Abstract
Diapause-destined embryos of the crustacean, Artemia franciscana, accumulate large amounts of an oligomeric, heat-stable, molecular chaperone termed artemin, a cysteine-enriched ferritin homologue. In this study, cysteines 22, 61, 166, and 172 of artemin were substituted with alanines, respectively yielding ArtC22A, ArtC61A, ArtC166A, and ArtC172A. Wild-type and modified artemins were synthesized in transformed bacteria and purified. As measured by heat-induced denaturation of citrate synthase in vitro, each substitution reduced chaperone activity, with ArtC172A the least active. Protein modeling indicated that C172 is close to a region of surface hydrophobicity, also present in ferritin, suggesting that this site contributes to chaperone activity. Only slight differences in oligomer molecular mass were apparent between artemin variants, but ArtC22A and ArtC61A displayed significantly reduced thermostability, perhaps due to the disruption of an inter-subunit disulphide bridge. In contrast, ArtC172A was thermostable, reflecting the location of C172 on the oligomer surface and that it contributes minimally to artemin stabilization. To our knowledge, this is the initial study of structure/function relationships within a ferritin homologue of importance in diapause and the first to indicate that a defined region of hydrophobicity contributes to artemin and ferritin chaperoning.
Keywords: Artemin, Ferritin homologue, Molecular chaperone, Diapause, Protein structure/function, Cysteine, Artemia franciscana
Introduction
Survival of the extremophile crustacean, Artemia franciscana, in high-salinity environments subject to fluctuations in temperature, oxygen concentration, and food availability depends on its unusual life history. Artemia embryos may undergo ovoviviparous development with release of swimming nauplii from females. In contrast, embryos can develop oviparously, arresting as gastrulae and becoming enclosed in a rigid chitinous shell (Liang and MacRae 1999; MacRae 2003). Upon exiting females, cysts enter diapause, a physiological state characterized by the cessation of growth and profound dormancy (Jackson and Clegg 1996; Clegg et al. 2000; MacRae 2003, 2005, 2010). Cysts are exceptionally resistant to physiological stress even when hydrated, and as one extraordinary example of this capability, they survive years of anoxia (Clegg 1994, 1997; Clegg et al. 1999, 2000; van Breukelen et al. 2000; Viner and Clegg 2001). Diapause continues, even under conditions favorable for growth, until terminated by environmental stimuli such as desiccation, light, and/or cold (Drinkwater and Crowe 1987; Van Der Linden et al. 1988; Drinkwater and Clegg 1991; Clegg et al. 2000; Robbins et al. 2010). Activated cysts either remain in quiescence (Clegg and Jackson 1998) or they resume development when hydrated at appropriate temperatures and oxygen levels (Drinkwater and Clegg 1991; Clegg et al. 1999).
The ferritin homologue artemin, synthesized only in diapause-destined Artemia embryos, constitutes 10–15% of post-ribosomal protein in cysts (Tanguay et al. 2004; Warner et al. 2004). Artemin and ferritin are very heat-stable (Kilic et al. 2003; Warner et al. 2004; Baraibar et al. 2008; Fan et al. 2009), and both proteins are molecular chaperones, preventing heat-induced denaturation of citrate synthase in vitro (Chen et al. 2007). Artemin also confers stress tolerance on transfected mammalian cells (Chen et al. 2007) and binds RNA at high temperature, perhaps protecting this macromolecule (Warner et al. 2004). During post-diapause development, artemin mRNA and protein degrade, suggesting, in concert with several of its other characteristics, a diapause-specific function for the protein.
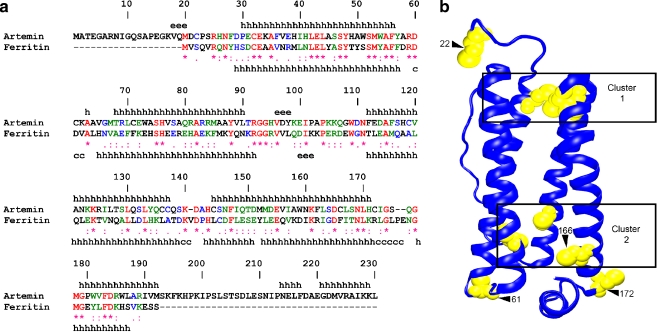
The artemin monomer, consisting of 230 amino acid residues including the initiator methionine, has a molecular mass of approximately 26 kDa, and it shares sequence similarity with ferritin (Fig. 1a; De Graaf et al. 1990; Chen et al. 2003). Excluding the artemin amino- and carboxy-terminal extensions, artemin and bullfrog ferritin are 27.4% identical and 54.9% similar. Computer modeling indicates comparable secondary (Fig. 1a) and tertiary (Fig. 1b) structures in regions of sequence similarity (Chen et al. 2003). Artemin and ferritin assemble into oligomers of about 600 kDa composed of 24 subunits (De Graaf et al. 1990; Kilic et al. 2003; Chen et al. 2003; Theil and Matzapetakis 2006). The amino-terminal of each artemin monomer protrudes from the oligomer, whereas the carboxy-terminus possesses a sequence of 45–50 residues not found in ferritin. The carboxy-terminal extension of artemin fills the oligomer cavity that in ferritin is used to store iron (De Graaf et al. 1990; Chen et al. 2003, 2007). Artemin is missing all but one residue constituting the ferritin di-iron ferroxidase center, another indication it does not sequester metals (De Graaf et al. 1990; Harrison and Arosio 1996; Chen et al. 2003, 2007; Theil and Matzapetakis 2006; Rasti et al. 2009).
Fig. 1.
