Abstract
The goal of this study was to evaluate prostate cancer gene expression signatures associated with elevated body mass index (BMI). Global gene expression profiles of prostate tumor cells and matching normal epithelial cells were compared between patients with features of normal- and high BMI at the time of radical prostatectomy. Knowledge-based analyses revealed an association of high BMI with altered levels of lipid metabolism and cholesterol homeostasis genes, such as stearoyl-CoA desaturase 1 (SCD1) and insulin-induced gene 1 (INSIG1), respectively, in prostate tumor cells. These genes were connected to known pathways of tumorigenesis revealed by the v-maf (musculoaponeurotic fibrosarcoma) oncogene homolog (MAF), notch receptor ligand, jagged 1 (JAG1), and the alanyl aminopeptidase (ANPEP/CD13) genes. This study highlighted that SCD1, a known target of statins, may play a mechanistic role in the recently noted beneficial effects of statin treatment in reducing biochemical recurrence of prostate cancer. An additional finding of our study is that some of the obesity related genes were upregulated in tumor-matched normal cells within the high BMI group, when compared to normal cells within the normal BMI cohort.
Keywords: body mass index, prostate cancer, gene expression, microarray, obesity, lipid metabolism, statins
Introduction
Studies have linked prostate cancer to age, race, family history, genetic risk alleles, diet and altered metabolic conditions, such as obesity and diabetes.1–4 Although prostate cancer is uncommon in younger men (age < 45), the risk of developing the disease increases with age.5 Early reports have warned about the potentially negative consequences of unhealthy dietary habits in increasing the risk of prostate cancer.6,7 However, the rigorous examination of metabolic conditions in relation to prostate cancer incidence and disease progression has just begun to unfold. Obesity has reached epidemic levels in America and is currently one of the largest medical challenges.8 Whether or not obese men are at increased risk for prostate cancer or mortality is complex and debatable.9,10 However, recent epidemiological and clinical studies indicate the association of obesity with disease aggressiveness.11,12 Consistent with these findings, strong correlation between obesity and disease progression is shown in other studies.4,13 Increased odds of high-grade disease at diagnosis of prostate cancer indicate that obesity-associated altered metabolic conditions may be linked to prostate cancer progression.14 Recent reports described the benefits of statins in reducing prostate cancer progression, which support the adverse role of altered metabolic conditions in prostate cancer.15–20 It has also been shown that molecular models of cancer progression indicate the mechanistic connection between obesity-related gene expression signatures and cancer-promoting genetic alteration.21 Thus, there is a need for better understanding the role of obesity-associated gene expression signatures in prostate cancer. There are numerous challenges in this endeavor. First, BMI information in existing prostate cancer gene expression data sets is often insufficient. Second, gene expression data from matching benign prostatic epithelial cells that can be used as internal references are frequently missing. Third, the lack of history of BMI can severely confound the interpretation of obesity gene expression signature in relation to prostate cancer progression. For example, prostate cancer patients whose high BMI existed from youth would likely present cases where prostate cancer emerged within a cellular environment characteristic to the high BMI conditions. We reasoned that the onset of obesity shows a wide range of variations with age within the population.22 In contrast, within recipients of the military health care, strict physical requirements restrict the onset of weight gain to retirement age. Thus, in this prostate cancer patient group, early weight gain is less likely to confound the onset of prostate cancer.23–25 The goal of this proof-of-principle study was to carefully evaluate the comparative gene expression signatures of tumor and normal prostate epithelial cells from patients stratified for similar clinicopathological features presented with normal or high BMI at the time of radical prostatectomy (RP). These data will provide the foundation for targeted studies focusing on molecular mechanisms of tumor progression in prostate cancer patients with high BMI.
Materials and methods
Specimen selection
Patients with prostatic adenocarcinoma were treated at the Walter Reed Army Medical Center Urology Service. RP specimens were graded and staged.26 Frozen tissue samples were obtained at the time of RP, according to the approved protocol. Frozen tissues, were stored in OCT at −80°C. From 40 patients reported in a previous microarray study,27 12 were selected for the current analysis. Well- and moderately-differentiated tumor cells were obtained from specimens with Gleason score 6–7 with no seminal vesicle invasion. Laser capture micro-dissection (LCM) compatible specimens from six patients with high BMI and six patients with normal BMI were selected by matching the age and race (Caucasians), assuring that patients were not treated with cholesterol-lowering drugs prior to RP (Table 1).
Table 1.
Clinico-pathological characteristics and selection criteria of high and normal BMI prostate cancer patients.
| Patient Group |
BMI (Mean±SD) |
Age (Mean±SD) |
N | Race | Tumor Differentiation |
PSA Recurrence |
Cholesterol Lowering Drug |
|---|---|---|---|---|---|---|---|
| High BMI | 27.55±1.67 | 58.23±10.09 | 6 | Caucasian | WD | None | None |
| Normal BMI | 21.85±2.20 | 61.95±3.25 | 6 | Caucasian | WD | None | None |
Laser Capture Microdissection and quality control
For the LCM selection of benign prostate epithelial cells with normal morphological appearance (N) and prostate tumor epithelial cells (T) from H&E-stained frozen tissue sections the PixCell® II Laser Capture Microdissection System (Arcturus, California, USA) was used. We obtained approximately 5000 cells from each cell types. Cells with well- and moderately-differentiated morphology were captured from tumor foci. Nonadjacent prostate epithelial cells were collected from morphologically normal fields. Optimal composition of the samples was defined as less than 10% contamination with stroma tissue. Captured cells were processed for RNA extraction. Quantitative RT-PCR analysis of the prostate cancer marker genes, ERG, AMACR and PCA3 were performed from LCM-derived RNA samples.28 Tumor-over-normal (T/N) gene expression rations were log10 transformed and were represented on a heat map.
