Abstract
The Ccr4–Not complex has been implicated in the control of multiple steps of mRNA metabolism; however, its functions in transcription remain ambiguous. The discovery that Ccr4/Pop2 is the major cytoplasmic mRNA deadenylase and the detection of Not proteins within mRNA processing bodies have raised questions about the roles of the Ccr4–Not complex in transcription. Here we firmly establish Ccr4–Not as a positive elongation factor for RNA polymerase II (RNAPII). The Ccr4–Not complex is targeted to the coding region of genes in a transcription-dependent manner similar to RNAPII and promotes elongation in vivo. Furthermore, Ccr4–Not interacts directly with elongating RNAPII complexes and stimulates transcription elongation of arrested polymerase in vitro. Ccr4–Not can reactivate backtracked RNAPII using a mechanism different from that of the well-characterized elongation factor TFIIS. While not essential for its interaction with elongation complexes, Ccr4–Not interacts with the emerging transcript and promotes elongation in a manner dependent on transcript length, although this interaction is not required for it to bind RNAPII. Our comprehensive analysis shows that Ccr4–Not directly regulates transcription, and suggests it does so by promoting the resumption of elongation of arrested RNAPII when it encounters transcriptional blocks in vivo.
Keywords: Ccr4–Not, RNA polymerase II, elongation, mRNA decay, transcription arrest
Regulation of gene expression is the result of the combined control of transcription and mRNA processing in the nucleus, as well as translation and mRNA decay in the cytoplasm. While most protein complexes have been assigned to function in a particular aspect of gene regulation, the Ccr4–Not complex has been proposed to have roles throughout the “life” of mRNAs and protein (for review, see Collart 2003; Denis and Chen 2003; Collart and Timmers 2004).
The Not proteins were originally identified in a screen for mutations that increased expression of the HIS3 gene, particularly through the noncanonical TATA promoter (Collart 2003; Denis and Chen 2003; Collart and Timmers 2004). The Not proteins were then shown to be a part of a larger transcriptional complex, which additionally contained Ccr4p and Pop2p, among others (Collart 2003; Denis and Chen 2003; Collart and Timmers 2004). The Ccr4–Not complex is an evolutionarily conserved complex composed of nine core subunits, of which only Not1 and Not2 are essential in yeast. The C terminus of Not1 associates with the “Not group” of proteins, which contains Not2–Not5 (Collart 2003; Denis and Chen 2003). Previous studies support a role for the “Not group” in transcription initiation, especially regulating TFIID (Badarinarayana et al. 2000; Deluen et al. 2002). Some subunits of this complex can cross-link to the promoters of stress and Gcn4-regulated genes, and Ccr4–Not mutants display reduced TFIID recruitment to promoters (Deluen et al. 2002; Swanson et al. 2003; Qiu et al. 2004). A role for Ccr4–Not in elongation has also been suggested based on the 6-AU and MPA sensitivity of mutants and genetic interactions with mutations in elongation factors such as DST1 (TFIIS) and SPT16 (Denis et al. 2001). Additionally, some Ccr4–Not complex genes were identified in a screen for mutants that affect expression in a gene length-dependent manner, but not all elongation factors show phenotypes in this assay, and many factors involved in diverse processes such as mRNP biogenesis, mRNA export, and translation also score in this assay (Gaillard et al. 2009). Despite early evidence supporting a role for this complex in some aspect of transcription, whether the genetic and physical interactions are direct is unclear.
The growing body of evidence for nontranscription activities of Ccr4–Not has generated questions about its true functions. One subunit of the complex, Not4, was shown to be an E3 ubiquitin ligase that regulates the nascent polypeptide-associated complex (NAC) in the cytoplasm (Panasenko et al. 2006). Mutations in Not4 or other Ccr4–Not complex subunits reduce H3K4 methylation levels in cells (Laribee et al. 2007; Mulder et al. 2007a), but recently this has been attributed to the stabilization of Jhd2, the H3K4me3 demethylase, in Not mutants (Mersman et al. 2009). Therefore, the proposed effect of Ccr4–Not mutations on elongation and elongation-linked histone modifications may be indirect. Thus, a direct involvement of this complex in elongation is not certain, and if it participates in elongation, its mechanism of action is not known.
Although initially characterized for its role in transcription, Ccr4 was identified as the major cytoplasmic mRNA deadenylase in yeast (Tucker et al. 2001; Chen et al. 2002). Additional support for the complex playing a role in mRNA degradation was the identification of Dhh1, an RNA helicase, as a protein associated with the N terminus of Not1 (Maillet and Collart 2002). Ccr4, Pop2, Dhh1, and other Not subunits regulate mRNA stability and localize to cytoplasmic processing bodies (P-bodies), which are the sites of mRNA degradation (Parker and Sheth 2007). Hence, some of the Ccr4–Not mutant phenotypes and genetic interactions with transcription factors might be an indirect result of its functioning in mRNA processing or decay.
In this study, we present strong evidence that the Ccr4–Not complex plays a direct role in transcription elongation. The Ccr4–Not complex cross-links in vivo to the ORF of genes in a transcription- and RNA polymerase II (RNAPII)-dependent manner. We show that Ccr4–Not interacts physically with RNAPII and, importantly, binds to functional RNAPII elongation complexes (ECs) in vitro. In vitro transcription run-on assays indicate that purified Ccr4–Not can stimulate the resumption of RNAPII elongation from an arrested state. Finally, inactivating the Ccr4–Not complex changes the RNAPII distribution along the gene in a way that suggests an elongation defect. Together, our data provide the first comprehensive analysis of how the Ccr4–Not complex functions in elongation, and firmly establish Ccr4–Not as a bona fide elongation factor.
Results
Ccr4–Not is recruited to genes during elongation by RNAPII
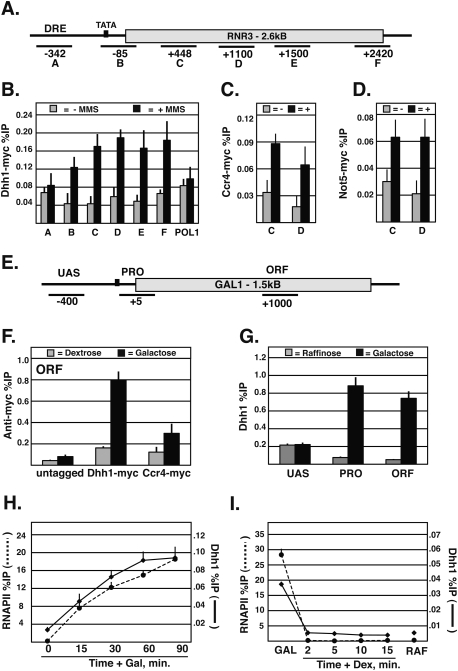
Ccr4–Not is required for the full expression of the RNR3 gene (Mulder et al. 2005). We verified this observation and showed that TFIID (TBP and TAF1) recruitment is impaired in Ccr4–Not mutants (Supplemental Fig. S1). The recruitment of Ccr4–Not to RNR3 has not been shown before, so we analyzed this to test if it directly regulates this gene. Following the induction of transcription by the addition of the DNA-damaging agent methyl methanesulfonate (MMS), Dhh1 cross-linking increased across RNR3 (Fig. 1A,B). In addition to cross-linking to the promoter region, Dhh1 was strongly recruited over the entire ORF, showing a fourfold increase in MMS-treated cells. Ccr4–Not was not recruited to the ORF of POL1, a gene not regulated by DNA damage (Fig. 1B). As a control, we examined the recruitment of TBP across RNR3, and, not surprisingly, it was only recruited to the promoter (Supplemental Fig. S1). Because Dhh1 may have functions independent of Ccr4–Not, we verified that other subunits of the complex are recruited to RNR3. We examined representatives of the Ccr4 group and the Not group (see above), and found that Ccr4 and Not5 are recruited to RNR3 in an MMS-dependent manner (Fig. 1C,D). The distribution of Ccr4 and Not5 cross-linking across RNR3 was similar to that of Dhh1 (data not shown). Of the subunits examined, Dhh1 cross-linked most robustly, and was used throughout the remaining studies. Interestingly, Ccr4–Not subunits were not recruited to the upstream regulatory region, which contains the binding sites for the repressor/activator Crt1 and the activator Rap1 (Zhang and Reese 2005; Tomar et al. 2008). This suggests that sequence-specific DNA-binding proteins do not recruit Ccr4–Not to RNR3, and that, in addition to regulating TFIID function at promoters, Ccr4–Not may regulate transcription elongation.
