and the other members of the genus Morbilliviruses, glycoprotein (G) for the Henipaviruses, and hemagglutinin-neuraminidase (HN) for most other genera. While the H and G proteins bind proteinaceous receptors, HN proteins bind sialic acid 12.
MV H is a type II transmembrane glycoprotein comprised of an amino-terminal cytoplasmic tail, a membrane-spanning segment, and an extracellular membrane-proximal stalk region connected to a large cuboidal head with six-blade beta-propeller structure contacting the receptors (Figure 1a-c). MV H proteins form dimers stabilized by two inter-subunit disulfide bonds 13 (Cys139 and Cys154, Figure 1a, d) located at the top of the long helical stalk 14, below the base of the cuboidal heads. It was recently shown that H-tetramers or higher oligomeric forms sustain fusion-support function 15.
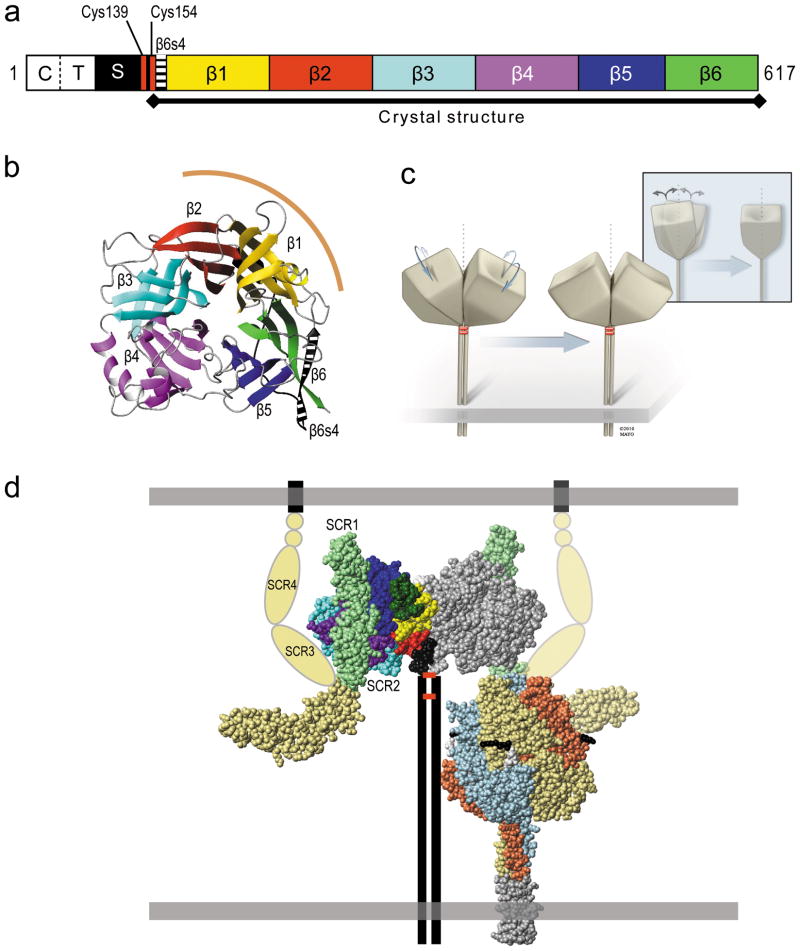
Figure 1. Structure of the MV H-dimer, and its interactions.
(a) Primary structure of the H-protein; C, cytoplasmic tail; T, transmembrane region; S, stalk; β1–β6, beta-propeller blades 1–6. (b) Top view of the six-bladed β-propeller sheet H-protein head crystal structure as a ribbon plot 19. Color-coding of beta-strands is consistent with panel a. The dimer interface is indicated by a curved brown line. (c) Schematic of the H-dimer, and illustration of one possible form of adjustment, subunit realignment. Inset: side view of the same adjustment. (d) Left, space-filling representation of the crystal structure of the H-protein dimeric head bound to CD46. The left monomer of the H-dimer is color-coded as in panel a. The four-domain CD46 crystal structure (SCR1–4) was modelled into the H–CD46 (2-domain; SCR1–2) co-crystal structure. The CD46 domains SCR1–2 are shaded light green and apposed as in the co-crystal 36, domains 3 and 4 are shaded pale yellow. The proposed “hook” alternate conformation is indicated using pale yellow ovals. The small STP domains are shown as pale yellow circles, and the transmembrane region and cytoplasmic tail as a black line. The stalk, transmembrane region and cytoplasmic tail of the H-dimer are represented by vertical lines and the disulfide bonds that hold the H-dimer together by horizontal red bars. Right, space-filling representation of the F-trimer crystal structure of the paramyxovirus, PIV5 47. The three monomers are shown with different colors for clarity. The five residues preceding the cleavage site are shown in black, the five following it in white. A trimeric coiled-coiled domain appended to the F-protein ectodomain is shown in grey. The membranes are illustrated as horizontal grey boxes.
Specific interactions of the attachment and F proteins of Paramyxoviridae are required for membrane fusion because heterotypic glycoprotein pairs cannot mediate this process 16,17. In particular, the stalk regions of different HN proteins determine the specificity for the cognate F proteins 4,7. Similarly, the MV H-stalk region interacts directly with the F protein head 18, an observation suggesting that the H-oligomer is much taller than the F-trimer (Figure 1d). These F-H interactions are relevant to late phases of the fusion triggering process.