Artemin and ferritin structures are related. a The amino acid sequences of artemin (AAL55397) and bullfrog ferritin (P07798) were aligned by CLUSTALW and numbered according to the artemin sequence. *, identical residues; :, conserved substitution; ., semi-conserved substitution. The secondary structure of ferritin was obtained from its crystal structure (1MFR), while the secondary structure of artemin was predicted by the MLRC method at NPS@server. The secondary structures of artemin and ferritin are indicated above and below their sequences. h α-helix, e extended strand, c random coil. b The location of cysteine residues within the artemin monomer was determined by MODELER 7 × 7 using the crystal structure of bullfrog ferritin and residues 21-192 and 2-189 of artemin and ferritin, respectively. Cysteines, yellow. Clustered cysteines are in numbered boxes and modified cysteines are shown by numbered arrowheads. The image was generated with VMD and Raster3D in new cartoon
As indicated by molecular modeling, each artemin monomer consists of a bundle of four parallel α-helices and a fifth shorter α-helix perpendicular to the bundle (Fig. 1b). The ferritin monomer has the same structure with the corresponding short α-helix participating in the interaction between four monomers and formation of the fourfold channel in the oligomer (Fan et al. 2009). Modeling implies that seven of the ten cysteines in artemin are buried in two clusters at opposite ends of the four-helix bundle (Fig. 1b). Cluster 1 is composed of C32, C136, C137, and C144, while cluster 2 is composed of C71, C119, and C166. On the other hand, C22, C61, and C172 are situated in unstructured regions connecting helixes. The cysteines may form disulfide bridges or interact with other residues, thereby stabilizing monomer and/or oligomer structure. Cysteine residues in other proteins modulate chaperone activity as shown for the redox-regulated chaperones bacterial Hsp33 and eukaryotic 2-Cys peroxiredoxins (Kumsta and Jakob 2009), and they influence switching of the disulfide reductase, thioredoxin, to a chaperone (Park et al. 2009). In this paper, alanines were substituted for cysteines at positions 22, 61, 166, and 172 of artemin. Analysis of these variants, in concert with molecular modeling, has increased our understanding of the contributions made by cysteine residues to artemin structure and chaperoning, the latter an important activity required for stress tolerance during diapause maintenance in Artemia embryos.
Materials and methods
Site-directed mutagenesis of artemin
Artemin cDNA was modified with the QuikChange®II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) using designated primers (Table 1) and pPROTet.E133-artemin-wt (Chen et al. 2003) as template. Fifty-microliter PCR mixtures included 5 μl of 10× reaction buffer, 2 μl (10 ng) of cDNA, 1.25 μl (125 ng) each of sense and anti-sense primers, 1 μl of dNTP mix, 3 μl of QuikSolution reagent, 1 μl of pfuUltra HF DNA polymerase (2.5 U/μl), and 35.5 μl of ddH2O. The reaction mixtures were incubated 1 min at 95°C followed by 18 cycles of 50 s at 95°C, 50 s at 60°C, 3 min at 68°C, and 7 min at 68°C, then placed on ice for 2 min prior to the addition of 1 μl of Dpn I (10 U/μl) and incubation at 37°C for 1 h. Escherichia coli XL10-Gold ultra-competent cells (VWR International, Mississauga, ON) were transformed with the DNA-containing reaction mixtures following manufacturer’s instructions. Bacterial clones were selected randomly after plating and incubation, plasmids were obtained, and the identity of each mutant was verified by sequencing (DNA Sequencing Facility, Center for Applied Genomics, Hospital for Sick Children, Toronto, ON). The modified artemin cDNAs generated from wild-type artemin (ArtWT) were ArtC22A, ArtC61A, ArtC166A, and ArtC172A.
Table 1.
Primers employed for the generation of nucleotide substitutions in artemin cDNA
| Artemin variant | Primer sequence |
|---|---|
| ArtC22A | 5′-GGCAAAGTTCAAATGGACGCCCCAAGCAGGCACAATT-3′ |
| 3′-AATTGTGCCTGCTTGGGGCGTCCATTTGAACTTTGCC-3′ | |
| ArtC61A | 5′-TTCTATGCCAGAGACGCCAAGGCTGCCGTGG-3′ |
| 5′-CCACGGCAGCCTTGGCGTCTCTGGCATAGAA-3′ | |
| ArtC166A | 5′-GGAATAAATTCCTATCCGACGCCTTGTCCAACCTACACTG-3′ |
| 5′-CAGTGTAGGTTGGACAAGGCGTCGGATAGGAATTTATTCC-3′ | |
| ArtC172A | 5′-GTCCAACCTACACGCCATCGGTTCACAAGG-3′ |
| 5′-CCTTGTGAACCGATGGCGTGTAGGTTGGAC-3′ |
Single-nucleotide substitutions of artemin cDNA were generated by site-directed mutagenesis using the listed primers with modified nucleotides in bold. Substitutions were named by starting with the mutated amino acid residue, ollowed by its position, and then the replacement residue
Artemin synthesis and purification
Wild-type and mutated artemin cDNAs cloned in the 6×HN-tagged prokaryotic expression vector pPROTet.E133 were transformed into E. coli BL21PRO (Clontech Laboratories, Inc., Mississauga, ON). Transformed bacteria incubated in Luria–Bertani medium containing 50 μg/ml spectinomycin (Sigma-Aldrich Canada Ltd., Oakville, ON, Canada) and 34 μg/ml chloramphenicol (Sigma) were induced with anhydrotetracycline (Spectrum Chemical Mfg. Corp. Gardena, CA, or Sigma) at 100 ng/ml for 8–16 h. Bacteria collected by centrifugation at 5,000×g for 15 min at 4°C were suspended in 2 ml of extraction/wash buffer (50 mM Na2HPO4, 300 mM NaCl, pH 7.0) containing 100 μg/ml lysozyme (Sigma), 1 mM phenylmethanesulphonyl fluoride (Sigma), and 1 μg/ml each of pepstatin A (Sigma), soybean trypsin inhibitor (Sigma), and leupeptin (Sigma). Bacterial suspensions underwent three freeze–thaw cycles before sonication three times for 10 s using a Branson Sonifier™ 150 (Branson Ultrasonics, Danbury, CT) at medium setting with intermittent cooling on ice for 30 s. Homogenates were centrifuged at 10,000×g for 20 min at 4°C and supernatant protein concentrations were ascertained via the Bradford assay (Bio-Rad, Hercules, CA). Artemin was purified on BD TALON metal affinity resin (BD Biosciences Clontech, Mississauga, ON) following manufacturer’s instructions, except the resin was washed with modified extraction/wash buffer (50 mM Na2HPO4, 500 mM NaCl, 15 mM imidazole, pH 7.5) before the recovery of artemin with elution buffer (50 mM Na2HPO4, 500 mM NaCl, 300 mM imidazole, pH 7.0). Artemin-containing fractions were desalted and transferred to chaperone assay buffer (40 mM HEPES/KOH, pH 7.5) with desalting spin columns (Pierce, Rockford, IL) and concentrated with Centriprep YM-10 centrifugal filter devices (Amicon Bioseparations, Billerica, MA).