Figure 1.
Ccr4–Not is recruited to genes during elongation by RNAPII. (A) Schematic of RNR3 and primer locations. (B–D) ChIP analysis of Dhh1-myc, Ccr4-myc, and Not5-myc across RNR3 in cells untreated or treated with MMS for 2.5 h. Background (untagged) percent immunoprecipitated (%IP) was subtracted from anti-myc %IP. %IP was calculated by dividing the immunoprecipitation DNA signal by the Input DNA signal using each primer set. POL1 was used as a control. (E) Schematic of GAL1 and primer locations. (F) Recruitment of Dhh1-myc and Ccr4-myc to the GAL1 ORF under dextrose or galactose conditions. (G) Location of Dhh1p at GAL1 in cells grown in raffinose or galactose media. (H) Cells were grown overnight in raffinose and induced with 2% galactose for 15, 30, 60, and 90 min. RNAPII (dotted line) and Dhh1 (solid line) densities at the GAL1 ORF. (I) Galactose-grown cells were treated with 4% dextrose and cross-linked at 2, 5, 10, and 15 min.
Next, we examined Ccr4–Not's cross-linking to the well-characterized GAL1 gene to take advantage of its fast on–off kinetics (Fig. 1E). The recruitment of Dhh1 and Ccr4 to GAL1 was analyzed under repressed (dextrose) and activated (galactose) conditions, and both subunits showed enrichment at the promoter and within the ORF when GAL1 was activated (Fig. 1F). As observed at RNR3, Dhh1 cross-linked more robustly to GAL1 than Ccr4 (Fig. 1F), and neither was recruited to the upstream activating sequence of GAL1 (Fig. 1G; data not shown). The cross-linking profile of Ccr4–Not to GAL1 and RNR3 suggests that RNAPII or its associated factors play a role in the recruitment of the complex to genes. To better correlate RNAPII and Ccr4–Not recruitment, we compared the kinetics of cross-linking of these factors during the activation and repression of transcription. The cross-linking of both RNAPII and Dhh1 gradually increased at the GAL1 ORF following the addition of galactose, reaching a maximum level by 90 min (Fig. 1H). Furthermore, adding dextrose to repress GAL1 led to a rapid loss of RNAPII and Dhh1 back to preinduction (raffinose) levels within 2 min (Fig. 1I). The correlative recruitment and disassociation of Dhh1 and RNAPII at GAL1 suggest that these two events are linked to one another, and implicate RNAPII in the recruitment of Ccr4–Not to genes.
Ccr4–Not associates with RNAPII
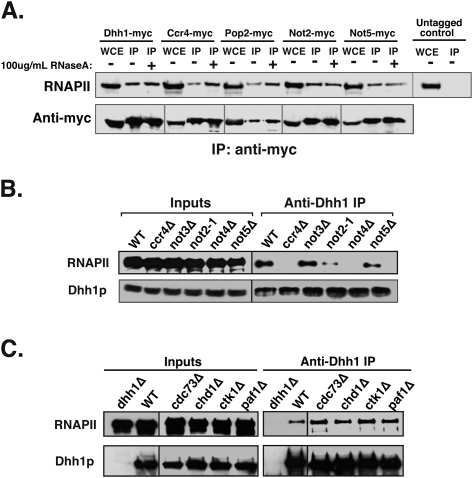
Ccr4–Not has been shown to have physical interactions with TBP, TAFIIs, and SAGA (Collart 2003; Denis and Chen 2003); however, it is not known if it associates with RNAPII. Ccr4p copurified with a PAF1C–RNAPII complex, but other Ccr4–Not subunits were not present in this complex, leading Chang et al. (1999) to conclude that it was distinct from the Ccr4–Not complex. We re-evaluated whether the Ccr4–Not complex interacts with RNAPII by coimmunoprecipitation. Our results show that all Ccr4–Not subunits examined coimmunoprecipitated with RNAPII (Fig. 2A). Because Dhh1, Ccr4, Pop2, and Not4 potentially bind RNA, RNase was added to the extracts. The addition of RNase did not disrupt the association between Ccr4–Not and RNAPII; therefore, it is unlikely that the interaction is mediated exclusively through RNA (Fig. 2A). These results, combined with the chromatin immunoprecipitation (ChIP) results, suggest that the Ccr4–Not complex is recruited to genes through an interaction with RNAPII during elongation.
Figure 2.
Ccr4–Not associates with RNAPII. (A) Coimmunoprecipitation of myc-tagged Ccr4–Not subunits with RNAPII. Whole-cell extracts (WCEs) were either treated or left untreated with 100 μg/mL RNase A at room temperature prior to addition of antibody. Rpb1 subunit of RNAPII (8WG16) was detected by Western blotting. The amount of immunoprecipitated protein recovered was analyzed by Western blotting using an anti-myc antibody. Since Ccr4–Not subunits run at different molecular weights, regions corresponding to the location of each myc-tagged protein were cut from their respective regions on the membrane and placed in a row. (B) Same as in A except immunoprecipitation was performed using a polyclonal Dhh1 antisera. Anti-Dhh1 was used to probe the blot as an immunoprecipitation control. (C) Same as in B except extracts from various transcription factor mutants were analyzed.
To test the specificity of the interaction and determine the requirement for the integrity of the Ccr4–Not complex for its binding to RNAPII, we examined the interaction between Dhh1 and RNAPII in Ccr4–Not mutants. The Ccr4–Not complex can be divided into two subcomplexes based on their association with Not1: the Ccr4 group and the Not group. The Not group, consisting of Not2–Not5, interacts with the C terminus of Not1; the Ccr4 group, comprised of Ccr4, Pop2, and Dhh1, associates with the N terminus (Bai et al. 1999). The association of Dhh1 with RNAPII was greatly reduced when either of the Ccr4 group subunits—Ccr4 (Fig. 2B) or Pop2 (data not shown)—was deleted. In addition, mutation of NOT2 or deletion of NOT4 reduces or abolishes the interaction, respectively. Finally, the interaction between Dhh1 and RNAPII did not require Not3 or Not5. These results show that the integrity of the Ccr4–Not complex is important for maintaining its association with RNAPII, and indicates that Dhh1 associates with RNAPII within the context of the Ccr4–Not complex.
As discussed above, Ccr4 copurifies with the Paf1c complex. Additionally, Paf1c mutants display a number of similar phenotypes as Ccr4–Not mutants (Chang et al. 1999; Mueller et al. 2004). We explored the possibility that the interaction of Ccr4–Not with RNAPII is mediated through Paf1c. We examined two strains with mutations in PAF1C: paf1Δ and cdc73Δ. Deletion of PAF1 reduces the levels of other PAF1C subunits in cells, while deletion of CDC73 eliminates the interaction of PAF1C with chromatin and RNAPII (Mueller et al. 2004). However, deleting either of these PAF1C subunits does not disrupt the association between Ccr4–Not and RNAPII (Fig. 2C). This indicates that the interaction described here is not the Paf1C–Ccr4–RNAPII complex isolated by others. A chd1Δ mutant was also examined as a control, as it is implicated in elongation but has no known link to Ccr4–Not. Deletion of CHD1 did not impact the association of Ccr4–Not with RNAPII (Fig. 2C). Finally, we examined whether Ctk1, the kinase that modifies Ser2 within the C-terminal domain (CTD) of the large subunit of RNAPII, is required for the association of Ccr4–Not with RNAPII. Ser2 phosphorylation coordinates the recruitment and release of mRNA processing factors with RNAPII (for review, see Bentley 2005; Buratowski 2009) and, since Ccr4–Not plays a role in mRNA metabolism, it may be recruited to RNAPII by phosphorylation of Ser2. However, deletion of CTK1 did not disrupt the association of Ccr4–Not with RNAPII (Fig. 2C).