Novel information revealing structural diversity of the attachment proteins of Paramyxoviridae prompted us to analyze early triggering phases. In particular, the interface of the H-dimer 19 was found to be much smaller than that of HN-dimers 20–22 (1070 Å2 versus 1800 Å2 in a direct comparison 23), and the H protein head crystallized as a monomer when Cys154 was omitted 24. Moreover, while the H-heads lean about 40° away from the symmetry axis, and tilt sideways slightly (schematics in Figure 1c; see also Figure 1d, left) 19, the HN-heads are more upright 20–22. The biology of these attachment proteins also differs: while H hetero-oligomerizes with F in the endoplasmic reticulum 25, HN and F travel separately to the cell surface 26.
As to fusion triggering, the small interface area of the H-dimer suggested to us that the heads might move relative to each other: Figure 1c illustrates a re-alignment movement, but adjustment or rotation movements are also possible. We set out to test the ‘movement’ hypothesis by adding covalent cross-links that stabilize the dimer in a single conformation. Indeed certain cross-links block membrane fusion.
We also noted that the footprints of the three MV receptors 27–35 are located distal to the dimer interface, such that two bound receptors per dimer would have enough leverage to force the heads to move. We set out to test this ‘twist’ hypothesis by directing a membrane-anchored receptor to bind tags inserted in strategic locations on the H-head. Indeed, only binding close to the locations used by the natural receptors, and distal to the H-dimer interface, efficiently triggers membrane fusion.
RESULTS
Stabilization of the H-dimer interface
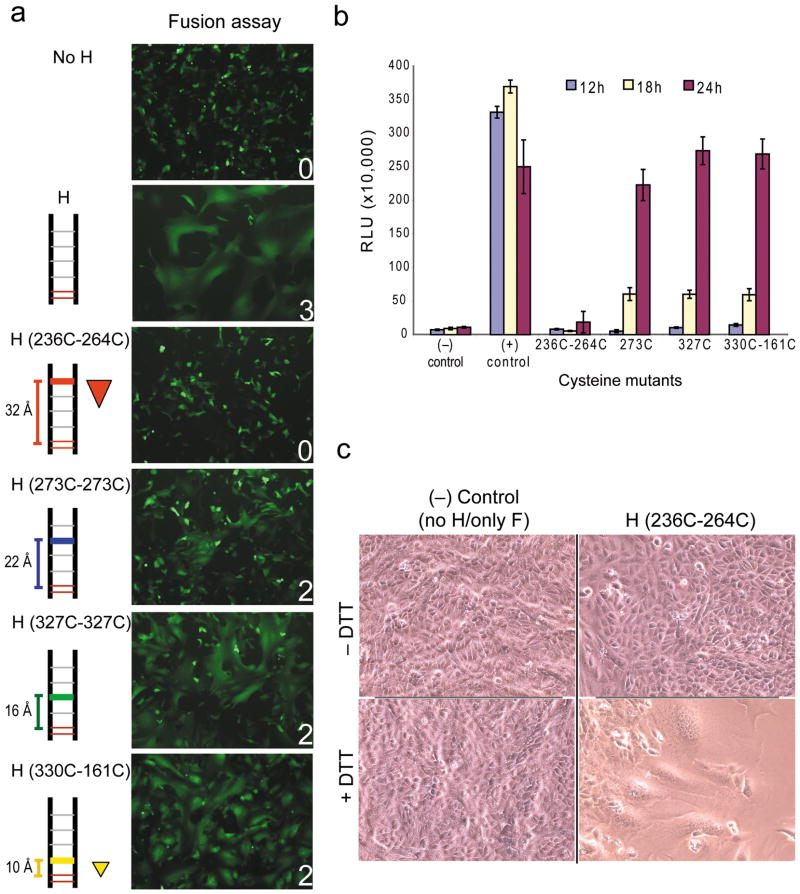
To test the ‘movement’ hypothesis we sought to stabilize the H-dimer interface by adding inter-subunit disulfide bonds. We note that the heads have a solid beta-propeller structure, and may thus move as rigid bodies. Moreover, CD46 binding does not induce substantial conformational changes of the H-head 36, consistent with a rigid structure suited to conduct torsion. We identified amino acid pairs across the interface whose C-alpha atoms are less than 7.5 Å apart (Table 1) and mutated these to cysteine. We sought to introduce two classes of disulfide bonds: single bonds located on the two-fold symmetry axis of the dimer and involving the same residue on both molecules (Table 1, top four lines), and twin bonds located on both sides of the axis and involving two different residues (Table 1, bottom four lines). Twin cysteine bonds, forming a triangle with Cys154, would provide more resistance to a force, increasing the energy barrier for fusion triggering; larger triangles would provide more resistance than smaller triangles. Figure 2a shows the location of the two key single bonds (T273C–T273C, blue and W327C–W327C, green) and the two key twin bonds (K236C–G264C, brown/red and P330C–L161C, orange/yellow) in a side view of the dimer interface. Figure 2b shows a front view of the dimer interface with the same structures.
Table 1.