Protein samples were resolved in 12.5% SDS polyacrylamide gels which were either stained with Coomassie Brilliant Blue R-250 (Sigma) or blotted to nitrocellulose (Bio-Rad). Protein transfer was confirmed by staining membranes with 2% Ponceau-S (Sigma) in 3% trichloroacetic acid. Western blots were probed with antibody raised to artemin (Chen et al. 2007) followed by washing and reaction with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG). Immunoconjugates were detected with the Western Lightening Enhanced Chemiluminescence Reagent Plus (PerkinElmer Life Sciences, Boston, MA) using Fuji X-ray film (Fuji Photo Film Co., Ltd, Tokyo).
Artemin chaperone activity
Dimeric citrate synthase (Sigma) at 150 nM in 40 mM HEPES/KOH buffer was heated in 96-well plates at 43°C in the absence and presence of artemin. Turbidity was monitored every minute for 1 h at 360 nm with a SPECTRAmax PLUS spectrophotometer (Molecular Devices). Data are presented as turbidity increase over time at artemin molarities calculated on the basis of monomer molecular mass and as the inhibition of turbidity development (%) after 60 min of incubation. Inhibition (%) was calculated as: 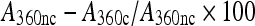 , where A360nc and A360c, respectively, represent turbidity in the absence and presence of artemin. Utilization of monomer molecular mass to calculate molarities of oligomeric chaperones is common and in this case simplifies comparisons between artemin variants that produce heterogeneous oligomers. It is unknown if the artemin monomer or oligomer is responsible for chaperoning. Representative results are shown for experiments done in duplicate with independently prepared artemin samples. Bovine serum albumin (BSA, Sigma) and IgG (Sigma) were used at 600 nM to control for non-specific citrate synthase protection.
, where A360nc and A360c, respectively, represent turbidity in the absence and presence of artemin. Utilization of monomer molecular mass to calculate molarities of oligomeric chaperones is common and in this case simplifies comparisons between artemin variants that produce heterogeneous oligomers. It is unknown if the artemin monomer or oligomer is responsible for chaperoning. Representative results are shown for experiments done in duplicate with independently prepared artemin samples. Bovine serum albumin (BSA, Sigma) and IgG (Sigma) were used at 600 nM to control for non-specific citrate synthase protection.
Artemin oligomerization
Two hundred microliters of protein extract from transformed bacteria was applied to a Sepharose CL-6B (Sigma) column (1.0 × 48 cm) equilibrated at 4°C with 0.1 M Tris/glycine buffer, pH 7.4. Protein was eluted at 20 ml/h and fractions of 0.8 ml were collected. Fifteen microliters from each fraction was resolved in SDS polyacrylamide gels followed by transfer to nitrocellulose and immunodetection of artemin. Column standardization was with the molecular mass markers carbonic anhydrase, 29 kDa; bovine serum albumin, 66 kDa; α-amylase, 200 kDa; apoferritin, 443 kDa; thyroglobulin, 669 kDa (Sigma).
Artemin heat stability
Fifty microliters of artemin-containing bacterial extracts at a protein concentration of 2.0–2.3 mg/ml were heated at 75°C for 13 min, followed by cooling on ice and centrifugation at 15,000×g for 10 min at 4°C. Supernatants were harvested, and after rinsing the tubes three times with 40 mM HEPES/KOH buffer, pellets were recovered in 50 μl of the same buffer. Supernatant and pellet samples of equal volume were resolved in SDS polyacrylamide gels and either stained with Coomassie blue or transferred to nitrocellulose and probed with antibody to artemin.
Modeling of artemin structure
The amino acid sequences of artemin (AAL55397) and ferritin (P07798) were aligned by CLUSTAL W (Thompson et al. 1994) with numbering of the alignment according to the artemin sequence. The secondary structure of ferritin was obtained from its crystal structure (1MFR), and the secondary structure of artemin was predicted using the multivariate linear regression combination (MLRC) method available at NPS@server. Residues 20-168 and 2-149, respectively, of artemin and ferritin were used for modeling. Artemin structures were generated with MODELER 7 × 7 (Šali and Blundell 1993) using bullfrog ferritin (pdb code 1MFR, chains B, C, F, L, M, O, S, and V) as template. Subunit interfaces between chain F and chains B, C, M, O, and V of bullfrog ferritin (IMFR) were analyzed with the Protein–protein interface analysis server (ProtorP; Reynolds et al. 2009) and compared with the corresponding artemin sequence using the alignment shown in Fig. 1a. Graphical representations of models were made with Visual Molecular Dynamics (VMD; Humphrey et al. 1996) and Raster3D (Merritt and Bacon 1997) with images either in new cartoon or surf.
