Abstract
Endogenous retroviruses (ERVs) constitute a substantial portion of mammalian genomes, and their retrotransposition activity helped to drive genetic variation, yet their expression is tightly regulated to prevent unchecked amplification. We generated a series of mouse mutants and embryonic stem (ES) cell lines carrying “deletable” and “rescuable” alleles of the lysine-specific demethylase LSD1/KDM1A. In the absence of KDM1A, the murine endogenous retrovirus MuERV-L/MERVL becomes overexpressed and embryonic development arrests at gastrulation. A number of cellular genes normally restricted to the zygotic genome activation (ZGA) period also become up-regulated in Kdm1a mutants. Strikingly, many of these cellular genes are flanked by MERVL sequences or have cryptic LTRs as promoters that are targets of KDM1A repression. Using genome-wide epigenetic profiling of Kdm1a mutant ES cells, we demonstrate that this subset of ZGA genes and MERVL elements displays increased methylation of histone H3K4, increased acetylation of H3K27, and decreased methylation of H3K9. As a consequence, Kdm1a mutant ES cells exhibit an unusual propensity to generate extraembryonic tissues. Our findings suggest that ancient retroviral insertions were used to co-opt regulatory sequences targeted by KDM1A for epigenetic silencing of cell fate genes during early mammalian embryonic development.
Keywords: lysine-specific demethylase 1 (LSD1/KDM1A), murine endogenous retrovirus MuERV-L/MERVL, chromatin, embryonic stem (ES) cell fate, zygotic genome activation (ZGA), gene regulation
Retrotransposable elements (REs) and their remnants account for ∼40% of mammalian genomes (Waterston et al. 2002). REs include members of the long terminal repeat (LTR)-containing endogenous retroviruses (ERVs), and the non-LTR-containing retrotransposons (LINE-1 and SINE-1). The expression of these elements is tightly regulated during embryonic development to balance the potentially damaging effects of widespread retrotransposition against the benefits of promoting genetic diversity and thus gene and regulatory innovation. The long-term silencing of REs has been attributed to DNA CpG methylation (Reik 2007). However, additional repressive machinery likely acts to silence retrotransposition during reprogramming in the germline and early embryonic development, since DNA is largely demethylated during these key time windows (Morgan et al. 2005; Golding et al. 2010; Popp et al. 2010).
Silencing of REs can also be controlled by the state of histone methylation. Recruitment of histone H3K9 methyltransferase ESET by the corepressor KAP1 is responsible for silencing intracisternal A particles (IAPs) (Matsui et al. 2010; Rowe et al. 2010). Histone methylation, however, can also contribute to gene activation. For example, trimethylation of histone H3K4 is associated with active or poised promoters (Bernstein et al. 2006), while monomethylation is associated with enhancers (Heintzman et al. 2007, 2009). The methylation of histone tails is regulated by not only the histone methyltransferases (HMTs) that catalyze the addition of methyl groups to lysine residues, but also the lysine demethylases (KDMs) that remove methyl groups. The first KDM identified was the lysine-specific demethylase 1 LSD1/KDM1A, which demethylates histone H3 mono- and dimethyl K4 (Shi et al. 2004). The demethylating activity of KDM1A can either enhance or repress gene expression, depending on the affected histone residue. KDM1A had been characterized previously as a component of the HDAC1/2-containing CoREST complex that mediates the repression of REST target genes outside of the nervous system (Andres et al. 1999; You et al. 2001). Delivery of siRNA against Kdm1a in cell lines leads to up-regulation of target genes with a concomitant increase in H3K4 methylation at their promoters (Shi et al. 2004). In contrast, KDM1A mediates activation of the androgen and estrogen receptors via demethylation of H3K9 (Metzger et al. 2005; Garcia-Bassets et al. 2007; Wang et al. 2007; Hu et al. 2008). Because of the divergent activities attributed to KDM1A, a genome-wide analysis of chromatin modifications in KDM1A mutants would help to clarify its function.
KDM1A is a highly conserved protein from fission yeast to humans. Genetic deletion of Kdm1a orthologs in model organisms leads to a range of phenotypes, including slow growth and sterility in fission yeast, progressive sterility in worms, and sterility and other developmental defects in flies (Nottke et al. 2009). Embryonic stem (ES) cells lacking KDM1A derived by gene targeting were reported to display hypomethylation of DNA, which was attributed to low levels of the maintenance DNA methyltransferase DNMT1 (Wang et al. 2009). Yet the embryonic lethality observed in Kdm1a mutants is unlikely to be caused solely by DNMT1 loss and hypomethylation of DNA, as Dnmt1 mutant embryos survive to midgestation, whereas Kdm1a mutants die earlier at gastrulation (Li et al. 1992; Wang et al. 2007, 2009). Thus, the basis for KDM1A's critical requirement in early mammalian development is unclear.