Cysteine substitution mutants with potential to form disulfide bonds at the H-dimer interface
| Bond(s) | Residue(s) | Distance from Cys154 (Å) | Area of triangle (Å2) | Alanine tolerated | Dimer formed | Disulfide bond formed | Cα–Cα distance (Å) |
|---|---|---|---|---|---|---|---|
| single | K328 | 16 | -na- | yes | yes | no | 6.96 |
| single | W327 | 16 | -na- | yes | yes | yes | 6.50 |
| single | T273 | 22 | -na- | yes | yes | yes | 5.54 |
| single | L203 | 32 | -na- | no | yes | no | 4.78 |
| twin | P330, L161 | 10 | 68 | yes | yes | yes | 6.68 |
| twin | P330, E162 | 10 | 75 | yes | yes | yes | 6.78 |
| twin | K236, L265 | 32 | 498 | yes | yes | no | 7.39 |
| twin | K236, G264 | 32 | 533 | yes | yes | yes | 6.32 |
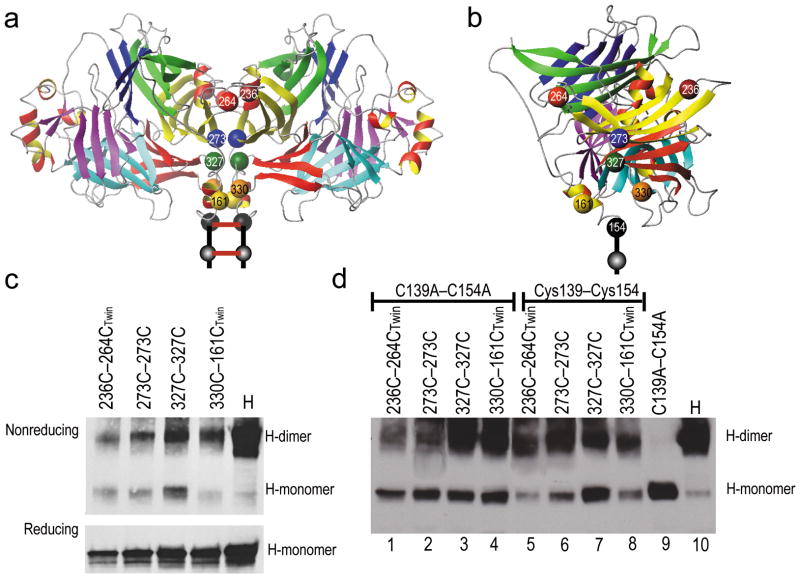
Figure 2. Location of cysteines introduced to constrain the H-dimer interface and evidence of disulfide bond formation.
(a) Ribbon representation of the front view of the H-dimer head crystal structure. Cysteines 154 and 139 are indicated with black spheres; a twin cysteine pair introduced near the top of the dimer by red and brown spheres; two single cysteine pairs by blue and green spheres, respectively and a twin cysteine pair near the stalk by yellow and orange spheres. The two red bars represent the intersubunit disulfide bonds. (b) A front view of H-dimer interface with the cysteine substitutions indicated by spheres. Color-coding is consistent with panel a. (c) Immunoblot analysis of the dimerization capacity (top) and protein expression levels (bottom) of the H-dimer cysteine substitution mutants. Protein extracts from transfected Vero cells were separated on a 4–15% SDS-PAGE gel in the absence (top) or presence (bottom) of β-mercaptoethanol. (d) Immunoblot analysis of the ability of the introduced cysteines to form intersubunit disulfide bonds, performed in the C139A–C154A mutant backbone (lanes 1–4) or the parental background (Cys139–Cys154). Protein was extracted from transfected Vero cells into lysis buffer containing 10 mM iodoacetamide and analyzed on a 4–15% SDS-PAGE gel in the absence of β-mercaptoethanol. β-actin expression levels were used as loading controls (data not shown).
The dimerization capacity of the mutated H-proteins was then assessed (Table 1 and Figure 2c). We note that the cysteine substitutions are distal from known receptor binding epitopes, but to control for the ability of each position to tolerate mutations we also made alanine substitutions, and assayed fusion triggering by the alanine mutants. Where twin bonds were involved, we ensured that neither one of the cysteine substitutions in the pair had an effect on fusion function. As listed in Table 1 (fifth column), only one of the alanine mutants, L203A was fusion deficient, and this residue was eliminated from further consideration. The fact that all the other alanine mutations were fusion-competent implies that the corresponding mutant proteins have intact receptor binding function.
Since total expression levels of certain mutants were reduced (Figure 2c), we sought to measure their cell surface expression. We measured the mean fluorescence intensity (MFI) by FACS analysis for each mutant (Supplementary Table 1), which was 47–68% as that of the standard H-NSe protein. Even with this reduction in cell surface protein levels, we note the MV fusion system is very efficient, and that decreased H surface expression may not necessarily translate in decreased fusion 13. We also probed the mutants with a conformation-specific monoclonal antibody that recognizes the H-noose epitope 37. The MFI measured with this antibody was similar to that recorded with the anti-H cocktail (Supplementary Table 1), indicating that the mutants had similar conformations to the unmodified H-protein.
We then assessed whether covalent bonds are formed, taking advantage of an H protein backbone with mutations in both cysteine residues (Cys139 and Cys154) that normally mediate covalent inter-subunit cross-linking. While this mutant forms minimal amounts of stable dimers, given time it does support low levels of membrane fusion 13. Figure 2d, ninth lane, shows that indeed the C139A–C154A mutant loses the ability to form detectable amounts of stable H-dimers, as assayed by non-reducing SDS-PAGE. Thus, in the C139A–C154A background the only inter-subunit bridges are those formed at positions with added cysteines. Figure 2d (lanes 1–4) show that cysteine substitutions at 236+264, 273, 327 or 330+161 restore the dimerization capacity of the H-protein, demonstrating that these substitutions allow inter-subunit disulfide bonds under physiological conditions. Comparison of the dimerization capacity of the mutants with either the C139A–C154A (lanes 1–4) or the normal H-backbone (lanes 5–8), indicated that the cysteine substitution mutants alone are less efficient at forming dimers. This is not surprising as the introduced cysteines may not promote covalent linkage as efficiently as the two stalk cysteines. This analysis also revealed that K328C and K236C+L265C did not yield cross-linked dimers, and these residues were eliminated from further consideration (Table 1, seventh column). Moreover, P330C+E162C dimerized but was not pursued further because it is similar to P330C+L161C. Thus, analysis focused on two single-bond and two twin-bond cysteine mutants.