RNA extraction labeling and gene expression analysis
Typically, 20 ng of total RNA was isolated from the LCM samples by using the Paradise MicroRNA kit (Arcturus, Mountain View, CA). The isolated RNA was quantified by using RiboGreen dye (Molecular Probes, Eugene, OR) and VersaFluor fluorimeter (BioRad, Hercules, CA). Linear RNA amplification was carried out by using the Paradise RNA amplification kit (Arcturus, Mountain View, CA). The first round of amplification was performed with two nanograms of total RNA. The second round of amplification reaction included the cDNA synthesis and biotinylation steps. Linearly amplified RNA samples were hybridized to a high-density oligonucleotide human genome array HG-U133A Affymetrix GeneChip Arrays.26
Gene expression data analysis
Bioinformatics analysis of the raw gene expression data output (CEL files) was performed by using the Robust Multi-array Analysis (RMA, http://rmaexpress.bmbolstad.com) and by the ChipInspector a single-probe analysis approach (Genomatix GmbH, Munich, Germany). ChipInspector software (http://www.genomatix.de) analyzes raw gene expression data at the single probe levels by matching single probes to transcripts, normalizing the total intensities and by the Significance Analysis of Microarrays (SAM) analysis and enrichment of significantly altered signal intensities. This approach improves the signal-to-noise ratio, increases the statistical stringency and eliminates probe mismatches or multiple matches.29 Probe signal intensities that met both RMA and ChipInspector normalization criteria yielded 3770 significantly up-regulated and 1885 significant down-regulated probes with a false discovery rate (FDR) of < 0.05%. Signal intensities below 30 in both the tumor and corresponding normal probe were excluded from further analyses. T/N ratios were calculated by applying 2X, 2.5X (data not shown) and 3X cut-off, and probes were matched to genes. The analysis revealed 53 unique genes within the high BMI prostate T/N data set, and 134 unique genes within the normal BMI prostate T/N data set. Venn diagram analysis showed that 34 genes were shared between the high and normal BMI data sets, indicating AMACR and ERG as top scoring common genes. Gene set unique to high BMI prostate cancer T/N was further analyzed (Figure 2).
Figure 2.
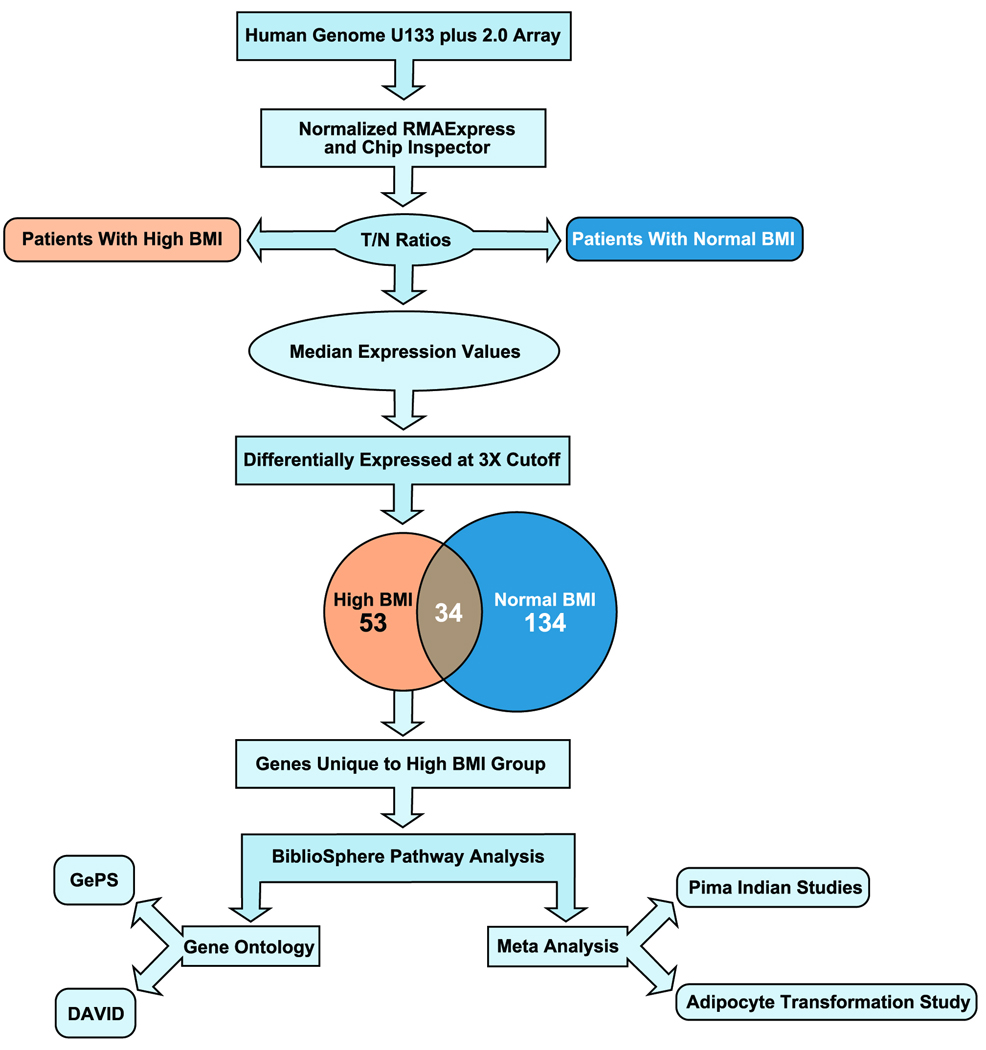
Schematic diagram of bioinformatic analysis.