Results
Preparation of artemin
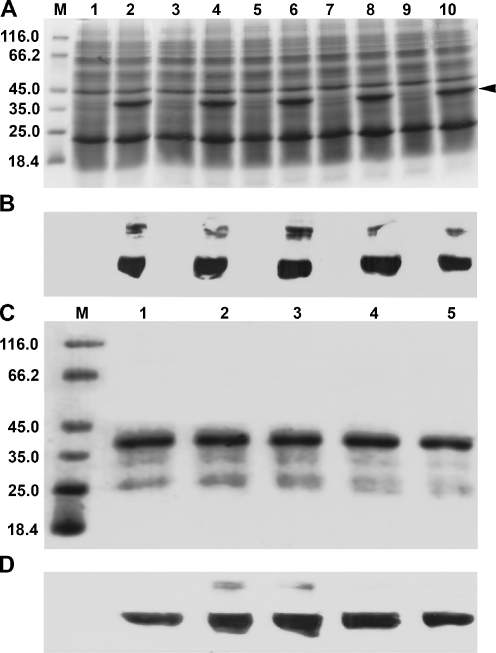
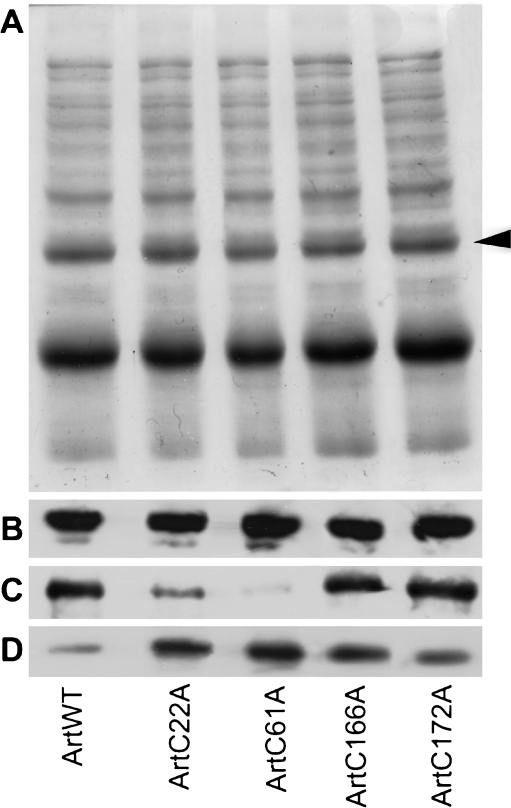
cDNA sequencing confirmed that ArtC22A, ArtC61A, ArtC166A, and ArtC172A each possessed the correct substitution and that extraneous nucleotide alterations were absent. Anhydrotetracycline-induced bacteria transformed individually with wild-type and mutated cDNAs produced an abundant polypeptide with a molecular mass corresponding to 6×HN-tagged artemin and which was recognized by antibody raised to artemin (Fig. 2a, b). The antibody-reactive band of higher molecular mass was not identified (Fig. 2b), but a similar polypeptide, thought to be a stable artemin doublet, was detected in earlier work (Chen et al. 2007). Affinity chromatography of protein extracts from transformed bacteria yielded polypeptides of the expected mass that reacted with anti-artemin antibody (Fig. 2c, d). The lower molecular mass polypeptides apparent on Coomassie blue-stained gels were possibly artemin degradation products, although they did not react with antibody (Fig. 2c, d).
Fig. 2.
Artemin synthesis and purification. Protein extracts from transformed bacteria were resolved in SDS polyacrylamide gels and either stained with Coomassie blue (a) or blotted to nitrocellulose and reacted with antibody to artemin (b). Lanes 1 and 2 ArtWT, 3 and 4 ArtC22A, 5 and 6 ArtC61A, 7 and 8 ArtC166A, 9 and 10 ArtC172A. Samples in odd- and even-numbered lanes were respectively from non-induced and induced bacteria. All lanes received equal amounts of protein. Artemin purified by affinity chromatography was electrophoresed in SDS polyacrylamide gels and either stained with Coomassie blue (c) or blotted to nitrocellulose and reacted with antibody to artemin (d). All lanes received equal amounts of protein. Arrowhead artemin. M molecular mass marker (kDa)
Mutagenesis decreased artemin chaperone activity
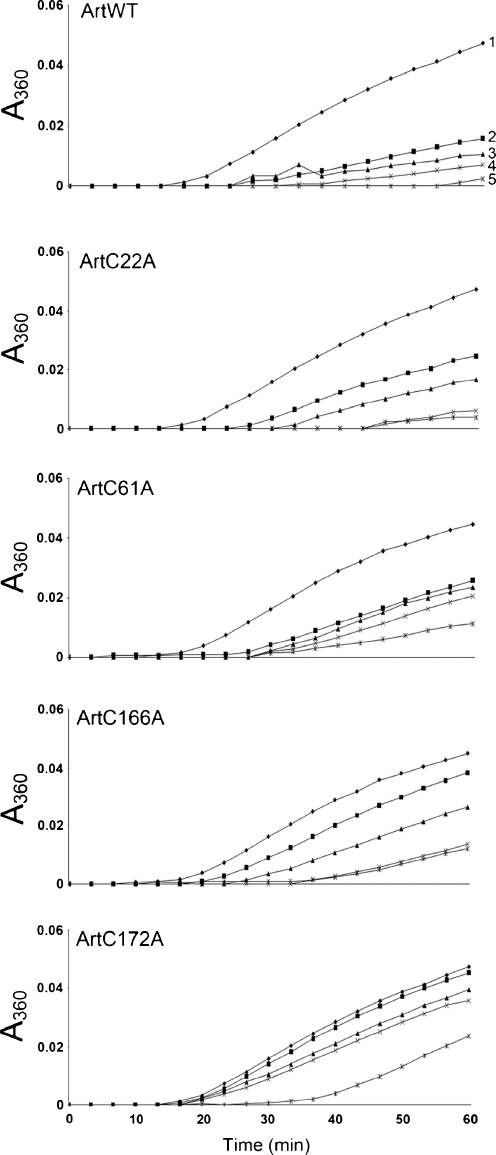
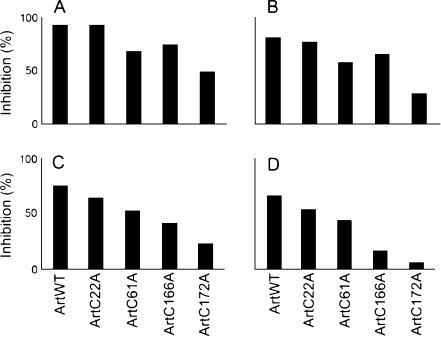
The mutated artemins protected citrate synthase against heat-induced precipitation in the order ArtWT > ArtC22A > ArtC61A > ArtC166A > ArtC172A (Figs. 3 and 4), although at higher concentrations, ArtC166A functioned more effectively that ArtC61A, a result seen most clearly in Fig. 4. Chaperone activity was inversely related to the amount of citrate synthase precipitation (Fig. 3) and directly related to the ability of artemin to inhibit substrate precipitation (Fig. 4). Chaperone activity was dose-dependent, and although the time course of citrate synthase precipitation in the presence of each artemin variant was similar, stronger chaperones tended to delay citrate synthase precipitation slightly longer than weaker chaperones (Fig. 3). BSA and IgG at 600 nM provided no protection against heat-induced citrate synthase precipitation (not shown).