We examined the requirement for the CTD of the largest subunit of RNAPII, Rpb1, for the RNAPII–Ccr4–Not interaction. The CTD of Rpb1 is known to serve as a docking site for a number of elongation and mRNA processing factors (Bentley 2005; Buratowski 2009). Because deletion of the entire CTD is lethal, we coexpressed a CTD-less version (ΔCTD) of Rpb1 in the presence of endogenous wild-type Rpb1 and immunoprecipitated Ccr4–Not subunits from the extracts (Supplemental Fig. S2). The presence of both forms of Rpb1 was detected using an antibody specific for the N-terminal region of Rpb1, and the ΔCTD version is distinguished from the full-length protein by its faster mobility in SDS-PAGE gels. Both the wild-type and CTD-less versions of Rpb1 copurified with myc-tagged Dhh1 or Not2. Similar results were obtained when immunoprecipitations were conducted in extracts from strains containing progressive truncations in the CTD as the only source of Rpb1 (data not shown). These results suggest that Ccr4–Not does not require the CTD to interact with RNAPII.
An alternative possibility for what could mediate the interaction between Ccr4–Not and RNAPII is the Rpb4/Rpb7 heterodimer. Rpb4 is a nonessential subunit of RNAPII and is required for the association of Rpb7 with the core of RNAPII (Pillai et al. 2001). Rpb4 regulates cotranscriptional RNA processing and accompanies the nascent transcript from the nucleus to cytoplasmic P-bodies during cell stress to regulate the turnover of certain RNAs (Lotan et al. 2005; Runner et al. 2008). These functional similarities between Ccr4–Not and Rpb4 prompted us to examine whether Rpb4 tethers Ccr4–Not to RNAPII. The interaction of Ccr4 and Dhh1 with RNAPII was examined in an rpb4Δ mutant, and they are capable of coimmunoprecipitating RNAPII lacking the Rpb4 subunit (Supplemental Fig. S2). Thus, the Rpb4/7 heterodimer is dispensable for the interaction of Ccr4–Not with polymerase, and suggests that Ccr4–Not is interacting with the “core” of RNAPII.
Ccr4–Not directly binds to RNAPII elongation complexes
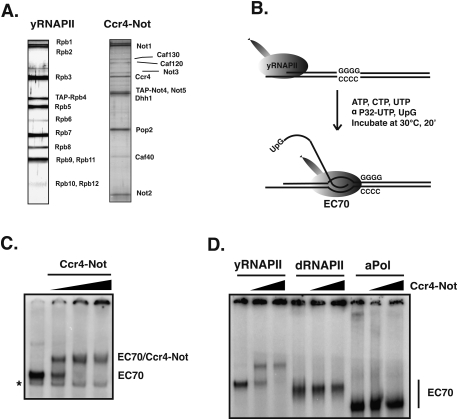
The Ccr4–Not complex copurifies with RNAPII from whole-cell extracts. However, these assays cannot determine if Ccr4–Not directly interacts with RNAPII or if it can bind elongating RNAPII. Yeast RNAPII (yRNAPII) was isolated to high purity, as described previously (Suh et al. 2005), and the Ccr4–Not complex was purified by tandem affinity purification (TAP) from a TAP-Not4 strain. Silver-staining reveals that all 12 subunits of RNAPII are present (Fig. 3A). Rbp1 is predominantly hypophosphorylated (Suh et al. 2005; data not shown). All of the known core subunits of the Ccr4–Not complex were detected in the TAP purification, and its composition matched that published by others (Mulder et al. 2007b; Azzouz et al. 2009). We assembled radiolabeled elongation complexes (EC70) on a tailed template, as described previously (Zhang et al. 2005) and illustrated in Figure 3B. Transcription from some tailed templates by eukaryotic RNA polymerases produces RNA:DNA hybrids with the transcribed strand, displacing the nontranscribed strand and affecting elongation (Dedrick and Chamberlin 1985). However, this was not detected using the EC70 template described here and yeast RNAPII (Zhang et al. 2005). Increasing amounts of the Ccr4–Not complex were incubated with the EC70 complexes, followed by resolution of the complexes on a native polyacrylamide gel. A slower-migrating species formed with the addition of Ccr4–Not, with a corresponding decrease in the faster-migrating EC70 complex (Fig. 3C). Similar results were obtained when Ccr4–Not was purified through a TAP tag in Ccr4 (Supplemental Fig. S3). This demonstrates that the Ccr4–Not complex directly interacts with elongating RNAPII.
Figure 3.
The Ccr4–Not complex interacts directly with yeast RNAPII elongation complexes. (A) Silver-stained SDS-PAGE gels showing the composition of yeast RNAPII purified from a TAP-Rpb4 strain (left) and the Ccr4–Not complex purified from a TAP-Not4 strain (right). (B) Schematic representation of the in vitro elongation system. RNAPII initiates transcription with UpG on the tailed template and stalls at the G-residues located at the end of the G-less cassette. (C) Native gel analysis of the interaction of Ccr4–Not with elongation complexes. RNAPII elongation complexes were formed on the tailed template (EC70) containing a 70-nt radiolabeled nascent transcript. One-hundred nanograms of template was used in each reaction. Approximately 100 ng (∼0.25 pmol) of RNAPII was present in each sample. Elongation complexes stalled at the end of the G-less cassette were incubated for 10 min with increasing amounts of purified Ccr4–Not complex (∼0.5, 1, and 1.5 pmol of Ccr4–Not complex). The concentrations of all proteins were estimated by comparing intensities of their bands with those of known amounts of BSA on a silver-stained SDS-PAGE gel. RNAPII-only lane contains 1 μg of BSA. (D) Yeast (yRNAPII), Drosophila (dRNAPII), and archaeal polymerase from Pyrococcus furiosus (aPol) were used to generate elongation complexes (EC70) on the tailed template. Ccr4–Not complex (0.5 pmol and 1 pmol) was added to each of the elongation complexes and reactions were analyzed on a 4% native gel.
Although excess competitor RNA was included in the reactions to suppress nonspecific binding to the nascent transcript, it is possible that Ccr4–Not associates with ECs exclusively through RNA or a transcription-dependent nucleic acid structure. To rule out this possibility, we repeated the assay using ECs formed from either Drosophila melanogaster RNAPII (dRNAPII) or archael RNA polymerase. ECs formed from these other sources of polymerase contain the same transcript and template DNA as those formed from yeast RNAPII. The results in Figure 3D indicate that the Ccr4–Not complex failed to shift ECs formed from dRNAPII or archael polymerase. Interestingly, even though yeast and Drosophila RNAPII are highly conserved, species specificity was observed. Similar species specificity was observed recently where Drosophila DSIF failed to shift ECs formed from yeast RNAPII (AG Missra and DS Gilmour, unpubl.). This result demonstrates that the interaction of Ccr4–Not with the yRNAPII EC is specific and requires direct contacts with the enzyme.
Ccr4–Not rescues arrested/backtracked RNAPII
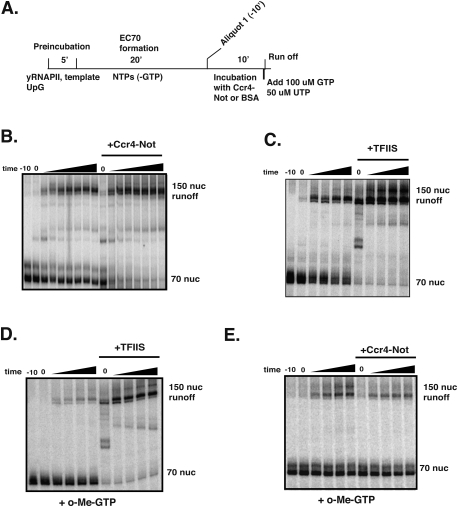
Next, we examined whether Ccr4–Not affects elongation. The use of the defined system allows us to bypass potentially confounding effects of Ccr4–Not on initiation and focus exclusively on its effect on RNAPII elongation. We set up elongation complexes on the 70-nucleotide (nt) G-less cassette template, added Ccr4–Not or carrier protein, and, after 10 min, GTP and excess cold UTP were added to the mixture to allow transcription to run off the template (Fig. 4A, schematic). The addition of excess cold UTP prevents the detection of newly initiated transcripts during the runoff period; thus, only products produced from the stalled EC are observed. In the absence of Ccr4–Not, only ∼40%–50% of the elongation complexes can be chased into runoff products (Fig. 4B). The ECs not being chased into runoff products by the addition of GTP display the characteristics of backtracked RNAPII complexes, rather than “dead-end” products, because they can be reactivated to produce runoff products by TFIIS (Fig. 4C). As shown in Figure 4B, elongation by RNAPII is stimulated by the Ccr4–Not complex by 1.5-fold to 2.0-fold, converting most of the arrested ECs into an elongation-competent form. Since Ccr4–Not was added after RNAPII was arrested, it can reactivate the backtracked RNAPII complexes. It should be noted that the length of the runoff product is only 150 nt, and any stimulation of elongation rate (nucleotides per second) would not be detected on such a short template; therefore, the effects we observe are likely the result of “reactivating” the arrested—and, in some cases, backtracked—RNAPII. Although carrier protein was added to the RNAPII-only reactions in approximately equal amounts to that of the Ccr4–Not complex, it is possible that stimulation of elongation by the Ccr4–Not complex may be a nonspecific effect caused by adding the complex to the reaction. To rule out this possibility, we carried out transcription run-on assays using Drosophila RNAPII in the presence and absence of Ccr4–Not. Consistent with the inability of Ccr4–Not to shift Drosophila ECs (Fig. 3D), addition of Ccr4–Not to the reaction failed to stimulate elongation of dRNAPII (Supplemental Fig. S3). Together, these results provide evidence that the Ccr4–Not complex directly and specifically interacts with elongating yRNAPII and stimulates transcription elongation.