Gene ontology and pathway analysis
The unique high BMI-associated genes (n=53; 20 up-regulated and 33 down-regulated) were queried by the Genomatix Pathway System (GePS) that utilizes expert-curated gene ontology information from public and proprietary databases (Genomatix GmbH, Munich, Germany). In an independent approach high BMI associated genes were queried by The Database for Annotation, Visualization and Integrated Discovery (DAVID) software (http://david.abcc.ncifcrf.gov).30 The Functional Classification Tool was used to assess the functional similarity between input genes. The clustering algorithm used by DAVID classifies highly related genes into functionally related groups. The “high clustering criteria” was used that revealed tight association of genes belonging to cholesterol and lipid metabolism. These genes ranked 2nd in the clustering with an enrichment score of 1.47. To evaluate functional connections between genes in the high BMI set we entered the gene ID of the 53 genes into the BiblioSphere knowledge-based pathway analysis software (Genomatix GmbH, Munich, Germany).31 The pathway analysis connects genes by co-citation within one sentence at abstract levels. Genes forming the central node, such as SCD1, INSIG1, ANPEP, MAF, and JAG1 were further analyzed to evaluate significant probe-signal intensities individually in tumor and benign samples. Signal intensities passing the greater than 30 cutoff in either or both of tumor and benign data sets were log2 transformed and presented in box-plot diagrams.
Results
Selection of patients with elevated and normal BMI
Prostate cancer patients treated with RP were selected in the high and low BMI groups by matching their age, race, differentiation status, absence of pretreatment with cholesterol-lowering drugs and family history of prostate cancer (Table 1). LCM-selected tumor cells from RP specimens were matched by the cellular differentiation status. From each specimen, tumor and matching morphologically normal prostate epithelial cells were isolated, total RNA were extracted, and gene expression levels were measured by microarray analysis.26,28
Expression of ERG, PCA3 and AMACR prostate cancer marker genes validate the precision of tumor and matching normal cell isolation
To assure that the tumor and matching normal gene expression data set represents the precise isolation of tumor cells and normal prostate epithelial cells by LCM, prostate cancer-associated expression of ERG, PCA3, and AMACR genes were examined by quantitative polymerase chain reaction (Q-PCR). Robust overexpression of these prostate cancer markers indicated the precise isolation of tumor and normal cells from the RP specimens (Figure 1).
Figure 1.
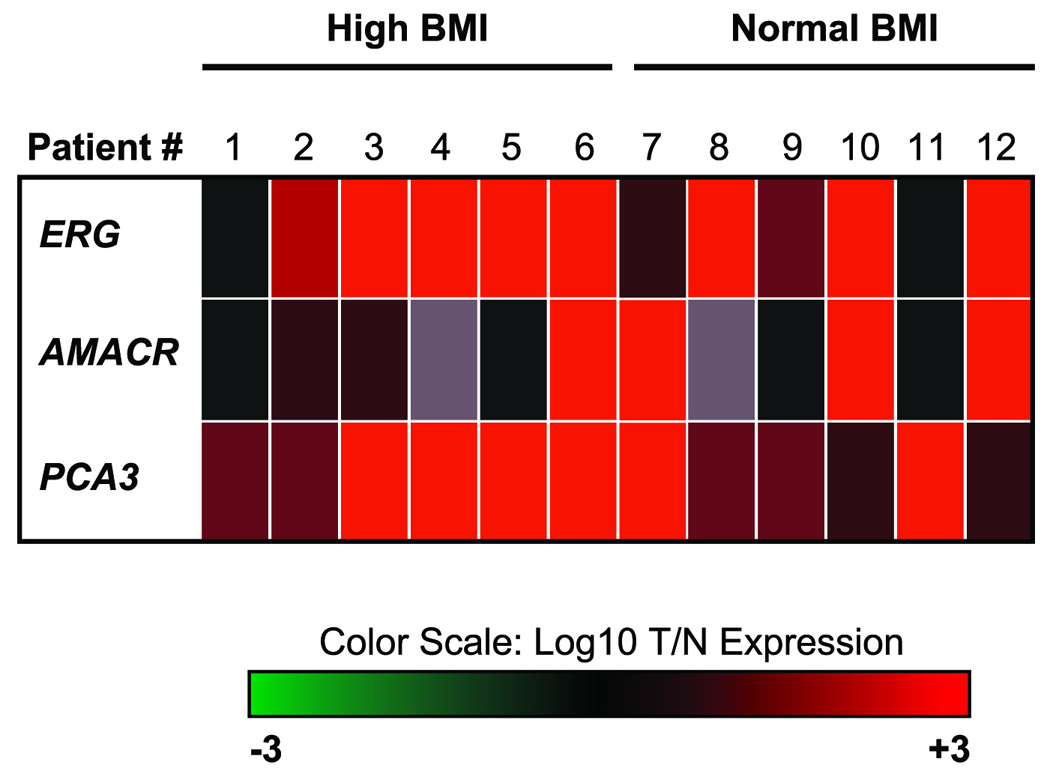
Quantitative RT-PCR analysis of ERG, AMACR and PCA3 prostate cancer marker genes. For the quality control of LCM selected tumor and matching benign epithelial cells, RNA was isolated and ERG, AMACR and PCA3 gene expression levels were defined by QRT-PCR. T/N ratios were calculated and were log10 transformed and are represented on the heat map. Extreme green (−3) and extreme red (+3) colors denote 1000X down or 1000X upregulation, respectively.
Defining the prostate cancer signature unique to high BMI by stringent bioinformatic criteria
Gene expression features were normalized by using RMAExpress and by ChipInspector softwares (false discovery rate of 0.05%) (Figure 2). Gene expression features with signal intensities less than 30 in both tumor and normal expressions were excluded from further evaluation. Gene expression features meeting this criterion were normalized within both high and low BMI groups by dividing the gene expression intensities of tumor cell-associated signals by the intensities of their matching normal pair (T/N expression ratio). Median values were calculated for both high and low BMI T/N ratios. A stringent 3X cut off was applied to enrich gene expression signatures of significant expression alterations. High BMI-unique gene expression features were further analyzed by excluding gene expression features that were either shared between the high and normal BMI groups or unique to the normal BMI group. Gene expression features unique to the high BMI group were matched to 53 genes, 20 up-regulated and 33 down-regulated genes, respectively (Table 2), and were evaluated by gene ontology (GO), pathway, and meta-analyses approaches.