Fig. 3.
Artemin chaperone activity in vitro. Artemin was heated at 43°C for 1 h with 150 nM citrate synthase, and solution turbidity (absorbance) was measured at 360 nm. The final artemin concentrations, based on monomer molecular mass, were: 1 0 nM, 2 150 nM, 3 300 nM, 4 450 nM, and 5 600 nM. The same artemin concentrations in different panels are represented by identical symbols
Fig. 4.
Artemin chaperone activity is concentration-dependent and reduced by cysteine modification. Artemin at 600 nM (a), 450 nM (b), 300 nM (c), and 150 nM (d) was heated for 60 min at 43°C with 150 nM citrate synthase, and solution turbidity was measured at 360 nm. Artemin molarities were based on monomer molecular mass. The chaperone activity of each artemin variant, measured as the ability to inhibit turbidity development (Inhibition%), was calculated as described in “Materials and methods”
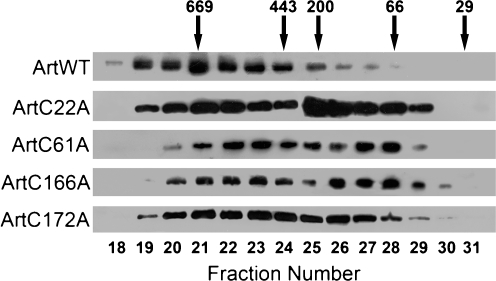
Oligomer formation was modified by cysteine substitution
All artemins generated large oligomers with differing amounts of smaller oligomers and perhaps monomers (Fig. 5). ArtWT assembled more readily into large oligomers than did any mutated artemin, and it formed oligomers of the greatest molecular mass. As observed previously for ArtWT (Chen et al. 2007), but not in the current experiments, the oligomer size of modified artemins exhibited biphasic distribution. ArtC22A was the most enriched in smaller oligomers of 200 kDa or less, whereas samples of ArtC166A and ArtC172A potentially contained small amounts of monomers.
Fig. 5.
Artemin oligomer formation is modified by mutagenesis. Bacterial protein extracts containing similar amounts of artemin were fractionated in Sepharose 6B columns. Fifteen-microliter samples from selected fractions were resolved in SDS polyacrylamide gels, blotted to nitrocellulose, and reacted with antibody to artemin followed by HRP-conjugated goat anti-rabbit IgG. Numbered arrows molecular mass markers
Differential effects of cysteine modifications on artemin thermostability
Starting with bacterial extracts individually containing similar amounts of each artemin variant (Fig. 6a, b), ArtC61A and ArtC22A exhibited almost complete loss of thermostability; supernatants were virtually depleted of artemin after heating and centrifugation (Fig. 6c), whereas pellets were enriched in the protein (Fig. 6d). Conversely, ArtC166A and ArtC172A tended to remain in solution upon heating with the poorest chaperone, ArtC172A, essentially as stable as ArtWT (Fig. 6c, d). Artemin stability upon heating decreased in the order ArtWT > ArtC172A > ArtC166A > ArtC22A > ArtC61A.
Fig. 6.