Figure 4.
Ccr4–Not stimulates the resumption of transcription from arrested RNAPII elongation complexes. (A) Outline of the in vitro elongation runoff assay. Arrested RNAPII elongation complexes (EC70) were formed in the absence of GTP. RNAPII is able to resume elongation and generate a 150-base runoff transcript after the addition of GTP and UTP. (B) In vitro runoff assay in the presence of Ccr4–Not. Arrested elongation complexes were incubated with 1.5 pmol of Ccr4–Not complex or 1 μg BSA for 10 min, and then GTP and UTP were added to allow runoff. A total of 1.5 pmol of complex was used because it is just above the amount required to fully shift polymerase in the gel shift assay (Fig. 3). (C) TFIIS rescues arrested EC70 complexes. TFIIS (0.5 pmol) was added to the reaction after forming the arrested EC70. (D,E) Ccr4–Not, unlike TFIIS, does not stimulate the nucleolytic activity of RNAPII. In vitro elongation system was set up as described above, with the exception that O-me GTP was added to form the arrested elongation complex. Prior to the addition of nucleotides, 0.5 pmol of TFIIS (D) or 1.5 pmol of Ccr4–Not complex (E) was added to the reactions.
The previous experiment assessed the ability of Ccr4–Not to stimulate the “reactivation” of an arrested RNAPII complex on a short runoff template. We altered the experimental design and conducted the assay on a longer runoff template, which allowed us to determine if Ccr4–Not stimulates the rate of transcription. RNAPII, initiating dinucleotide, and the template were incubated to form initiation complexes. Ccr4–Not was then added, followed by all NTPs, enabling runoff to occur (Supplemental Fig. S4). Interestingly, Ccr4–Not did not stimulate the production of runoff products when RNAPII was not stalled at a G-tract (Supplemental Fig. S4). This was observed with high (100 μM) or low (10 μM) amounts of GTP. The assay was also repeated in parallel on the longer template, where stalled elongation complexes were preformed prior to the addition of nucleotides (Supplemental Fig. S4). Under these conditions, stimulation (approximately twofold to 2.5-fold) was observed. These results suggest that Ccr4–Not predominantly stimulates the reactivation of arrested RNAPII, rather than affecting the rate of elongation per se.
Ccr4–Not mechanism of action is distinct from that of TFIIS
Our data suggest that Ccr4–Not stimulates the reactivation of stalled and backtracked RNAPII in vitro. This activity is similar to that of the well-characterized elongation factor TFIIS (Dst1). These observations prompted us to test whether Ccr4–Not functions by a similar mechanism. TFIIS binds to RNAPII and stimulates the intrinsic nuclease activity of the enzyme, leading to cleavage of the displaced 3′ end of the RNA, realignment of the RNA in the active site, and the resumption of elongation (Fish and Kane 2002; Arndt and Kane 2003). This activity allows TFIIS to reactivate ECs terminally stalled by the incorporation of chain-terminating nucleotides into the 3′ end of the transcript. We exploited this property of TFIIS to compare its mechanism of action with that of Ccr4–Not. ECs arrested by the incorporation of O-me-GTP into the 3′ end of the transcript were formed and then incubated with either TFIIS or the Ccr4–Not complex, followed by the addition of excess GTP and UTP to allow transcription runoff (Fig. 4A). In the absence of TFIIS, a small percentage of ECs ran off (Fig. 4D), indicating that the majority of the ECs remained arrested. Adding TFIIS greatly stimulated the resumption of elongation, and significant readthrough was detected immediately after the addition of GTP (t = 0) (Fig. 4D). In contrast, adding Ccr4–Not did not stimulate the resumption of elongation of ECs containing blocked 3′ ends (Fig. 4E). These results indicate that Ccr4–Not cannot stimulate the intrinsic nuclease activity of RNAPII; thus, Ccr4–Not and TFIIS stimulate elongation by a different mechanism.
Ccr4–Not activity is transcript length-dependent
In order to reactivate the backtracked complexes, Ccr4–Not must effectively realign the 3′ end of the RNA back into register with the active site of the enzyme without stimulating the nucleolytic activity of RNAPII. One way it could do so is by binding to the transcript and promoting forward translocation of RNAPII. The association of Ccr4–Not with RNAPII is not dependent on RNA (Figs. 2A, 3D); however, this does not preclude the possibility that it binds to the RNAPII EC, interacts with the RNA emerging from the exit channel, and exerts its functions in a transcript-dependent manner.
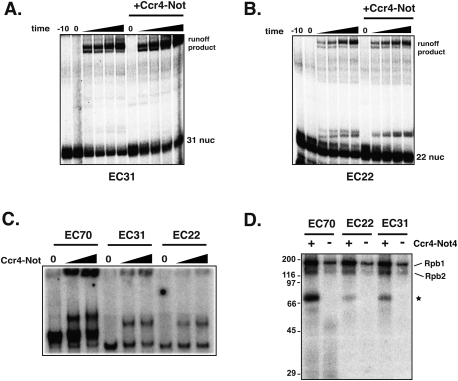
Elongation complexes were formed on templates containing a 31- and 22-nt G-less cassette, producing EC31 and EC22, respectively. These ECs have the same 3′-end sequences as EC70, but differ in length by the incorporation of different nucleotides at the 5′ end. Transcription runoff assays were performed in the presence and absence of Ccr4–Not on EC31 and EC22 complexes. Unlike what was observed with EC70 complexes (Fig. 4B), adding Ccr4–Not failed to stimulate the activation of the stalled EC31 and EC22 complexes (Fig. 5A,B). The extension of the stalled EC31 and EC21 complexes into runoff products could be stimulated by TFIIS, indicating that they are not prematurely terminated (data not shown). The transcription length requirement for the activity of Ccr4–Not suggests that its ability to stimulate elongation is dependent on the emerging transcript.
Figure 5.
Ccr4–Not functions in a transcript length-dependent manner and binds the transcript. (A,B) Transcription elongation assays from EC31 and EC22 complexes. Assay conditions and experimental design are described in the legend for Figure 4. (C) Native PAGE of EC complexes. The assay was carried out as described in Figure 3C, except that Ccr4–Not was added at 1:1 and 2:1 to RNAPII. (D) UV cross-linking of Ccr4–Not to the transcript in elongation complexes. EC complexes were formed from templates containing a 70-, 31-, and 22-nt G-less cassette, forming EC70, EC31, and EC22, respectively. Transcription was carried out in the presence of radiolabeled CTP and bromouridine. After the formation of the ECs, Ccr4–Not was added to the complexes and the mixture was cross-linked by UV light as described in the Materials and Methods. DNase and RNase were added prior to electrophoresis. The migration of molecular weight markers (in kilodaltons) are indicated on the left, and the Ccr4–Not-specific band is marked by a star on the right.