Table 2.
Tumor over normal gene expression ratios of the high BMI prostate cancer signature.
| UP REGULATED | |
|---|---|
| Gene Symbol |
High BMI Median T/N Fold Expression |
| C8orf4 | 5.14 |
| TMC5 | 4.83 |
| PDLIM5 | 4.51 |
| JAG1 | 4.36 |
| HOXA9 | 4.25 |
| C4A | 3.98 |
| STEAP4 | 3.92 |
| HLA-DMB | 3.85 |
| COL12A1 | 3.81 |
| DHRS8 | 3.81 |
| C10orf137 | 3.62 |
| SLC7A1 | 3.44 |
| PDZRN3 | 3.30 |
| F3 | 3.26 |
| ACER3 | 3.17 |
| POLB | 3.16 |
| PXDN | 3.12 |
| PROM2 | 3.11 |
| GUCY1A3 | 3.10 |
| SOX4 | 3.02 |
| DOWN REGULATED | |||
|---|---|---|---|
| Gene Symbol |
High BMI Median T/N Fold Expression |
Gene Symbol |
High BMI Median T/N Fold Expression |
| ACADL | 0.33 | ZNF532 | 0.26 |
| PHLDA2 | 0.33 | SCD1 | 0.25 |
| GPD1L | 0.33 | PTN | 0.24 |
| NTAN1 | 0.33 | SC4MOL | 0.24 |
| LOC389048 | 0.32 | MAF | 0.22 |
| SFN | 0.32 | NPAL3 | 0.22 |
| HSRG1 | 0.32 | CYP3A5 | 0.21 |
| PAK1IP1 | 0.32 | NOV | 0.20 |
| EBP | 0.31 | ALOX15B | 0.17 |
| ANPEP | 0.31 | LOC399959 | 0.17 |
| ANTXR2 | 0.29 | VGL-3 | 0.16 |
| COL4A6 | 0.29 | INSIG1 | 0.16 |
| LOC400880 | 0.29 | Rean3 | 0.15 |
| RHOU | 0.29 | Poteg | 0.14 |
| GREB1 | 0.28 | P704P | 0.13 |
| FLJ30428 | 0.27 | EFS | 0.13 |
| C3orf14 | 0.27 | ||
Gene expression signature of prostate tumors in high BMI patients is enriched in lipid metabolic process and oxidative stress response genes
The high BMI-associated gene set was queried for GO by the GePS software (Genomatix GmbH, Munich, Germany) (Table 3). GePS analysis indicated enrichment of genes belonging to lipid and steroid metabolic process by querying the biological process category. The highest enrichment score in GePS highlighted oxidation-reduction and lipid metabolic process as the top scoring GO categories. To confirm these findings by an independent approach the high BMI gene signature was also queried by DAVID by using the Functional Classification Tool (data not shown).30 The clustering algorithm used in DAVID classifies highly related genes into functionally related groups. The high clustering stringency used in this analysis confirmed the enrichment of lipid metabolism-associated genes.
Table 3.
Gene ontology categories of genes identified within the high BMI prostate cancer signature.
| GO-term | P-value | Genes (High BMI Tumor/Normal) |
Observed Genes | ||
|---|---|---|---|---|---|
| Observed | Expected | Total | |||
| Oxidation Reduction | 2.35E-05 | 9 | 1.62 | 606 | ACDAL, SCD, SC4MOL, STEAP4, GPDIL, HSD17B11, PXDN, ALOX15B |
| Cellular Lipid Metabolic Process |
4.54E-05 | 9 | 1.76 | 659 | ACADL, SCD, SC4MOL, ACER3, HSD17B11, CYP3A5, ALOX15B |
| Steroid Metabolic Process |
1.80E-04 | 5 | 0.54 | 200 | EBP, SC4MOL, HSD17B11, INSIG1, CYP3A5 |
| Lipid Metabolic Process |
2.27E-04 | 9 | 2.17 | 813 | ACADL, SCD, SC4MOL, ACER3, HSD17B11, CYP3A5, ALOX15B |
Knowledge-based pathway analysis of the high BMI signature suggest a gene network linking fatty acid synthesis regulatory genes to genes associated with oncogenesis
We have evaluated the high BMI unique signature gene set (53 genes) in a knowledge-based pathway analysis method by using the Genomatix BiblioSphere software (Figure 3).31 The resulted network consisted of 11 genes connected at the co-citation level within one sentence at abstract levels. The pathway revealed that genes involved in the regulation of lipid metabolism, such as INSIG1 and SCD1, are connected through the ANPEP (CD13) gene involved in the metabolism of regulatory peptides, to MAF protooncogene and JAG1, a ligand for notch receptors, forming the central regulatory node.
Figure 3.