Twin pivot-distal disulfide bonds reversibly inhibit fusion
After confirming disulfide bond formation, H-protein mutants were co-expressed with F-protein to test the hypothesis that movement of the subunits is required to allow membrane fusion. Figure 3a shows that the most distal twin bonds (K236C+G264C, 32Å from fusion pivot, 533 Å2 stabilized area) abrogated fusion. On the other hand, the two single bonds (22Å and 16Å from the pivot) and the pivot proximal twin bond (10Å from pivot, 68 Å2 stabilized area) had little effect on fusion, in this assay performed 24 hours after transfection.
Figure 3. Constraining the H-dimer interface disrupts fusion.
(a) Visual fusion assay to monitor effect of H-dimer disulfide bonds on membrane fusion. This visual fusion assay is based on the co-transfection of Vero cells with plasmids expressing H-protein, F-protein and GFP. Fusion scores were determined 24 hours post-transfection and are averages of 3 replicates. A fusion score of 3 denotes wild type fusion levels; 2 and 1, intermediate fusion; and 0, no fusion. The relative locations of introduced disulfide bonds are indicated on the left, as is the distance from the proposed Cys154 pivot. The triangle denotes a twin bond. (b) Quantitative fusion assay at 12-, 18- and 24-hours post transfection. Mutants are indicated on the horizontal axis. (−), negative control, F-protein expression plasmid only. (+) control, standard H- and F-protein expression plasmids. Vertical axis, RLU, relative luciferase units. Results represent an average of at least three replicates (± s.d.). (c) Fusion induction in cells transfected with the K236C+G264C mutant by reduction of disulfide bonds. Vero cells were co-transfected with the standard F-expression plasmid and the K236C–G264C H mutant expression plasmid (right) or not (left). Twenty-four hours post-transfection cells were treated with 12 mM DTT in PBS (bottom) or PBS alone (top). Fusion was recorded 1 hour post DTT treatment.
This analysis was repeated using a luciferase-based quantitative fusion assay that provided additional 12- and 18-hour time points. At 12-hours the positive control showed high fusion levels, while all the mutants had fusion levels at or near background (Figure 3b). At 18-hours the fusion functions of the T273C, W327C and P330C–L161C mutants were reduced by a factor of 6 compared to the positive control. At 24-hours the fusion function of the K236C+G264C mutant was reduced by a factor of 13 compared to the wild type protein, while fusion efficiency of the other three mutants was comparable to wild type H, which syncytia had already begun to lyse. These results indicate that the single bonds at 273 and 327, and the twin bond at 330+161 interfere with fusion temporarily. However, given time fusion is observed, possibly because occasionally disulfide bonds are reduced.
We then assessed whether eliminating the disulfide bonds with a reducing agent restores fusion function. Cells expressing the K236C+G264C mutations were treated with 1,4-Dithio-DL-threitol (DTT). In the presence of DTT, fusion function was restored in the K236C+G264C H mutant that previously exhibited no fusion (Figure 3c, compare lower left and right panels). Similar results were observed for the T273C and W327C mutants when treated with DTT 12-hours post-transfection (data not shown), indicating that this protein is available in sufficient quantities and retains the capacity to interact with receptors. Thus, movement of the H-dimer is required for membrane fusion.
Directing receptor binding with hexahistidine tags
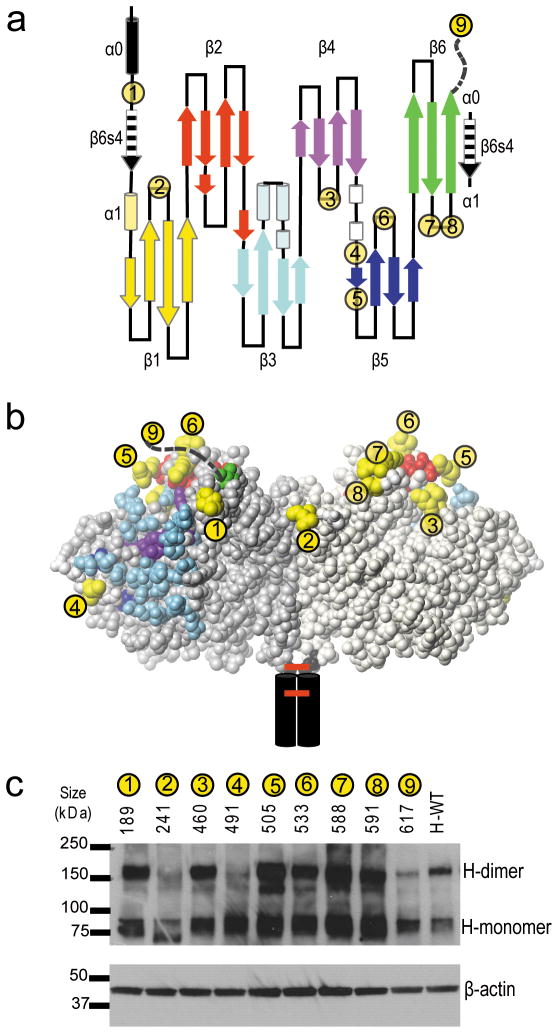
We next assessed whether the efficiency of fusion triggering depends on interface-distal receptor binding on the H-dimer. We took a systematic approach and inserted a short hexahistidine tag in different positions at the top half of the H-dimer. The hexahistidine tag directs binding to Vero-His cells which express a membrane-anchored single-chain antibody that recognizes the six-histidine peptide38. The insertion points were chosen taking into consideration structural constraints, including N-linked oligosaccharide chains at N168, N187, N200 and N215, and availability of surface accessible loops (Figure 4a, b).