A trivial explanation for this result is that Ccr4–Not does not bind to the shortened ECs. The association of Ccr4–Not with ECs of various lengths was compared by native gel electrophoresis. An amount of Ccr4–Not that shifts ∼30%–50% of the EC70 was examined in the assay to allow for the detection of changes in its binding to the shorter ECs. The results shown in Figure 5C indicate that Ccr4–Not supershifts EC70, EC31, and EC22 complexes to approximately equivalent levels. The greater intensity of the signal from the EC70 complex results from more label incorporated into the longer transcript, rather than a quantitative difference in the amount of EC complex formed.
Next, we examined whether Ccr4–Not binds to the transcript using a UV cross-linking assay. UV cross-linking assays were conducted on ECs containing 22-, 31-, and 70-base radiolabeled transcripts in the presence or absence of Ccr4–Not. In the absence of Ccr4–Not, Rpb1 was the predominantly cross-linked species, with Rpb2 cross-linking to a lesser degree (Fig. 5D, lane 1). This agrees well with the results of others (Ujvari and Luse 2006; Missra and Gilmour 2010). No incorporation of label into proteins was detected in the absence of UV cross-linking (Supplemental Fig. S5, data not shown). Adding Ccr4–Not to the ECs resulted in the detection of two cross-linked species in the gel (Fig. 5D, lane 1 vs. 2). The small amount of the faster-migrating species varied between experiments and may be a breakdown product of the larger cross-linked protein (data not shown). The estimated molecular weight of the major cross-linked product is ∼65–70 kDa, and runs in a region of the gel devoid of any subunits of RNAPII. Allowing for some alteration in the migration due to the incorporation of RNA into the protein (not all of the RNA can be digested), the cross-linked protein in Ccr4–Not could be Pop2, Not4, Not5, or Dhh1. Pop2 migrates near 50 kDa, while the latter three migrate very closely to each other between 65–70 kDa. Interestingly, the level of cross-linking of Rpb1 and Rpb2 was enhanced in the presence of Ccr4–Not (Fig. 5D, cf. even vs. odd lanes). This result is highly reproducible (data not shown), and would be consistent with Ccr4–Not stabilizing or altering the conformation of RNAPII in the ECs. As the transcript length was shortened, the amount of the Ccr4–Not-dependent cross-linked product decreased with transcript length. Since the cross-linked material was extensively treated with nucleases prior to SDS-PAGE analysis, the greater intensity of the band observed when Ccr4–Not was cross-linked to ECs of different lengths is not explained by more label incorporation into the longer transcripts. However, the reduced level of cross-linking could be explained, at least partially, by the number of photo-activatable groups incorporated into the transcript. When corrected for the cross-linking to Rpb1, shortening the transcripts to 31- and 22-nt reduced the cross-linking of Ccr4–Not to ∼40% and 25% of that observed on EC70, respectively. Some cross-linking is still observed with EC22, which is predicted to have ∼4 nt emerging from RNAPII (Andrecka et al. 2008), including two bromouridine bases. The detection of some level of cross-linking to the transcript in the EC22 complex suggests that Ccr4–Not subunits are located near the RNA exit channel.
Ccr4–Not stimulates elongation in vivo
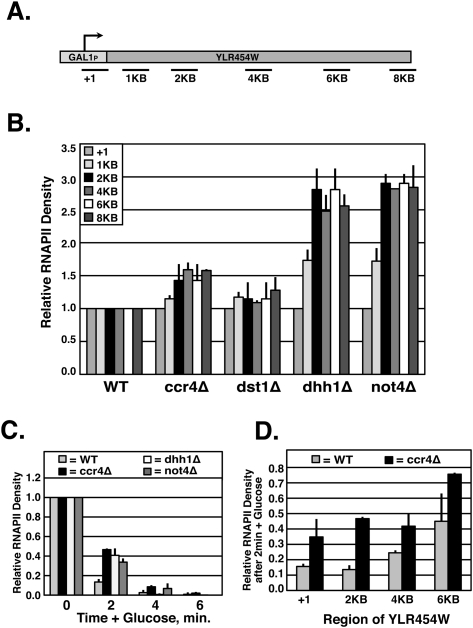
We used an assay that measures RNAPII density across a long GAL1p-regulated gene (GAL1P-YLR454W) to test whether Ccr4–Not affects elongation in vivo. First, we analyzed processivity in Ccr4–Not mutants by measuring RNAPII density at multiple locations across the 8-kb gene under galactose-inducing conditions (Fig. 6A). Previous studies described a processivity defect as showing a gradual decrease in RNAPII density as you move across the gene, which suggests that a certain fraction of the RNAPII loaded onto the promoter is released prior to the completion of the gene (Mason and Struhl 2005). Surprisingly, deletion of DHH1, NOT4, and, to a lesser extent, CCR4 resulted in the opposite phenotype—the density of RNAPII increased further along into the gene (Fig. 6B). This implies that, although RNAPII remains engaged with the template, it is not efficiently traversing the gene. As a control, we analyzed processivity in a TFIIS mutant, dst1Δ, and, consistent with the results of others (Mason and Struhl 2005), it did not show a significant change in processivity. The difference in phenotypes between dst1Δ and Ccr4–Not mutants in this assay further supports the idea that they stimulate transcription elongation through distinct mechanisms.
Figure 6.
Ccr4–Not promotes elongation in vivo. (A) Schematic of GAL1P-YLR454W and primer locations. (B) ChIP analysis of RNAPII density across GAL1P-YLR454W in wild-type and mutant strains under galactose-inducing conditions. RNAPII%IP in mutant strains was normalized to the corresponding region in wild type, and the promoter region for each strain was then set to 1. (C) ChIP of relative RNAPII density for wild-type and mutant strains at the 2-kb region under inducing and repressing conditions for the indicated times. RNAPII density in galactose-grown cells was set to 1, and time points after addition of glucose were normalized to the RNAPII%IP prior to repression. (D) ChIP of relative RNAPII density in wild-type and ccr4Δ strains following 2 min of treatment with dextrose. A ratio of (RNAPII%IP at 2 min + glucose)/(RNAPII%IP in galactose) was calculated for wild-type and ccr4Δ strains at the indicated primer regions.
In order to confirm that the increase in RNAPII density is due to the slowed progression of RNAPII across the gene, we adapted this assay to monitor the kinetics of the “last wave” of polymerase after repressing GAL1p with dextrose (Mason and Struhl 2005). In wild-type cells, RNAPII levels at 2 kb within the gene are reduced to near background levels by 2 min after adding dextrose (Fig. 6C). In contrast, there is a higher density of RNAPII over the gene in the mutants at the same time point, suggesting a delay in RNAPII progression across the gene (Fig. 6C). Ccr4–Not mutants have as much as fourfold higher levels of RNAPII after 2 min of transcription shutoff, and at 4 min, there is still a higher level of RNAPII in the ccr4Δ and not4Δ cells. Since the level of RNAPII is so low at 4 min in wild-type cells, an accurate estimation of the increase in RNAPII in the Ccr4–Not mutants is not possible.
To examine this differently, we analyzed the amount of RNAPII across the entire length of GAL1p-YLR454W at the 2-min time point. In wild-type cells, RNAPII levels are reduced to near background levels at the 5′ end of the gene (+1 and 2 kb). However, progressively higher levels are detected in the middle (4 kb) and toward the end (6 kb) of the gene because the RNAPII loaded onto the promoter has not completed its “last wave” (Fig. 6D). All regions examined in the ccr4Δ mutant show significantly higher RNAPII density relative to wild-type cells (Fig. 6D). These results support the hypothesis that, in the absence of the Ccr4–Not complex, elongation of RNAPII across the gene is impaired, resulting in more RNAPII remaining within the body of the gene. As a whole, the data presented herein clearly show that the Ccr4–Not complex functions directly during transcription elongation, and solidify it as a bona fide elongation factor.