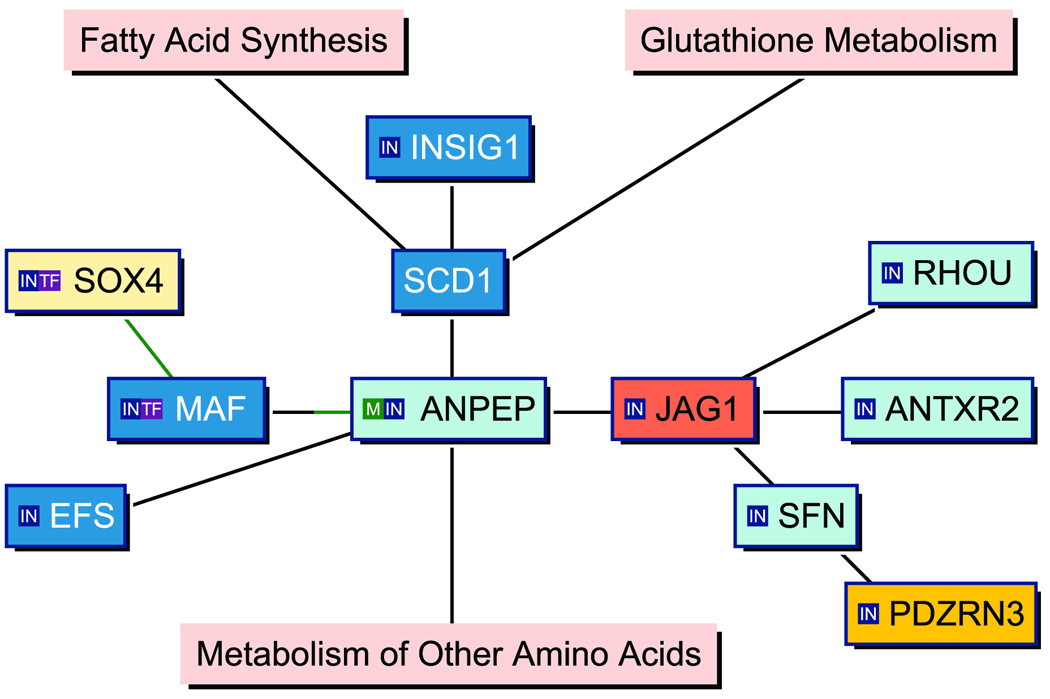
Knowledge-based pathway analysis (Genomatix BiblioSphere Software) indicating the network of high BMI-associated genes. The displayed network is constructed from 53 input genes. Connection lines are drawn as a result of co-citation of two genes within one sentence. Green or partially green connection lines indicate a transcription factor matrix match in the promoter of the gene connected by the green line. Orange or yellow indicates up-regulated, shades of blue mark down-regulated genes in the T/N data sets. Major biochemical pathways are highlighted with pink color.
The high BMI prostate cancer signature is a result of elevated gene expression in normal epithelial cells within the prostate tumor microenvironment
For this study, both tumor and matching normal gene expression values were available. Thus, we addressed the patient-to-patient variations in the informatic analyses by using T/N normalized expression ratios. Within the group of genes forming the central regulatory node, we noted frequently decreased T/N ratios. We reasoned that reduced T/N may be the result of elevated gene expression in the matching normal cells within the high BMI signature. To address this possibility, we examined patient-by-patient the signal intensities of genes contributing to the central regulatory node (Figure 4a–e). Signal intensities from RMA and ChipInspector normalized data were log2 transformed and the data were summarized in box-plot graphs showing the median value, distribution, and spread of gene expression signal intensities in tumor and matching normal cells. The results suggested that elevated gene expression in normal prostate epithelial cells from high BMI patients may significantly contribute to the high BMI signature. The observed upregulation of INSIG1 and SCD1 genes in normal prostate epithelial cells is consistent with the elevated BMI status of patients. These results highlight the marked elevation of genes in fatty acid and steroid metabolic pathways within the benign prostate epithelial environment of prostate tumors.
Figure 4.
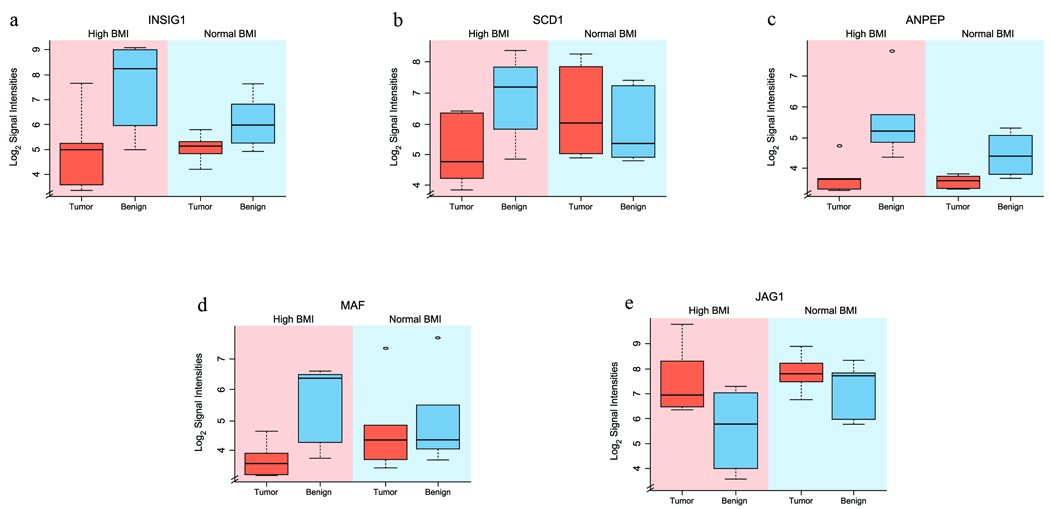
a–e: Expression of the individual genes forming the central node of high BMI prostate cancer signature. Genes forming the central node were individually analyzed to assess the differential expression in tumor and matching benign cells. Log2 transformed gene expression signal intensities of (a) INSIG1 (p=0.0038), (b) SCD1 (p=0.0113), (c) ANPEP (p=0.0150), (d) MAF (p=0.0131) and (e) JAG1 (p=0.0081) genes are shown in box-plot diagrams representing expression signal intensity values in tumor and normal prostate epithelial cells in high and normal BMI patients.
The high BMI prostate gene expression signature is consistent with signatures of obesity-related gene expression data sets
We have examined the consistency between genes observed in the high BMI-associated prostate expression signature and adipocyte gene expression datasets obtained from Pima Indians study32 and from an adipocyte differentiation model (Table 4).21 The meta-analysis revealed that out of the 53 genes identified in our study, 26 overlapped with either or both of the examined datasets. Remarkably, all genes of the central node (SCD1, ANPEP, MAF, and JAG1) were identified by the meta-analysis.
Table 4.