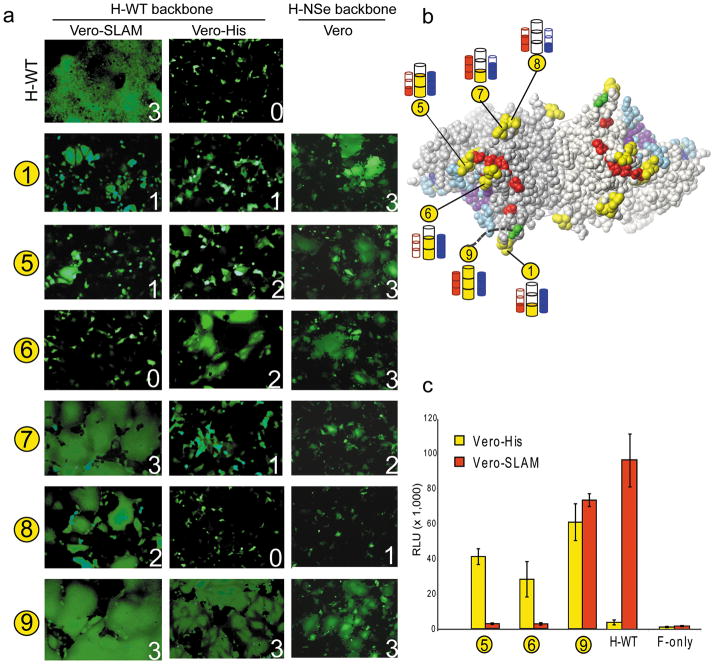
Figure 4. Positioning hexahistidine tags to direct receptor binding.
(a) Secondary structure of the H-head domain beginning at cysteine 154 and ending with the last residue of the ectodomain. Hexahistidine tag insertion sites are indicated with numbered yellow circles. The unstructured final 10 amino acids of the H ectodomain to which the hexahistidine is linked in mutant 9 are indicated with an interrupted line. The amino acid residue preceding the insertion site of each tag is indicated above panel c. (b) Front view of the H-dimer head domain with the residues preceding and following each hexahistidine insertion site shown in yellow. The footprints of the receptors are indicated: red, SLAM-specific residues; blue, CD46-specific residues and purple, EpR-specific residues. (c) Protein expression analysis and dimerization capacity of the hexahistidine tag insertion mutants (top). On the top of panel c, the residue of insertion and the number of each hexahistidine tag are indicated. H-WT: non-mutated H-protein. Molecular weight markers are indicated on the left. β-actin expression levels were used as loading controls (bottom).
The tags were inserted in eight exposed loops (Figure 4a, b, yellow residues). We used mutant 9 with a carboxy-terminal tag as the positive control 38. Mutants 5 and 6 are located on the top of the H-dimer, between the H-residues in close contact with CD46, as identified based on the H-CD46 co-crystal 36 (highlighted in light blue) and those identified by functional assays to be relevant for SLAM-dependent fusion 27–29 (highlighted in red). Mutants 1, 3, 7 and 8 are all located near the top of the H-dimer. While mutant 1 is close to the CD46 footprint, and mutant 7 close to the SLAM footprint, mutants 3 and 8 are located away from both receptor binding areas. Mutant 2 is also located away from these binding areas and close to the interface. Mutant 4 is the only one in the bottom half of the dimer.
To identify functional mutants, dimerization capacity, expression levels, and accessibility of the His-tag were analysed. Protein extracts from transfected Vero cells were separated on SDS-PAGE under non-reducing conditions, and immunoblotted. Since mutants 2 and 4 dimerized poorly (Figure 4c), they were excluded from further analysis. Surface expression levels of the remaining mutants, measured by FACS, were at 39–99% of standard H-NSe (Supplementary Table 2, third column), which was considered acceptable for all mutants. To determine the accessibility of the His-tag we compared the MFI generated by an anti-His antibody to that generated by an anti-H cocktail (Supplementary Table 2, last column). All but one mutant had MFI ratios close to 1, indicating that their His-tags were equivalently accessible. Since mutant 3 had a lower (0.65) MFI ratio, it was not considered when drawing conclusions. Thus, hexahistidine tagging of exposed protein loops is compatible with efficient protein folding and dimerization, and allows display of accessible tags in a majority of the constructs.
Only interface-distal binding efficiently triggers fusion
We then measured the fusion efficiency of the dimerization-competent mutants and the control mutant, 9, in cell lines expressing either the anti-hexahistidine antibody as the designated receptor, or the natural receptor SLAM, or the vaccine strain receptor, CD46. Since none of the hexahistidine tags were predicted to interfere with CD46-binding, these fusion assays were considered positive controls. On the other hand, SLAM-dependent fusion assays were expected to further define the SLAM footprint. Finally, hexahistidine receptor-dependent fusion assays were expected to reveal the H-dimer surface area poised for fusion triggering.
For consistency in host cell type, we used Vero cells, or Vero cells expressing anti-hexahistidine (Vero-His), or SLAM (Vero-SLAM). To focus exclusively on entry through one receptor, we generated a second set of tag mutants in an H-protein backbone exclusively recognizing SLAM (H-WT, Figure 5a, first and second column). The H-NSe vaccine lineage backbone was used only to measure CD46-dependent fusion (H-NSe, Figure 5a, third column). This analysis indicated that, with the exception of mutants 7 and 8, which were reduced in CD46-specific fusion (fusion scores of 2 and 1, respectively), all the mutants fully retained CD46-dependent fusion function. The results are also illustrated in Figure 5b with blue cylinders.