Cysteine modifications affect artemin thermostability differently. Bacterial protein extracts were resolved in SDS polyacrylamide gels and either stained with Coomassie blue (a) or transferred to nitrocellulose and probed with antibody to artemin followed by HRP-conjugated goat anti-rabbit IgG (b). Equivalent amounts of the same extracts were then heated, cooled on ice, and centrifuged. The resulting supernatants (c) and pellets (d) were electrophoresed in SDS polyacrylamide gels, transferred to nitrocellulose, and probed with antibody to artemin followed by HRP-conjugated goat anti-rabbit IgG. Arrowhead artemin
Artemin structure
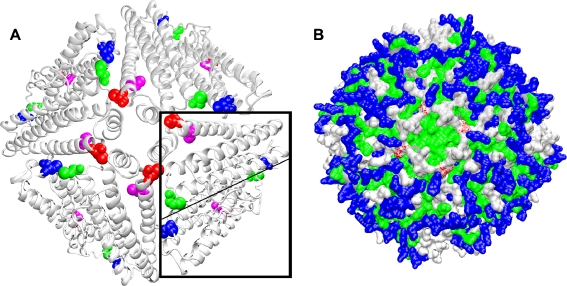
The sequence similarity between artemin and ferritin, exclusive of the amino- and carboxy-terminal extensions, provided a base for the investigation of artemin structure. Modeling revealed that artemin and ferritin possess similar secondary structures (Fig. 1) and that seven of the ten cysteines in an artemin monomer are buried in two clusters at opposite ends of four-helix bundles (Fig. 1b). C166 is in cluster 2, whereas C22, C61, and C172 are in loops external to helixes. ProtorP analysis using the structure of bullfrog ferritin showed that 87 residues, or about half of the polypeptide, contact neighboring subunits. Fifty-two residues are identical or conserved substitutions in artemin equalling approximately 60% amino acid conservation within interacting interfaces of ferritin and artemin (Table 2). Given these similarities between primary, secondary, and tertiary structures, monomer number in oligomers, subunit interfaces, thermostability, and chaperoning, it is reasonable to propose that the quaternary structures of artemin and ferritin are comparable. Based on this proposal, an artemin octomer was modeled on the structure of bullfrog ferritin, revealing that C21 and C61 of neighboring monomers are close to one another at a subunit interface (Fig. 7a), possibly forming a disulfide bridge. In contrast, C172 is exposed on the oligomer surface and the closest of the modified residues to a hydrophobic subunit interface formed by four helices of neighboring subunits (Fig. 7a, b). C166, although buried within cluster 2 at the end of the monomer four-helix bundle (Fig. 1b), is the nearest of the remaining three modified residues to C172 (Fig. 7a).
Table 2.
Amino acid conservation at comparable ferritin and artemin subunit interfaces
| Ferritin interacting chains | Number of amino acids at ferritin subunit interfaces | Number of conserved amino acids at artemin subunit interfaces | Amino acid conservation (%) |
|---|---|---|---|
| F:B | 35 | 18 | 51 |
| F:V | 15 | 8 | 53 |
| F:M | 3 | 2 | 67 |
| F:C | 14 | 11 | 78 |
| F:O | 20 | 13 | 65 |
Subunit interfaces included in the analysis are between chain F and chains B, V, M, C, and O of bullfrog ferritin. Amino acid residues at the ferritin subunit interfaces were identified with ProtorP and compared with the corresponding artemin sequence using the alignment shown in Fig. 1a. Identical amino acids and conserved substitutions are included in the calculation for amino acid conservation (%)
Fig. 7.
Cysteine localization within an artemin octomer model. An octomer of artemin monomers was modeled with MODELER 7 × 7 using the crystal structure of bullfrog ferritin. Residues 21-192 and 2-189, respectively, of artemin and ferritin were used for modeling. a Artemin octomer showing the positions of C22 (blue), C61 (green), C166 (magenta), and C172 (red). An artemin dimer is boxed and the monomer–monomer interface is indicated by the light line. b C172 (red) is shown within a hydrophobic region formed by four helices of neighboring subunits as shown in a. Polar charged residues (blue), polar non-charged residues (silver), hydrophobic residues (green). Images generated with VMD and Raster3D are in new cartoon (a) and surf (b)
Discussion
Stress tolerance contributing to diapause maintenance in Artemia cysts is thought to require molecular chaperones, and in support of this proposal, diapause-destined embryos display at least three sHSPs not synthesized in ovoviviparous embryos (Qiu and MacRae 2008a, b; Qui et al. 2007; MacRae 2003; Liang and MacRae 1999; Jackson and Clegg 1996). Artemin, produced only in diapause-destined Artemia embryos, is also a molecular chaperone, preventing heat-induced precipitation of citrate synthase in vitro and protecting transfected mammalian cells against heat and oxidative damage (Chen et al. 2007). These findings indicate that artemin affects stress resistance in Artemia embryos, but details of artemin function and structure are limited. To address this issue, the consequences of substituting alanine for cysteine at positions 22, 61, 166, and 172 of artemin were determined. Cysteines were chosen because artemin is enriched in this residue. Moreover, the oxidative state of cysteines modulates the activity of redox-regulated chaperones (Kumsta and Jakob 2009) and influences switching of the disulfide reductase, thioredoxin, to a chaperone (Park et al. 2009).
Artemin chaperoning, as quantified in vitro by turbidimetric assay, was compromised by cysteine modification. ArtC172A displayed the greatest functional impairment followed in order by ArtC166A, ArtC61A, and ArtC22A, although the comparative activities of ArtC166A and ArtC61A varied with protein concentration. C172 is the most exposed of the modified artemin cysteines and spatially closest to a hydrophobic oligomeric interface containing four helices from neighboring monomers. The corresponding fourfold interface in ferritin is also hydrophobic, although sequence conservation with artemin is lacking in this region. The loss of chaperone activity upon modification of C166, C61, and C22 was inversely proportional to their distance from the hydrophobic region. These observations indicate that surface hydrophobicity mediates artemin chaperoning and that altering different cysteines confers as yet unidentified structural changes which affect function to varying extents. It is possible that the surface-exposed Cys172 interacts directly with denaturing proteins independent of the hydrophobic region, but the role of hydrophobicity in the protective activities of other chaperones such as the sHSPs (Sun and MacRae 2005) argues against this.