Discussion
Many functions have been attributed to the Ccr4–Not complex. Early on, it was proposed to regulate transcription initiation by controlling the function of the general transcription factor TFIID (Denis and Chen 2003; Collart and Timmers 2004). Later, the complex was shown to regulate mRNA decay and protein ubiquitylation (Tucker et al. 2001; Panasenko et al. 2006). Here we show that Ccr4–Not is recruited to DNA damage-dependent genes and regulates TFIID recruitment to these genes. More importantly, we demonstrate that Ccr4–Not is a direct regulator of RNAPII elongation. Below, we describe a model for how Ccr4–Not regulates elongation, and reconcile its seemingly contradictory functions in promoting transcription and mRNA decay.
Ccr4–Not directly associates with elongating RNAPII
Our results support a mechanism in which Ccr4–Not directly functions in elongation by interacting with RNAPII as it transcribes the gene. Ccr4–Not associates with the body of the gene in a transcription-dependent manner (Fig. 1). The pattern of Ccr4–Not cross-linking to genes is very similar to that of RNAPII and other factors that function during elongation, consistent with it being loaded and disassembled with RNAPII at GAL1 in response to gene activation and repression. Cross-linking of Ccr4–Not complex subunits was observed at stress and Gcn4-dependent promoters, but the presence of the complex within the ORF was not examined (Deluen et al. 2002; Swanson et al. 2003). The cross-linking of these subunits to promoters could have been attributed to its function in regulating TBP recruitment through TFIID or SAGA.
Numerous proteins in yeast have been defined as elongation factors if they possess a number of characteristics (see below), but in many cases these factors have not been shown to directly affect elongation. Although previous studies on Ccr4–Not have implied roles in elongation, no direct evidence for this function has been provided. Evidence supporting a role in elongation include genetic interactions between Ccr4–Not subunits and elongation factors and 6-AU sensitivity (Denis et al. 2001). While mutations in elongation factors cause 6-AU and MPA sensitivity and show reciprocal genetic interactions with other elongation factor mutants (Arndt and Kane 2003), many of these putative elongation factors also display genetic interactions with initiation and chromatin remodeling factors (Davie and Kane 2000; Simic et al. 2003), and mutations of many genes not known to be involved in transcription cause 6-AU and MPA sensitivity (Desmoucelles et al. 2002; Riles et al. 2004). Significantly, some of these genes are involved in mRNA processing and metabolism. Other tests are to demonstrate that they cross-link within the coding region of genes or can copurify with RNAPII (for review, see Arndt and Kane 2003). Although the latter test demonstrates a potential association with RNAPII, it fails to directly demonstrate that the factor associates with RNAPII in the context of an elongation complex.
Ccr4–Not mutants were recently identified in a screen for factors involved in mRNA biogenesis using the gene length-dependent accumulation of mRNA (GLAM ratio) (Gaillard et al. 2009). Ccr4–Not mutants failed to accumulate mRNA with long 3′ untranslated regions (UTRs) containing the lacZ gene. It is difficult to conclude from this assay alone if a factor is involved in elongation control because factors involved in all stages of mRNA biosynthesis and processing show reduced GLAM ratios (Morillo-Huesca et al. 2006; Gaillard et al. 2009). In addition, analysis of a not5Δ mutant showed that RNAPII density was reduced weakly at the end of the GLAM reporter gene in this mutant, and an extract prepared from a not5Δ mutant could not transcribe through a long lacZ-containing reporter gene in vitro (Gaillard et al. 2009). Thus, while these assays individually have limitations, the body of evidence suggested that Ccr4–Not is involved in elongation. However, the mechanism of action and whether the effects observed in the mutants are direct were not clear at the time. Surprisingly few putative elongation factors have been shown to have a direct effect on RNAPII elongation, and fewer yet have been shown to assemble into elongation complexes. Paf1c, long suspected to be an elongation factor in yeast, was only recently shown biochemically to affect elongation, and this study involved human Paf1c (Kim et al. 2010), however. The ability of Ccr4–Not to bind to functional elongation complexes and directly stimulate elongation provides the strongest possible evidence that it is a bona fide elongation factor.
Factors that bind to elongating RNAPII often do so through the CTD. We expected this to be true of Ccr4–Not because Not5 was identified in a screen for proteins retained on an affinity matrix of synthetic CTD peptides phosphorylated at Ser2 and Ser5 (Phatnani et al. 2004). However, other Ccr4–Not subunits were not identified in this screen, so the association of Not5 with the modified CTD peptides may have been indirect, or Not5 may bind to the CTD outside of the context of the Ccr4–Not complex. We found that Ccr4–Not does not require the CTD to interact with RNAPII and does not require phosphorylation on Ser2 on the CTD (Fig. 2C; Supplemental Fig. S2). Furthermore, Ccr4–Not interacted well with the hypophosphorylated form of RNAPII used in our elongation complex association assay (Fig. 4), and phosphorylating the CTD in vitro did not affect the association of Ccr4–Not with elongation complexes (A Dutta and JC Reese, unpubl.). The idea that Ccr4–Not binds to polymerase independent of its modification states is further substantiated by its cross-linking across the entire length of the gene, and cross-linking across the gene supports its role as a general regulator of elongation (Fig. 1B). It cannot be ruled out that changes in the CTD phosphorylation pattern throughout the transcription cycle modulate the activity of Ccr4–Not independent of recruitment.
Mechanism for Ccr4–Not function during elongation
Here we employed a simple, but powerful, assay to directly monitor elongation and characterize the function of the Ccr4–Not complex. Biochemical assays indicate that the Ccr4–Not complex interacts directly with elongating RNAPII and promotes elongation (Figs. 3, 4). Interestingly, the effect of Ccr4–Not on elongation is not detectable if RNAPII is not arrested (Supplemental Fig. S4). This suggests that Ccr4–Not is not affecting, or having a lesser effect on, the rate of transcription, but instead is affecting RNAPII's ability to resume transcription from an arrested state. Since TFIIS also stimulates the arrested RNAPII to resume elongation, it is likely that the arrested RNAPII has backtracked on the DNA template under the conditions used here. TFIIS overcomes the transcription block by stimulating the intrinsic nuclease activity of RNAPII. Ccr4–Not must be acting by a different mechanism because it was unable to overcome the transcription block caused by a chain terminator. In this regard, Ccr4–Not corrects backtracked complexes by a novel mechanism that possibly complements that of TFIIS. Such a mechanism provides an explanation for the synthetic growth defect when a ccr4Δ mutation is combined with dst1Δ mutation (Denis et al. 2001).
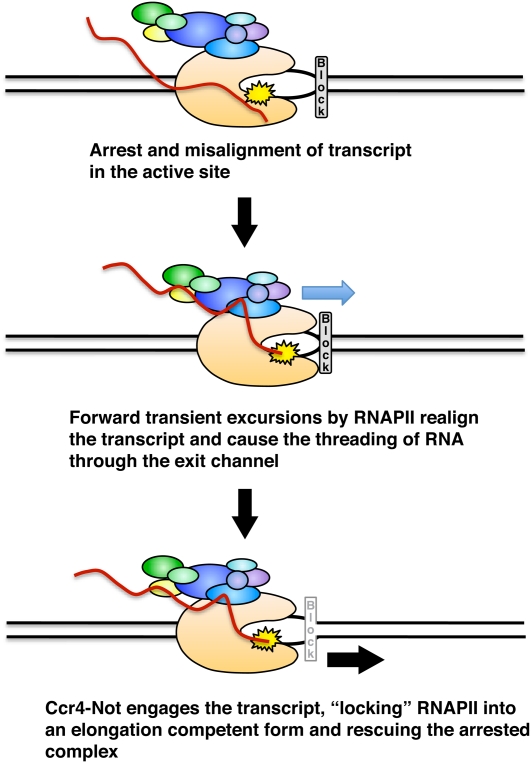
The experiments demonstrating that Ccr4–Not requires a minimal-length transcript to reactivate arrested RNAPII and that it cross-links to the transcript strongly suggest that an interaction with the emerging transcript is necessary for Ccr4–Not to function. Forward translocation of RNAPII occurs via Brownian motion, and stalled ECs are believed to undergo excursions in the forward and reverse directions (Cramer et al. 2008; Nudler 2009). Arrested RNAPII can move along the template in the forward and backward directions, causing the threading of the transcript through the RNA exit channel. The binding of proteins to the transcript could prevent the translocation of RNAPII by preventing the movement of the transcript in and out of the RNA exit channel. There is evidence that the binding of proteins to the emerging transcript can favor elongation by disfavoring backward translocation of the polymerase (Reeder and Hawley 1996; Roberts et al. 2008; Nudler 2009; Proshkin et al. 2010). We propose that Ccr4–Not stimulates elongation by promoting realignment of the 3′ end of the transcript in the active site by trapping RNAPII during its forward excursions along the template by binding to the transcript and preventing backward transitions. As RNAPII moves forward without nucleotide synthesis, more transcript emerges from the RNA exit channel, and Ccr4–Not undergoes reiterative cycles of transcript release and rebinding down the transcript in the 3′ direction and pushes RNAPII forward through a “ratcheting-like” mechanism (Fig. 7). This would lead to the realignment of the 3′ end of the transcript in backtracked complexes and promote elongation.