Meta-analyses of high BMI associated prostate cancer gene signature comparing gene expression datasets of the Pima Indian abdominal subcutaneous adipocyte (Meta 1 Genes) and adipocyte transformation (Meta 2 Genes) studies. Out of 53 gene expression alterations in the high BMI prostate cancer signature (Table 2), 26 genes matched either to Meta 1 or to Meta 2.
| Meta 1 Genes |
High BMI Signature |
Meta 2 Genes |
|
|---|---|---|---|
| Genes in Pathway | ANPEP | ANPEP | |
| SCD1 | SCD1 | SCD1 | |
| JAG1 | JAG1 | JAG1 | |
| ANTXR2 | ANTXR2 | ||
| INSIG1 | INSIG1 | ||
| SOX4 | SOX4 | ||
| MAF | MAF | ||
| Other High BMI Signature Gene Matches | C8orF4 | C8orF4 | |
| STEAP4 | STEAP4 | ||
| COL12A1 | COL12A1 | ||
| DHRS8 | DHRS8 | ||
| C10orF137 | C10orF137 | C10orF137 | |
| SLC7A1 | SLC7A1 | SLC7A1 | |
| F3 | F3 | ||
| POLB | POLB | ||
| GUCY1A3 | GUCY1A3 | ||
| ACADL | ACADL | ||
| PHLDA2 | PHLDA2 | PHLDA2 | |
| GPDIL | GPDIL | ||
| SFN | SFN | ||
| PAK11P1 | PAK11P1 | ||
| EBP | EBP | ||
| COL4A6 | COL4A6 | ||
| C3orF14 | C3orF14 | ||
| PTN | PTN | ||
| SC4MOL | SC4MOL | ||
| C3orF14 |
Discussion
Comparative evaluation of gene expression signatures of prostate tumor and matching normal cells from patients with high and normal BMI, indicated the enrichment of cholesterol and lipid metabolism associated genes within fatty acid synthesis and oxidation-reduction GO categories. Knowledge-based pathway analysis of high BMI-associated genes suggested a connection to genes involved in lipid metabolism, cholesterol homeostasis, and tumorigenesis. Our results highlight the SCD1 gene, a stearoyl-CoA desaturase associated with obesity.33 SCD1 enzyme regulates the synthesis of unsaturated fatty acids thereby altering membrane fluidity. Importantly, atorvastatin can reduce levels of SCD1, suggesting for a potential mechanistic link between the benefits of statins in prostate cancer progression.34 SCD1-enzyme inhibitors have been proposed for the therapy of obesity with cautions for the potential pro-inflammatory response to complete blockade of SCD1.35 Comparative evaluation of SCD1 gene expression levels in tumor and matching normal prostate epithelial cells from patients with high and normal BMI revealed the elevated gene expression of SCD1 in normal prostate epithelial cells. This finding is consistent with reported obesity-associated gene expression alterations. In prostate tumor cells, we found that SCD1 expression levels were decreased, which may aggravate pro-inflammatory processes in prostate cancer.35
In an earlier report over expression of SCD1 was shown in human prostate cancer by analyzing macro-dissected tumors with Gleason ≥7.36 In our study prostate cancer cells were micro-dissected from tumors with Gleason ≤7. Thus, the two studies may not be directly comparable.
The insulin-induced gene, INSIG1, plays central roles in the feedback control of cellular cholesterol levels. Frequent up-regulation of INSIG1 is associated with metabolic disease conditions including elevated BMI. JAG1 is a receptor of notch signals37 and down-regulation of JAG1 results in inhibited prostate cancer growth.38 The association of JAG1 with prostate cancer recurrence and metastasis has been previously shown.39 In prostate cancer, MAF expression was associated with the receptor tyrosine kinase, PDGFR-beta status. Expression of ANPEP (also known as CD13), a broad specificity aminopeptidase, is frequently lost or reduced in prostate tumor cells.40 We individually analyzed the gene expression levels of these genes in prostate tumor cells and in matching normal epithelial cells. The analysis revealed elevated expression of INSIG1, SCD1, MAF, and ANPEP genes in tumor-matched normal epithelial cells. These findings indicate that the prostatic cellular environment harbors a gene expression signature that is a hallmark of elevated BMI. Whether this signature contributes to a permissive cellular environment similarly to the recently described role of FASN41 in prostate cancer progression will require further investigation.
A limitation of the present study is the smaller sample size, which may have resulted in model overfitting. However, high-quality GeneChip data set from RNA specimens of micro-dissected benign and malignant prostate epithelium; careful patient selection for matching age, race, and pathological stage; and meta-analysis in independent gene expression data sets underscores the strength of major findings of this study. Another limitation is that gene expression alterations below the stringent three-fold cut-off were potentially missed. However, the goal of using stringent criterion was to pinpoint robust gene expression alterations with potential translational utility.
Although the relationship of obesity and prostate cancer incidence is complex, and in many aspects unclear,8 previous reports have shown increased prostate cancer-related mortality with higher BMI.42,43 The association of elevated BMI with increased incidences of aggressive prostate cancer suggests that high BMI may contribute to hormone production and activation of cholesterol synthesis and inflammatory pathways, affecting prostate cancer growth.44–46 An intriguing, new observation in our study is the identification of SCD1 gene in high BMI prostate cancer signature, which is also a target for atorvastatin. The clinical use of statins has been reported to reduce prostate cancer recurrence and mortality, suggesting that lipids are important in prostate cancer progression.20,47,48 Whether SCD1 is indeed a therapeutic target for statins in reducing prostate cancer progression warrants further investigations.
Acknowledgements
Authors express sincere thanks to Ms. Lakshmi Ravindranath and Ms. London Toney for the superb technical assistance, to Mr. Zhe Chang and Ms. Sally Elsamanoudi for the medical informatics support, to Mr. Stephen Doyle for the outstanding art work, and to Ms. Tia Morris for editorial comments. This work was funded by the CPDR through an ongoing grant from the U.S. Army Medical Research and Materiel Command and the NIH grant RO1 DK065977 to S.S and G.P. The views expressed in this manuscript are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or the U.S. Government.
Footnotes
Conflict of interest
The authors declare no competing financial interests.