Wild-type artemin synthesized in cysts and bacteria forms oligomers of 24 monomers with a molecular mass of approximately 669 kDa, although smaller aggregates are observed (De Graaf et al. 1990; Chen et al. 2007). Establishing respective oligomer masses demonstrated that individually, the modified cysteines have limited roles in maintaining artemin quaternary structure at normal temperature, signaling the importance of multi-residue interactions at subunit interfaces in oligomerization. Moreover, all artemin variants formed oligomers but their chaperone activities were reduced, substantially in some cases, showing that oligomerization is not adequate to guarantee optimal levels of chaperoning.
ArtC172A, although exhibiting the least chaperone activity, was the most stable of the mutated artemins, effectively resisting heat-induced precipitation, a characteristic shared by ArtWT and almost equally by ArtC166A. In contrast, ArtC22A and ArtC61A tended to precipitate when heated, demonstrating that they were less stable at high temperature than other variants. Modeling suggests that substitution of alanine for cysteine either disrupted a disulphide bridge formed between C22 and C61 or induced other local changes within subunit interfaces perhaps disturbing salt bridges reminiscent of those that stabilize ferritin oligomers (Kilic et al. 2003). Interestingly, the changes induced by modifying C22 and C61, although destabilizing the protein at high temperature, had little consequence for chaperoning. Substitution of C172 which is exposed on the oligomer surface and unlikely to participate in subunit interactions had limited impact on stability. Changing C166, which is buried within cluster 2, had only a minor influence on oligomer stability, either suggesting no involvement of C166 in disulphide linkages or that interactions between other residues compensate for the loss of a disulphide bridge in ArtC166A.
In summary, modification of C172 had the greatest detrimental impact on chaperoning, but the least influence on protein stability at elevated temperature, whereas changing C22 and C61 had comparatively less impression on chaperone activity and more effect on thermostability. These data demonstrate that artemin chaperone activity correlates neither with protein stability nor oligomer formation. Molecular modeling allows reasonable interpretations of these experimental results by positioning cysteine residues within artemin while indicating that artemin and ferritin, both of which are heat-stable, have similar oligomer structures stabilized by residue interactions at strategic interfaces. The work advances our understanding of the structure/function relationships in artemin, a novel protein thought to augment diapause maintenance in Artemia embryos by enhancing stress tolerance.
Acknowledgments
This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant to THM. Dr. Tao Chen generated mutations ArtC22A and ArtC61A.
Footnotes
Yan Hu and Svetla Bojikova-Fournier contributed equally to the paper.
References
- Baraibar MA, Barbeito AG, Muhoberac BB, Vidal R. Iron-mediated aggregation and a localized structural change characterize ferritin from a mutant light chain polypeptide that causes neurodegeneration. J Biol Chem. 2008;283:31679–31689. doi: 10.1074/jbc.M805532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Amons R, Clegg JS, Warner AH, MacRae TH. Molecular characterization of artemin and ferritin from Artemia franciscana. Eur J Biochem. 2003;270:137–145. doi: 10.1046/j.1432-1033.2003.03373.x. [DOI] [PubMed] [Google Scholar]
- Chen T, Villeneuve TS, Garant KA, Amons R, MacRae TH. Functional characterization of artemin, a ferritin homolog synthesized in Artemia embryos during encystment and diapause. FEBS J. 2007;274:1093–1011. doi: 10.1111/j.1742-4658.2007.05659.x. [DOI] [PubMed] [Google Scholar]
- Clegg JS. Unusual response of Artemia franciscana embryos to prolonged anoxia. J Exp Zool. 1994;270:332–334. doi: 10.1002/jez.1402700312. [DOI] [Google Scholar]
- Clegg JS. Embryos of Artemia franciscana survive four years of continuous anoxia: the case for complete metabolic rate depression. J Exp Biol. 1997;200:467–475. doi: 10.1242/jeb.200.3.467. [DOI] [PubMed] [Google Scholar]
- Clegg JS, Jackson SA. The metabolic status of quiescent and diapause embryos of Artemia franciscana (Kellogg) Arch Hydrobiol Spec Issues Adv Limnol. 1998;52:425–439. [Google Scholar]
- Clegg JS, Willsie JK, Jackson SA. Adaptive significance of a small heat shock/α-crystallin protein (p26) in encysted embryos of the brine shrimp, Artemia franciscana. Am Zool. 1999;39:836–847. [Google Scholar]
- Clegg JS, Jackson SA, Popov VI. Long-term anoxia in encysted embryos of the crustacean, Artemia franciscana: viability, ultrastructure, and stress proteins. Cell Tiss Res. 2000;301:433–446. doi: 10.1007/s004410000249. [DOI] [PubMed] [Google Scholar]
- Graaf J, Amons R, MÖller W. The primary structure of artemin from Artemia cysts. Eur J Biochem. 1990;193:737–750. doi: 10.1111/j.1432-1033.1990.tb19394.x. [DOI] [PubMed] [Google Scholar]
- Drinkwater LE, Clegg JS. Experimental biology of cyst diapause. In: Browne RA, Sorgeloos P, Trotman CNA, editors. Artemia biology. Boca Raton: CRC; 1991. pp. 93–117. [Google Scholar]