Figure 7.
Model for the rescue of arrested elongation complexes by Ccr4–Not. (Top) Transcription blocks lead to arrest and backtracking of polymerase. The 3′ end of the transcript is out of register with the active site (yellow starburst), preventing productive elongation. (Middle) Transient forward excursions of polymerase threads transcript out of the RNA exit channel, which can associate with Ccr4–Not. (Bottom) Cycles of transcript binding and release by Ccr4–Not during forward excursions promote elongation by locking RNAPII into an elongation-competent form.
Ccr4–Not affects RNAPII elongation across a gene
The development of assays to measure RNAPII elongation rates and processivity in vivo across a large model gene, GAL1-YLR454W, has shed some light on the roles of transcription factors in elongation. This assay has the advantage that it measures RNAPII density, and any effects of a mutation on other aspects of mRNA metabolism do not confound the results. Deletion of CCR4, DHH1, or NOT4 results in a change in the distribution of RNAPII across GAL1p-YLR454W that is unique among elongation factors mutants described thus far. Mutation of most elongation factors leads to either no phenotype or reduced processivity, which appears in this assay as a loss of RNAPII across the gene under steady-state conditions (Mason and Struhl 2005). In contrast, RNAPII density increases across the ORF in Ccr4–Not mutants (Fig. 6B). The Ccr4–Not mutant phenotype suggests that the polymerase loaded onto the promoter is slow to complete transcription of the gene (rate) or is not resuming transcription after transient stalling or arrest. Our in vitro analysis is consistent with the latter. Deleting Ccr4–Not subunits results in a buildup of RNAPII; in contrast, deleting DST1, the gene encoding TFIIS, does not cause the same phenotype (Fig. 6; see Mason and Struhl 2005). Therefore, both the in vitro and in vivo elongation assays support the notion that Ccr4–Not and TFIIS relieve transcriptional blocks by a different molecular mechanism. TFIIS must be used when cleavage of the 3′ end is required, such as during misincorporation or extensive misalignment of the transcript, while Ccr4–Not promotes forward translocation of arrested RNAPII by interacting with the transcript.
Physiological significance of Ccr4–Not promoting elongation
RNAPII can pause or arrest stochastically during transcription, but particularly in response to sequence-specific pause sites, nucleotide limitations, DNA lesions, negative elongation factors, and DNA-bound proteins (Svejstrup 2007). Multiple transient stalling events are likely to occur in vivo when it encounters these situations, or simply when RNAPII encounters an intact nucleosome. RNAPII stalls within a nucleosome until chromatin remodeling factors alter nucleosome structure to allow the passage of polymerase (Kireeva et al. 2005; Carey et al. 2006). After pausing for remodeling, RNAPII must resume elongation, which may be influenced by elongation factors such as Ccr4–Not. The example described here is specific for nucleosomes, but the general mechanism may be applied to many different transcriptional blocks encountered by RNAPII. For example, pauses can occur during various transcription and cellular stresses. The Ccr4–Not complex is known to play roles in stress resistance (Collart and Timmers 2004), and one of its functions to aid in the cellular stress response may be to promote the resumption of transcription.
The interesting question that looms is why a complex that regulates mRNA decay also functions in promoting transcription. As stated above, the Ccr4–Not complex is implicated in regulating gene expression in response to stress. Most stress responses are transient, displaying an initial rapid induction phase followed by a fading or inactivation of the initial response. Ccr4–Not can positively affect the transcription of genes during the induction phase and then, upon changes to cellular signals, turn and promote the deadenylation and destruction of the mRNA it interacts with. By functioning in this way, Ccr4–Not could impose decay on the transcripts it used to promote synthesis for, and would thus contribute to the cell's overall mechanism for precisely controlling the level of mRNAs from “birth to death.”
Materials and methods
Strains and media
Strains used in this study are listed in Supplemental Table 1. Cells were grown in yeast extract-peptone at 30°C with an appropriate carbon source, either 2% or 4% dextrose (where indicated), 3% raffinose, or 2% galactose. MMS-induced cells were treated with 0.03% MMS and incubated for 2.5 h prior to analysis. Gene deletions and tagging were performed as described previously using PCR-mediated gene disruption and modification (Brachmann et al. 1998; Longtine et al. 1998). The RPB1 plasmids (full-length and ΔCTD) were described in a previous study (McCracken et al. 1997).
ChIP
ChIP assays were performed as described previously (Sharma et al. 2007). Whole-cell extracts were prepared by glass bead disruption and sheared into fragments averaging 200 to 600 base pairs (bp) in size using a Bioruptor (Diagenode). Whole-cell extracts were immunoprecipitated with the antibodies indicated in the Supplemental Material. Real-time PCR was performed with SYBR Green detection (Quanta Biosciences) using a StepOne Plus qPCR thermocycler (Applied Biosystems). The percent immunoprecipitated was calculated as follows: (immunoprecipitated signal/input signal) × 100. Primer sequences are available on request. Error bars represent the standard errors of at least three repetitions. The in vivo elongation assays were conducted and data were analyzed as described in another study (Mason and Struhl 2005).
Coimmunoprecipitation and protein purification
Protein extracts for coimmunoprecipitation were prepared as described previously (Reese et al. 1994). Two milligrams of protein extract was incubated either with or without 100 μg/mL RNase A. Extracts were incubated with antibody for 1–3 h prior to the addition of Protein A Sepharose CL-4B, followed by an overnight incubation at 4°C (GE Healthcare). After washing, the bound proteins were analyzed by Western blotting. Both TAP-Not4 and TAP-Ccr4 complexes were purified from strains containing a deletion of DST1. Purification of the complex in a dst1Δ strain is essential because trace amounts of TFIIS activity were detected in preparations from DST1+ strains (A Dutta and JC Reese, unpubl.). The protocol for TAP purifications was adapted as described previously (Rigaut et al. 1999), with some minor modifications. TFIIS was expressed in Escherichia coli and purified as a histidine-tagged protein (Kim et al. 2007). Yeast RNAPII was purified as described in a previous study (Suh et al. 2005).
Preparation of elongation complexes and runoff transcription assays
Elongation complexes and reagents were prepared similar to those described for Drosophila elongation complexes (Zhang et al. 2005) and are described in the Supplemental Material. Transcription and EC complex assembly were carried out in 15-μL volumes with 100 ng of template and ∼100 ng (∼0.25 pmol) of purified yeast RNAPII. The template was preincubated with RNAPII for 5 min in the transcription buffer, and then transcription was initiated by adding an NTP mix, yielding final concentrations of 0.1 mM ATP, 0.1 mM CTP, 5 μM UTP, 5 μM 3′O-methyl GTP, and 4 μCi per reaction of [a-32P] UTP. Each reaction was incubated for 20 min at 30°C. Elongation complexes with Pyrococcus furiosus archaeal polymerase (a kind gift of Katsu Murakami, Pennsylvania State University) were generated at 75°C, and then returned to 25°C. Purified Ccr4–Not complex (or carrier protein) was added to the stalled elongation complexes in the presence of 1 μg of yeast RNA. The samples were run on 4% native gels. To measure runoff transcription, elongation complexes were formed as described above, with the exception that 3′O-methyl GTP was not added to the reactions. Then, UTP and GTP were added to 50 μM and 100 μM, respectively, and the samples were removed at the indicated time points. RNA was purified and analyzed on urea-containing denaturing gels. The gels were dried and analyzed using a PhosphorImager and scanned using the Typhoon system (Molecular Dynamics).