Disclaimer
The views expressed in this manuscript are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or the U.S. Government. Released for publication by the WRAMC Public Affairs Office.
References
- 1.Moul JW, Sesterhenn IA, Connelly RR, Douglas T, Srivastava S, Mostofi FK, et al. Prostate-specific antigen values at the time of prostate cancer diagnosis in African-American men. JAMA. 1995;274:1277–1281. [PubMed] [Google Scholar]
- 2.Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004;13:103–121. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- 3.Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zadra G, Priolo C, Patnaik A, Loda M. New strategies in prostate cancer: targeting lipogenic pathways and the energy sensor AMPK. Clin Cancer Res. 2010;16:3322–3328. doi: 10.1158/1078-0432.CCR-09-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hankey BF, Feuer EJ, Clegg LX, Hayes RB, Legler JM, Prorok PC, et al. Cancer surveillance series: interpreting trends in prostate cancer--part I: Evidence of the effects of screening in recent prostate cancer incidence, mortality, and survival rates. J Natl Cancer Inst. 1999;91:1017–1024. doi: 10.1093/jnci/91.12.1017. [DOI] [PubMed] [Google Scholar]
- 6.Coffey RN, Watson RW, Fitzpatrick JM. Signaling for the caspases: their role in prostate cell apoptosis. J Urol. 2001;165:5–14. doi: 10.1097/00005392-200101000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Coffey DS. New insights and methodologies are needed to solve the many epidemiologic enigmas of prostate cancer. Epidemiol Rev. 2001;23:1. doi: 10.1093/oxfordjournals.epirev.a000772. [DOI] [PubMed] [Google Scholar]
- 8.Freedland SJ, Williams CD, Masko EM. Adiponectin and prostate cancer mortality: to be or not to be skinny? Clin Chem. 2010;56:1–3. doi: 10.1373/clinchem.2009.137406. [DOI] [PubMed] [Google Scholar]
- 9.Freedland SJ, Platz EA. Obesity and prostate cancer: making sense out of apparently conflicting data. Epidemiol Rev. 2007;29:88–97. doi: 10.1093/epirev/mxm006. [DOI] [PubMed] [Google Scholar]
- 10.Davies BJ, Smaldone MC, Sadetsky N, Dall'era M, Carroll PR. The impact of obesity on overall and cancer specific survival in men with prostate cancer. J Urol. 2009;182:112–117. doi: 10.1016/j.juro.2009.02.118. discussion 117. [DOI] [PubMed] [Google Scholar]
- 11.Michalakis K, Williams CJ, Mitsiades N, Blakeman J, Balafouta-Tselenis S, Giannopoulos A, et al. Serum adiponectin concentrations and tissue expression of adiponectin receptors are reduced in patients with prostate cancer: a case control study. Cancer Epidemiol Biomarkers Prev. 2007;16:308–313. doi: 10.1158/1055-9965.EPI-06-0621. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Stampfer MJ, Mucci L, Rifai N, Qiu W, Kurth T, et al. A 25-year prospective study of plasma adiponectin and leptin concentrations and prostate cancer risk and survival. Clin Chem. 2010;56:34–43. doi: 10.1373/clinchem.2009.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedland SJ. Obesity and prostate cancer: a growing problem. Clin Cancer Res. 2005;11:6763–6766. doi: 10.1158/1078-0432.CCR-05-1305. [DOI] [PubMed] [Google Scholar]
- 14.Nobes JP, Langley SE, Laing RW. Metabolic syndrome and prostate cancer: a review. Clin Oncol (R Coll Radiol) 2009;21:183–191. doi: 10.1016/j.clon.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Murtola TJ, Tammela TL, Maattanen L, Huhtala H, Platz EA, Ala-Opas M, et al. Prostate cancer and PSA among statin users in the Finnish prostate cancer screening trial. Int J Cancer. 2010 doi: 10.1002/ijc.25165. e-pub ahead of print 13 Jan 2010; doi: 10.1002/ijc.25165. [DOI] [PubMed] [Google Scholar]
- 16.Getzenberg RH. Statins and the risk of prostate cancer or benign prostatic hyperplasia: biological plausibility. J Urol. 2010;184:415–416. doi: 10.1016/j.juro.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang L, Kim J, Adam RM, Solomon KR, Freeman MR. Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. J Clin Invest. 2005;115:959–968. doi: 10.1172/JCI200519935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekine Y, Furuya Y, Nishii M, Koike H, Matsui H, Suzuki K. Simvastatin inhibits the proliferation of human prostate cancer PC-3 cells via down-regulation of the insulin-like growth factor 1 receptor. Biochem Biophys Res Commun. 2008;372:356–361. doi: 10.1016/j.bbrc.2008.05.043. [DOI] [PubMed] [Google Scholar]
- 19.Banez LL, Klink JC, Jayachandran J, Lark AL, Gerber L, Hamilton RJ, et al. Association between statins and prostate tumor inflammatory infiltrate in men undergoing radical prostatectomy. Cancer Epidemiol Biomarkers Prev. 2010;19:722–728. doi: 10.1158/1055-9965.EPI-09-1074. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton RJ, Banez LL, Aronson WJ, Terris MK, Platz EA, Kane CJ, et al. Statin medication use and the risk of biochemical recurrence after radical prostatectomy: results from the Shared Equal Access Regional Cancer Hospital (SEARCH) Database. Cancer. 2010;116:3389–3398. doi: 10.1002/cncr.25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch HA, Iliopoulos D, Joshi A, Zhang Y, Jaeger SA, Bulyk M, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention. Overweight and Obesity. U.S. Obesity Trends. [accessed 11 Aug 2010]; www.cdc.gov/obesity/data/trends.html.