- Drinkwater LE, Crowe JH. Regulation of embryonic diapause in Artemia: environmental and physiological signals. J Exp Zool. 1987;241:297–307. doi: 10.1002/jez.1402410304. [DOI] [Google Scholar]
- Fan R, Boyle AL, Cheong VV, Ng SL, Orner BP. A helix swapping study of two protein cages. Biochemistry. 2009;48:5623–5630. doi: 10.1021/bi900387t. [DOI] [PubMed] [Google Scholar]
- Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jackson SA, Clegg JS. Ontology of low molecular weight stress protein p26 during early development of the brine shrimp, Artemia franciscana. Dev Growth Differ. 1996;38:153–160. doi: 10.1046/j.1440-169X.1996.t01-1-00004.x. [DOI] [PubMed] [Google Scholar]
- Kilic MA, Spiro S, Moore GR. Stability of a 24-meric homopolymer: comparative studies of assembly-defective mutants of Rhodobacter capsulatus bacterioferritin and the native protein. Prot Sci. 2003;12:1663–1674. doi: 10.1110/ps.0301903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Jakob U. Redox-regulated chaperones. Biochemistry. 2009;48:4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, MacRae TH. The synthesis of a small heat shock/α-crystallin protein in Artemia and its relationship to stress tolerance during development. Dev Biol. 1999;207:445–456. doi: 10.1006/dbio.1998.9138. [DOI] [PubMed] [Google Scholar]
- MacRae TH. Molecular chaperones, stress resistance and development in Artemia franciscana. Semin Cell Dev Biol. 2003;14:251–258. doi: 10.1016/j.semcdb.2003.09.019. [DOI] [PubMed] [Google Scholar]
- MacRae TH. Diapause: diverse states of developmental and metabolic arrest. J Biol Res. 2005;3:3–14. [Google Scholar]
- MacRae TH. Gene expression, metabolic regulation and stress tolerance during diapause. Cell Mol Life Sci. 2010;67:2405–2424. doi: 10.1007/s00018-010-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3D photorealistic molecular graphics. Meth Enzymol. 1997;277:505–524. doi: 10.1016/S0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Park SK, Jung YJ, Lee JR, Lee YM, Jang HH, Lee SS, Park JH, Kim SY, Moon JC, Lee SY, Chae HB, Shin MR, Jung JH, Kim MG, Kim WY, Yun D-J, Lee KO, Lee SY. Heat-shock and redox-dependent functional switching of an h-type Arabidopsis thioredoxin from a disulfide reductase to a molecular chaperone. Plant Physiol. 2009;150:552–561. doi: 10.1104/pp.109.135426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, MacRae TH. ArHsp21, a developmentally regulated small heat-shock protein synthesized in diapausing embryos of Artemia franciscana. Biochem J. 2008;411:605–611. doi: 10.1042/BJ20071472. [DOI] [PubMed] [Google Scholar]
- Qiu Z, MacRae TH. ArHsp22, a developmentally regulated small heat shock protein produced in diapause-destined Artemia embryos, is stress inducible in adults. FEBS J. 2008;275:3556–3566. doi: 10.1111/j.1742-4658.2008.06501.x. [DOI] [PubMed] [Google Scholar]
- Qui Z, Tsoi SCM, MacRae TH. Gene expression in diapause-destined embryos of the crustacean, Artemia franciscana. Mech Dev. 2007;124:856–867. doi: 10.1016/j.mod.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Rasti B, Shahangian SS, Sajedi RH, Taghdir M, Hasannia S, Ranjbar B. Sequence and structural analysis of artemin based on ferritin: a comparative study. Biochim Biophys Acta. 2009;1794:1407–1413. doi: 10.1016/j.bbapap.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Reynolds C, Damerell D, Jones S. ProtorP: a protein–protein interaction analysis server. Bioinformatics. 2009;25:413–414. doi: 10.1093/bioinformatics/btn584. [DOI] [PubMed] [Google Scholar]
- Robbins HM, Stappen G, Sorgeloos P, Sung YY, MacRae TH, Bossier P. Diapause termination and development of encysted Artemia embryos: roles for nitric oxide and hydrogen peroxide. J Exp Biol. 2010;213:1464–1470. doi: 10.1242/jeb.041772. [DOI] [PubMed] [Google Scholar]
- Šali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguay JA, Reyes RC, Clegg JS. Habitat diversity and adaptation to environmental stress in encysted embryos of the crustacean Artemia. J Biosci. 2004;29:489–501. doi: 10.1007/BF02712121. [DOI] [PubMed] [Google Scholar]
- Theil EC, Matzapetakis M. Ferritins: iron/oxygen biominerals in protein nanocages. J Biol Inorg Chem. 2006;11:803–810. doi: 10.1007/s00775-006-0125-6. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breukelen F, Maier R, Hand SC. Depression of nuclear transcription and extension of mRNA half-life under anoxia in Artemia franciscana embryos. J Exp Biol. 2000;203:1123–1130. doi: 10.1242/jeb.203.7.1123. [DOI] [PubMed] [Google Scholar]
- Linden A, Blust R, Laere AJ, DeCleir W. Light-induced release of Artemia dried embryos from diapause: analysis of metabolic status. J Exp Zool. 1988;247:131–138. doi: 10.1002/jez.1402470204. [DOI] [Google Scholar]
- Viner RI, Clegg JS. Influence of trehalose on the molecular chaperone activity of p26, a small heat shock/α-crystallin protein. Cell Stress Chaperones. 2001;6:126–135. doi: 10.1379/1466-1268(2001)006<0126:IOTOTM>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner AH, Brunet RT, MacRae TH, Clegg JS. Artemin is an RNA-binding protein with high thermal stability and potential RNA chaperone activity. Arch Biochem Biophys. 2004;424:189–200. doi: 10.1016/j.abb.2004.02.022. [DOI] [PubMed] [Google Scholar]