Protein–RNA UV cross-linking
Elongation complexes were formed as described above in the presence of NTP mix containing 0.1 mM ATP, 0.1 mM Br-UTP (UV cross-linkable UTP analog), 5 μM CTP, and 4 μCi per reaction of a-32P CTP. Purified Ccr4–Not complex was added to the stalled elongation complexes and allowed to bind for 5 min. Yeast RNA (0.3 μg) was then added to the reactions to reduce nonspecific binding of proteins to the nascent transcript. Protein-bound elongation complexes were UV cross-linked for 10 min on ice using a 300-nm-wavelength lamp. The samples were then treated with RNase A (400 ng) and DNase I (0.2 U) for 15 min at room temperature. The reactions were boiled in SDS-PAGE buffer and proteins were separated by a SDS-PAGE gel. Gels were dried and exposed to a PhosphorImager screen and analyzed using a PhosphorImager. Gels were then rehydrated in water and silver-stained.
Acknowledgments
We thank Martine Collart, Alan Hinnebusch, and David Bentley for strains and plasmids used to complete this work. We are grateful to Katsu Murakami for the gift of archeal polymerase and Anamika Missra for advice on establishing the EC70 system in our laboratory. The members of the Center for Eukaryotic Gene Regulation and Center for RNA Biology are recognized for their comments during the completion of this work. This research was supported by funds from National Institutes of Health to J.C.R. (GM58672) and D.S.G. (GM47477).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2020911.
Supplemental material is available for this article.
References
- Andrecka J, Lewis R, Bruckner F, Lehmann E, Cramer P, Michaelis J 2008. Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci 105: 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt KM, Kane CM 2003. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet 19: 543–550 [DOI] [PubMed] [Google Scholar]
- Azzouz N, Panasenko OO, Colau G, Collart MA 2009. The CCR4–NOT complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS ONE 4: e6760 doi: 10.1371/journal.pone.00067060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badarinarayana V, Chiang YC, Denis CL 2000. Functional interaction of CCR4–NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics 155: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL 1999. The CCR4 and CAF1 proteins of the CCR4–NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol Cell Biol 19: 6642–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Li B, Workman JL 2006. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell 24: 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol 19: 1056–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chiang YC, Denis CL 2002. CCR4, a 3′–5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 21: 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA 2003. Global control of gene expression in yeast by the Ccr4–Not complex. Gene 313: 1–16 [DOI] [PubMed] [Google Scholar]
- Collart MA, Timmers HT 2004. The eukaryotic Ccr4–Not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol 77: 289–322 [DOI] [PubMed] [Google Scholar]
- Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, et al. 2008. Structure of eukaryotic RNA polymerases. Annu Rev Biophys 37: 337–352 [DOI] [PubMed] [Google Scholar]
- Davie JK, Kane CM 2000. Genetic interactions between TFIIS and the Swi–Snf chromatin-remodeling complex. Mol Cell Biol 20: 5960–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RL, Chamberlin MJ 1985. Studies on transcription of 3′-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry 24: 2245–2253 [DOI] [PubMed] [Google Scholar]
- Deluen C, James N, Maillet L, Molinete M, Theiler G, Lemaire M, Paquet N, Collart MA 2002. The Ccr4–Not complex and yTAF1 (yTafII130p/yTafII145p) show physical and functional interactions. Mol Cell Biol 22: 6735–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis CL, Chen J 2003. The CCR4–NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol 73: 221–250 [DOI] [PubMed] [Google Scholar]
- Denis CL, Chiang YC, Cui Y, Chen J 2001. Genetic evidence supports a role for the yeast CCR4–NOT complex in transcriptional elongation. Genetics 158: 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmoucelles C, Pinson B, Saint-Marc C, Daignan-Fornier B 2002. Screening the yeast ‘disruptome’ for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J Biol Chem 277: 27036–27044 [DOI] [PubMed] [Google Scholar]
- Fish RN, Kane CM 2002Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta 1577 287–307 [DOI] [PubMed] [Google Scholar]
- Gaillard H, Tous C, Botet J, Gonzalez-Aguilera C, Quintero MJ, Viladevall L, Garcia-Rubio ML, Rodriguez-Gil A, Marin A, Arino J, et al. 2009. Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4–Not in transcription-coupled repair. PLoS Genet 5: e1000364 doi: 10.1371/journal.pgen.1000364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA 2007. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci 104: 16068–16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG 2010. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140: 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M 2005. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell 18: 97–108 [DOI] [PubMed] [Google Scholar]
- Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, Weissman JS, Briggs SD, Krogan NJ, Strahl BD 2007. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc Natl Acad Sci 104: 5836–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev 19: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L, Collart MA 2002. Interaction between Not1p, a component of the Ccr4–Not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J Biol Chem 277: 2835–2842 [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840 [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD 2009. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev 23: 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra AG, Gilmour DS 2010. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci 107: 11301–11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo-Huesca M, Vanti M, Chavez S 2006. A simple in vivo assay for measuring the efficiency of gene length-dependent processes in yeast mRNA biogenesis. FEBS J 273: 756–769 [DOI] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 14: 447–456 [DOI] [PubMed] [Google Scholar]
- Mulder KW, Winkler GS, Timmers HT 2005. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4–Not complex. Nucleic Acids Res 33: 6384–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KW, Brenkman AB, Inagaki A, van den Broek NJ, Timmers HT 2007a. Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4–Not complex. Nucleic Acids Res 35: 2428–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KW, Inagaki A, Cameroni E, Mousson F, Winkler GS, De Virgilio C, Collart MA, Timmers HT 2007b. Modulation of Ubc4p/Ubc5p-mediated stress responses by the RING-finger-dependent ubiquitin-protein ligase Not4p in Saccharomyces cerevisiae. Genetics 176: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E 2009. RNA polymerase active center: the molecular engine of transcription. Annu Rev Biochem 78: 335–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko O, Landrieux E, Feuermann M, Finka A, Paquet N, Collart MA 2006. The yeast Ccr4–Not complex controls ubiquitination of the nascent-associated polypeptide (NAC–EGD) complex. J Biol Chem 281: 31389–31398 [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Jones JC, Greenleaf AL 2004. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 43: 15702–15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai B, Sampath V, Sharma N, Sadhale P 2001. Rpb4, a non-essential subunit of core RNA polymerase II of Saccharomyces cerevisiae is important for activated transcription of a subset of genes. J Biol Chem 276: 30641–30647 [DOI] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328: 504–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Yoon S, Natarajan K, Swanson MJ, Hinnebusch AG 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol Cell Biol 24: 4104–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder TC, Hawley DK 1996. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell 87: 767–777 [DOI] [PubMed] [Google Scholar]
- Reese JC, Apone L, Walker SS, Griffin LA, Green MR 1994. Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature 371: 523–527 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Riles L, Shaw RJ, Johnston M, Reines D 2004. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast 21: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JW, Shankar S, Filter JJ 2008. RNA polymerase elongation factors. Annu Rev Microbiol 62: 211–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runner VM, Podolny V, Buratowski S 2008. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol Cell Biol 28: 1883–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VM, Tomar RS, Dempsey AE, Reese JC 2007. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol Cell Biol 27: 3199–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 22: 1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MH, Ye P, Zhang M, Hausmann S, Shuman S, Gnatt AL, Fu J 2005. Fcp1 directly recognizes the C-terminal domain (CTD) and interacts with a site on RNA polymerase II distinct from the CTD. Proc Natl Acad Sci 102: 17314–17319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup JQ 2007. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem Sci 32: 165–171 [DOI] [PubMed] [Google Scholar]
- Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim SJ, Natarajan K, Yoon S, Hinnebusch AG 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol Cell Biol 23: 2800–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar RS, Zheng S, Brunke-Reese D, Wolcott HN, Reese JC 2008. Yeast Rap1 contributes to genomic integrity by activating DNA damage repair genes. EMBO J 27: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Ujvari A, Luse DS 2006. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat Struct Mol Biol 13: 49–54 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Reese JC 2005. Molecular genetic analysis of the yeast repressor Rfx1/Crt1 reveals a novel two-step regulatory mechanism. Mol Cell Biol 25: 7399–7411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Fu J, Gilmour DS 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev 19: 1572–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]