- 23.Giovannucci E, Rimm EB, Stampfer MJ, Colditz GA, Willett WC. Height, body weight, and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1997;6:557–563. [PubMed] [Google Scholar]
- 24.Okasha M, McCarron P, McEwen J, Smith GD. Body mass index in young adulthood and cancer mortality: a retrospective cohort study. J Epidemiol Community Health. 2002;56:780–784. doi: 10.1136/jech.56.10.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burton A, Martin R, Galobardes B, Davey Smith G, Jeffreys M. Young adulthood body mass index and risk of cancer in later adulthood: historical cohort study. Cancer Causes Control. 2010 doi: 10.1007/s10552-010-9625-3. e-pub ahead of print 1 Aug 2010; doi: 10.1007/s10552-010-9625-3. [DOI] [PubMed] [Google Scholar]
- 26.Furusato B, Shaheduzzaman S, Petrovics G, Dobi A, Seifert M, Ravindranath L, et al. Transcriptome analyses of benign and malignant prostate epithelial cells in formalin-fixed paraffin-embedded whole-mounted radical prostatectomy specimens. Prostate Cancer Prostatic Dis. 2008;11:194–197. doi: 10.1038/sj.pcan.4501007. [DOI] [PubMed] [Google Scholar]
- 27.Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 29.Cohen CD, Lindenmeyer MT, Eichinger F, Hahn A, Seifert M, Moll AG, et al. Improved elucidation of biological processes linked to diabetic nephropathy by single probe-based microarray data analysis. PLoS One. 2008;3:e2937. doi: 10.1371/journal.pone.0002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 31.Scherf M, Epple A, Werner T. The next generation of literature analysis: integration of genomic analysis into text mining. Brief Bioinform. 2005;6:287–297. doi: 10.1093/bib/6.3.287. [DOI] [PubMed] [Google Scholar]
- 32.Lee YH, Nair S, Rousseau E, Allison DB, Page GP, Tataranni PA, et al. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia. 2005;48:1776–1783. doi: 10.1007/s00125-005-1867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flowers MT, Ntambi JM. Stearoyl-CoA desaturase and its relation to high-carbohydrate diets and obesity. Biochim Biophys Acta. 2009;1791:85–91. doi: 10.1016/j.bbalip.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Fuentes P, Garcia-Otin AL, Calvo L, Gomez-Coronado D, Civeira F, Cenarro A. Atorvastatin decreases stearoyl-CoA desaturase gene expression in THP-1 macrophages incubated with oxidized LDL. Lipids. 2009;44:115–123. doi: 10.1007/s11745-008-3255-5. [DOI] [PubMed] [Google Scholar]
- 35.Brown JM, Rudel LL. Stearoyl-coenzyme A desaturase 1 inhibition and the metabolic syndrome: considerations for future drug discovery. Curr Opin Lipidol. 2010;21:192–197. doi: 10.1097/MOL.0b013e32833854ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fritz V, Benfodda Z, Rodier G, Henriquet C, Iborra F, Avances C, Allory Y, de la Taille A, Culine S, Blancou H, Cristol JP, Michel F, Sardet C, Fajas L. Abrogation of de novo lipogenesis by stearoyl-CoA desaturase 1 inhibition interferes with oncogenic signaling and blocks prostate cancer progression in mice. Mol Cancer Ther. 2010;9:1740–1754. doi: 10.1158/1535-7163.MCT-09-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leong KG, Gao WQ. The Notch pathway in prostate development and cancer. Differentiation. 2008;76:699–716. doi: 10.1111/j.1432-0436.2008.00288.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Li Y, Banerjee S, Kong D, Ahmad A, Nogueira V, et al. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J Cell Biochem. 2010;109:726–736. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 39.Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 40.Liu AY, Roudier MP, True LD. Heterogeneity in primary and metastatic prostate cancer as defined by cell surface CD profile. Am J Pathol. 2004;165:1543–1556. doi: 10.1016/S0002-9440(10)63412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen PL, Ma J, Chavarro JE, Freedman ML, Lis R, Fedele G, et al. Fatty Acid Synthase Polymorphisms, Tumor Expression, Body Mass Index, Prostate Cancer Risk, and Survival. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.27.0793. e-pub ahead of print 2 August 2010; doi: 10.1200/JCO.2009.27.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez C, Patel AV, Calle EE, Jacobs EJ, Chao A, Thun MJ. Body mass index, height, and prostate cancer mortality in two large cohorts of adult men in the United States. Cancer Epidemiol Biomarkers Prev. 2001;10:345–353. [PubMed] [Google Scholar]
- 44.Mink SR, Hodge A, Agus DB, Jain A, Gross ME. Beta-2-microglobulin expression correlates with high-grade prostate cancer and specific defects in androgen signaling. Prostate. 2010;70:1201–1210. doi: 10.1002/pros.21155. [DOI] [PubMed] [Google Scholar]
- 45.Neuhouser ML, Till C, Kristal A, Goodman P, Hoque A, Platz EA, et al. Finasteride modifies the relation between serum C-peptide and prostate cancer risk: results from the Prostate Cancer Prevention Trial. Cancer Prev Res (Phila Pa) 2010;3:279–289. doi: 10.1158/1940-6207.CAPR-09-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaaks R, Stattin P. Obesity, endogenous hormone metabolism, and prostate cancer risk: a conundrum of "highs" and "lows". Cancer Prev Res (Phila Pa) 2010;3:259–262. doi: 10.1158/1940-6207.CAPR-10-0014. [DOI] [PubMed] [Google Scholar]
- 47.Gutt R, Tonlaar N, Kunnavakkam R, Karrison T, Weichselbaum RR, Liauw SL. Statin use and risk of prostate cancer recurrence in men treated with radiation therapy. J Clin Oncol. 2010;28:2653–2659. doi: 10.1200/JCO.2009.27.3003. [DOI] [PubMed] [Google Scholar]
- 48.Platz EA, Leitzmann MF, Visvanathan K, Rimm EB, Stampfer MJ, Willett WC, et al. Statin drugs and risk of advanced prostate cancer. J Natl Cancer Inst. 2006;98:1819–1825. doi: 10.1093/jnci/djj499. [DOI] [PubMed] [Google Scholar]