Figure 5. Location of receptor-binding site determines efficiency of fusion triggering.
(a) Fusion trigger function of the hexahistidine tag mutants on cell lines expressing different receptors. This assay is based on the co-transfection of plasmids expressing H-protein, F-protein and GFP. Two sets of tag mutants were used, one in the wild-type H-backbone (H-WT, first two columns) and the other in the vaccine lineage H-backbone (H-NSe, last column). Fusion assays were conducted in Vero-SLAM cells (first column), or Vero-His cells (middle column) or Vero cells (last column). (b) Results of experiments in panel a visualized on the top view of the H-dimer head. SLAM-specific fusion trigger function is indicated with red cylinders, His-specific fusion trigger function with yellow cylinders and CD46-specific fusion trigger function with blue cylinders. Full cylinder, wild-type like fusion trigger function; two-thirds and one-third full cylinders, intermediate fusion trigger function; empty cylinder, no fusion. (c) Quantitative fusion assay of selected hexahistidine tag mutants, as indicated on the horizontal axis. H-WT, standard H-protein. Vertical axis, RLU, relative luciferase units. Results represent an average of three replicates (± s.d.).
Analysis of SLAM-dependent fusion revealed that the H-SLAM contact area may extend to the region defined by mutants 5 and 6, because these two mutants are specifically impaired in this fusion function (Figure 5a, first column and Figure 5b, red cylinders; fusion score of 1 and 0, respectively). The SLAM contact area on the top of the H-dimer may extend also to the position defined by mutant 1 (fusion score of 1).
We then assessed the efficiency of membrane fusion triggering through the membrane-anchored anti-hexahistidine antibody. Figure 5a, center column and Figure 5b, yellow cylinders, show that both mutants (5 and 6) in which the tags are located between the SLAM and CD46 footprints, had fusion scores of 2 each. Mutants 1 and 7, with tags located close to only one of the receptor footprints, and closer to the interface than mutants 5 and 6, had fusion scores of 1 each. Mutant 8 did not trigger fusion. Thus, both hexahistidine tags inserted within the SLAM-footprint trigger fusion through an artificial receptor, almost as efficiently as the control mutant.
To better compare the fusion efficiency of mutants 5 and 6, the luciferase-based quantitative fusion assay was used. Figure 5c documents fusion efficiency in Vero cells expressing either the membrane-anchored anti-hexahistidine antibody (yellow bars) or SLAM (red bars). Mutant 5 was about 65% as effective as positive control mutant 9 in inducing fusion of Vero-His cells, and mutant 6 was about 50% as effective. Thus, an artificial receptor must bind proximal to the locations used by the natural receptors to effectively trigger fusion. While preferential accessibility of this area of the H-head may influence this striking co-localization of functional binding sites, our findings are also fully consistent with the “twist” hypothesis proposing that receptors have more leverage to force the heads to move while engaging an interface-distal location.
DISCUSSION
We have used information from the static crystal structures of the attachment proteins of four Paramyxoviruses to formulate and test two hypotheses aiming at inferring the mechanism of a necessarily dynamic process, such as the triggering of membrane fusion. Our findings are consistent with the following model of H-based fusion triggering: first, two membrane-anchored receptor molecules bind the H-dimer heads opposite to the interface. Second, they force the H-heads to move relative to each other. This movement transmits a signal to the F protein, triggering irreversible F-trimer refolding and membrane fusion.
Our model provides a framework for interpreting recently published structural information documenting one type of interaction between two external CD46 short consensus repeats (SCR) and the H-heads 36 (Figure 1d; SCR1–2 are shown in pale green). Surprisingly, CD46 binds H in an “upside-down” orientation: SCR2 points towards the viral membrane. Very recently, almost the entire CD46 ectodomain (SCR 1–4) was co-crystallized with the Adenovirus knob. In this complex CD46 assumes a “hockey stick” conformation 39. When this structure is modelled into the H-dimer, SCR 4 is parallel to the membranes (Figure 1d). Remarkably, the SLAM footprint suggests that its H-binding domain is positioned tangentially to the H-dimer interface (Figure 5b) and thus also parallel to the membrane. We propose that tangential engagement sustains efficient transfer of forces arising from the receptor’s lateral movement in the plasma membrane to H-heads.
These observations also suggest that CD46 and the H-oligomer initially interact at another angle, or in a different conformation, than those shown in Figure 1d. Indeed, there are indications that these molecules have structural plasticity and assume different conformations during the binding and fusion processes. In particular, it was observed that CD46 domains 3 and 4 enhance binding of domains 1 and 2 to MV particles, but impair binding to soluble H 40. Since the domain 1–2 interface possesses some flexibility 41, and flexibility may also be a feature of the similarly-sized domain 2–3 interface 39, it is conceivable that before H-binding CD46 is shaped as a “hook” (Figure 1d; SCR 1–2 shown in pale green, linked to yellow ovals representing SCR 3–4). The “hockey stick” could be an alternative conformation occurring after binding and facilitating membrane fusion.
In addition, the H-oligomers could be flexible. A precedent for this is the PIV5 HN-tetrameric ectodomain: electron microscopy revealed a staggered arrangement of the heads, and it was proposed that flexibility of the stalk is important in membrane fusion 42. Similarly, H-proteins may form flexible tetramers with staggered heads. H-tetramers may bind CD46 molecules in the “hook” conformation, and subsequently these complexes may adjust to form a scaffold of optimal height to support fusion. Such a scaffold was previously proposed based on the analysis of the fusion-support efficiency of receptor molecules of variable length 43.
Based on the analysis of the H-oligomer’s intracellular transport, it was proposed that Paramyxoviruses binding either proteinaceous receptors (H-class), or sialic acid (HN-class), regulate fusion by different mechanisms 25. This suggestion was strengthened by the documentation of additional structural and functional differences between H- and HN-class proteins 12. We now propose that H-heads switch partners to transmit the fusion-triggering signal, while HN-heads must use another mechanism.
We know that receptors engage the cuboidal H-heads from the side (Fig. 1d) 19, and we propose that they make them rotate 90° about their stalk. H-heads will then switch partners while the stalk remains covalently linked with the stalk of the original partner, cross-linking two H-dimers into an irregular H-tetramer. Since the stalks of these H-tetramers would have anomalous contacts with F-trimers 18, their accumulation would progressively de-stabilize the viral envelope, eventually triggering membrane fusion.
Our model considers that fusion is supported by tetrameric or higher order H-oligomers 15, and implies that alteration of residues critical for formation of the interface between dimers would interfere with transmission of the fusion-triggering signal. However, we do not know whether this interface is formed between only one subunit in each dimer, analogous to the staggered arrangements of HN pairs-of-dimers revealed by electron microscopy 42, or between both subunits in each dimer, analogous to the tightly packed, quadratic arrangements of HN-head crystals 20–22.
Towards identifying residues in this interface and testing this aspect of the model we generated 10,000 docked poses of H-head pairs-of-dimers using the program ZDOCK 44 (N.O. and W.B., unpublished). We observed many poses with staggered, and few with tightly packed arrangements. We expressed mutated H-proteins, focusing initially on mutants representing tightly packed poses, but all mutated proteins maintained fusion function, and formed tetramers as documented by Blue Native gel electrophoresis (C.K.N., Amber Kirk, Robbyn Weaver and R.C., unpublished). We are now considering staggered poses. Since we have identified many potential interface residues, we plan iterative cycles of mutagenesis and functional tests.
HN-based triggering cannot be accounted by head partner switch because intermolecular disulfide bonds in the HN-dimer interface do not block fusion and thus both heads of a HN-dimer must move as a unit 45. Since receptors engage these units from the top 20–22, HN-head dimeric units may slide relative to another rather than rotate. Sliding would not cross-link subunits within HN-tetramers, but may catenate the tetramers, which would liberate individual F-trimer spikes and trigger fusion 46.
Supplementary Material
Acknowledgments
We thank Jim Maher, David Katzmann, Benhur Lee, Charles Russell and Ronald Iorio for comments on the manuscript, Steve Russell (Mayo Clinic) for the Vero-His cell line, Claude Muller (University of Trier) for the BH6 antibody, Amber Kirk (Mayo Clinic) and Robbyn Weaver (Mayo Clinic) for help with H-tetramer interface mutagenesis, Thilo Stehle (University of Tuebingen) for communicating unpublished information, and Mary Bennett for excellent secretarial assistance. This work was supported by grant NIH R01 CA90636, and by the Mayo Clinic Cancer Center. Part of L.K.’s and L.R.’s stipends was supported by the Mayo Graduate School SURF program.
Footnotes
AUTHOR CONTRIBUTIONS
R.C. and C.K.N. conceived the projects. N.O. and W.B. planned cysteine mutagenesis, C.K.N. and V.H.J.L. planned histidine tagging. C.K.N., L.R. and L.K. generated the expression plasmids and characterized the function of all the mutated proteins. C.K.N., V.H.J.L., W.B. and R.C. contributed to the analysis of the data and prepared the manuscript.
Yes there is potential Competing Interest.
Patent applications on which RC is an inventor have been licensed to NISCO, Inc. Mayo has an equity position in NISCO; Mayo has not yet received royalties from products developed by the company, but may receive these in the future.
References
- 1.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–8. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marsh M, Helenius A. Virus entry: open sesame. Cell. 2006;124:729–40. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–5. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamb RA, Parks GD. Paramyxovirus: The viruses and their replication. In: Fields B, Knipe D, Howley P, editors. Fields Virology. Vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1305–1340. [Google Scholar]
- 5.Griffin DE. Measles Virus. In: Knipe DM, Howley PM, editors. Fields’ Virology. Vol. 1. Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 1551–1585. [Google Scholar]
- 6.Moss WJ. Measles control and the prospect of eradication. Curr Top Microbiol Immunol. 2009;330:173–89. doi: 10.1007/978-3-540-70617-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith EC, Popa A, Chang A, Masante C, Dutch RE. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS Journal. 2009;276:7217–27. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cattaneo R, Miest T, Shashkova EV, Barry MA. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat Rev Microbiol. 2008;6:529–40. doi: 10.1038/nrmicro1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell SJ, Peng KW. Measles virus for cancer therapy. Curr Top Microbiol Immunol. 2009;330:213–241. doi: 10.1007/978-3-540-70617-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cattaneo R. Paramyxovirus entry and targeted vectors for cancer therapy. PLoS Pathogens. 2010;6:e1000973. doi: 10.1371/journal.ppat.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navaratnarajah CK, Leonard VHJ, Cattaneo R. Measles virus glycoprotein complex assembly, receptor attachment and cell entry. Curr Top Microbiol Immunol. 2009;329:59–76. doi: 10.1007/978-3-540-70523-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iorio R, Melanson V, Mahon P. Glycoprotein interactions in paramyxovirus fusion. Future Virology. 2009;4:335–351. doi: 10.2217/fvl.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plemper R, Hammond A, Cattaneo R. Characterization of a region of measles virus hemagglutinin sufficient for its dimerization. J Virol. 2000;74:6485–6493. doi: 10.1128/jvi.74.14.6485-6493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JK, et al. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem. 2008;283:16561–72. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brindley MA, Plemper RK. Blue Native-PAGE and biomolecular complementation reveal tetrameric or higher order oligomer organization of the physiological measles virus attachment (H) protein. J Virol. doi: 10.1128/JVI.01222-10. (published online ahead of print on 22 September 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cattaneo R, Rose JK. Cell fusion by the envelope glycoproteins of persistent measles viruses which caused lethal human brain disease. J Virol. 1993;67:1493–1502. doi: 10.1128/jvi.67.3.1493-1502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu XL, Ray R, Compans RW. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol. 1992;66:1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paal T, et al. Probing the spatial organization of measles virus fusion complexes. J Virol. 2009;83:10480–93. doi: 10.1128/JVI.01195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashiguchi T, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci U S A. 2007;104:19535–40. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence MC, et al. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–57. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 22.Yuan P, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–15. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Bowden T, Crispin M, Harvey D, Jones EY, Stuard D. Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J Virol. 2010;84:6208–6217. doi: 10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nature Structural & Molecular Biology. 2007;14:1227–8. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 25.Plemper RK, Hammond AL, Cattaneo R. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem. 2001;276:44239–44246. doi: 10.1074/jbc.M105967200. [DOI] [PubMed] [Google Scholar]
- 26.Connolly SA, Leser GP, Jardetzky TS, Lamb RA. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J Virol. 2009;83:10857–68. doi: 10.1128/JVI.01191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vongpunsawad S, Oezgun N, Braun W, Cattaneo R. Selectively receptor-blind measles viruses: identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J Virol. 2004;78:302–313. doi: 10.1128/JVI.78.1.302-313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masse N, et al. Measles virus (MV) hemagglutinin: evidence that attachment sites for MV receptors SLAM and CD46 overlap on the globular head. J Virol. 2004;78:9051–9063. doi: 10.1128/JVI.78.17.9051-9063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navaratnarajah CK, et al. Dynamic interaction of the measles virus hemagglutinin with its receptor signaling lymphocytic activation molecule (SLAM, CD150) J Biol Chem. 2008;283:11763–71. doi: 10.1074/jbc.M800896200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonard VHJ, et al. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J Clin Invest. 2008;118:2448–2458. doi: 10.1172/JCI35454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tahara M, et al. Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J Virol. 2008;82:4630–4637. doi: 10.1128/JVI.02691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naniche D, et al. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorig RE, Marcil A, Chopra A, Richardson CD. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 34.Tatsuo H, Ono N, Tanaka K, Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 35.Ono N, et al. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol. 2001;75:4399–4401. doi: 10.1128/JVI.75.9.4399-4401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol. 2010;17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 37.El Kasmi K, et al. Neutralization of measles virus wild-type isolates after immunization with a synthetic peptide vaccine which is not recognized by neutralizing passive antibodies. J Gen Virol. 2000;81:729–735. doi: 10.1099/0022-1317-81-3-729. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura T, et al. Rescue and propagation of fully retargeted oncolytic measles viruses. Nat Biotechnol. 2005;23:209–214. doi: 10.1038/nbt1060. [DOI] [PubMed] [Google Scholar]
- 39.Persson BD, et al. Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens. PLoS Pathogens. 2010;6:e1001122. doi: 10.1371/journal.ppat.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devaux P, et al. CD46 short consensus repeats III and IV enhance measles virus binding but impair soluble hemagglutinin binding. J Virol. 1997;71:4157–4160. doi: 10.1128/jvi.71.5.4157-4160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casasnovas JM, Larvie M, Stehle T. Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J. 1999;18:2911–2922. doi: 10.1093/emboj/18.11.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology. 2008;378:282–91. doi: 10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchholz CJ, Schneider U, Devaux P, Gerlier D, Cattaneo R. Cell entry by measles virus: long hybrid receptors uncouple binding from membrane fusion. J Virol. 1996;70:3716–3723. doi: 10.1128/jvi.70.6.3716-3723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 45.Mahon PJ, Mirza AM, Musich TA, Iorio RM. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein: implications for the mechanism of fusion promotion. J Virol. 2008;82:10386–96. doi: 10.1128/JVI.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludwig K, et al. Electron cryomicroscopy reveals different F1 + F2 protein states in intact parainfluenza virions. J Virol. 2008;82:3775–3781. doi: 10.1128/JVI.02154-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ono N, Tatsuo H, Tanaka K, Minagawa H, Yanagi Y. V domain of human SLAM (CDw150) is essential for its function as a measles virus receptor. J Virol. 2001;75:1594–1600. doi: 10.1128/JVI.75.4.1594-1600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–9. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cathomen T, Buchholz CJ, Spielhofer P, Cattaneo R. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology. 1995;214:628–632. doi: 10.1006/viro.1995.0075. [DOI] [PubMed] [Google Scholar]
- 51.Radecke F, et al. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeda M, et al. Recovery of pathogenic measles virus from cloned cDNA. J Virol. 2000;74:6643–6647. doi: 10.1128/jvi.74.14.6643-6647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comp Chem. 1998;19:319. [Google Scholar]
- 54.Cathomen T, Naim HY, Cattaneo R. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J Virol. 1998;72:1224–1234. doi: 10.1128/jvi.72.2.1224-1234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.