In this study, we isolated three different types of homozygous ES lines from blastocysts for functional studies of Kdm1a: (1) a conditional “rescuable” gene trap allele, (2) a “deletable” floxed allele with an inducible Cre, and (3) a null allele. Unlike previously reported Kdm1a mutant ES lines generated by two rounds of gene targeting (Wang et al. 2009), our ES lines isolated from embryos displayed normal levels of DNA methylation. Using high-throughput mRNA sequencing and microarray analyses, we discovered that MERVL retroviruses become expressed in Kdm1a mutant ES cells and blastocysts. A small group of cellular genes were also highly derepressed in Kdm1a mutant ES lines. Strikingly, the majority of these up-regulated genes had remnants of LTRs within 2 kb of their transcription initiation sites, and the LTRs were sufficient to confer KDM1A-dependent repression on reporter genes. Genome-wide epigenetic studies revealed multiple chromatin modifications associated with the derepression, including hypermethylation of histone H3K4, hyperacetylation of H3K27, and hypomethylation of H3K9. These data suggest that KDM1A plays an essential role in repressing gene expression in early development by opposing activating histone marks at LTR sequences flanking MERVL REs and cellular genes. Moreover, our findings indicate that endogenous retroviral LTR sequences have been adapted to establish repressive chromatin modifications at the promoters of genes normally silenced in early embryos.
Results
Kdm1a mutant ES cells express DNMT1 and display normal global DNA methylation
To examine KDM1A's function, we characterized several mouse lines with different types of Kdm1a mutations, including a “Cre-rescuable” gene trap mutation (GT) (Supplemental Fig. S1A), a floxed allele (FL) (Wang et al. 2007), and a null allele (KO) (Wang et al. 2007). Intercrosses between heterozygous mice with the wild-type/GT mutation and the wild-type/KO mutation revealed that the GT/GT, GT/KO, and KO/KO embryos failed to develop properly due to gastrulation defects (Supplemental Fig. 1B; Supplemental Table 1; Wang et al. 2007, 2009). Thus, the GT and KO mutations are genetic alleles. The GT insertion construct was designed with LOX sites flanking a splice acceptor that disrupts Kdm1a RNA splicing so that Cre can be used to rescue KDM1A expression, generating the GTrescue allele. We intercrossed GT mice with Sox2::Cre animals to produce KO/GTrescue embryos and found that normal development was restored (Supplemental Table 1). Furthermore, mating Kdm1a +/KO Sox2::Cre males with Kdm1a FL/FL females resulted in no viable Kdm1a KO/KO mice, indicating that KDM1A is required in the inner cell mass derivatives where Sox2 is expressed (Supplemental Table 1). Despite the post-implantation lethality of embryos lacking KDM1A, homozygous ES clones were derived for all of the Kdm1a mutations.
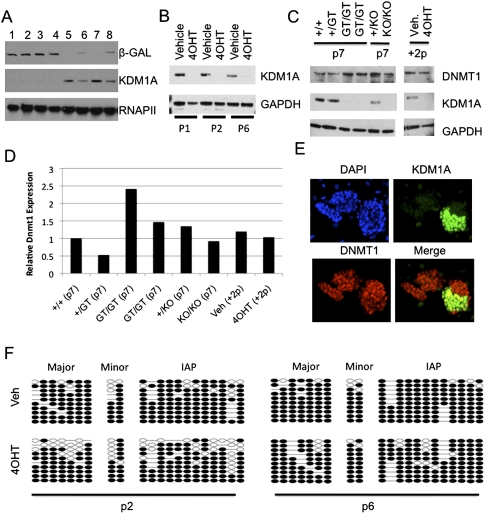
Kdm1a GT/GT ES cells lacked detectable KDM1A protein, as expected, but had normal colony morphology and proliferation rates and expressed normal levels of the pluripotency marker Oct3/4 (Fig. 1A; Supplemental Fig. 1C). It has been reported that ES cells derived by successive rounds of Kdm1a gene targeting progressively lose global DNA methylation, which was attributed to destabilization of the DNMT1 protein (Wang et al. 2009). However, acute deletion of Kdm1a in ES cells is reported to cause DNA hypomethylation only after 25 d (Foster et al. 2010). We found that DNMT1 levels were not significantly reduced in our Kdm1a GT/GT and KO/KO ES cell lines through at least seven passages (Fig. 1C,D). Bisulfite sequencing and methylated DNA immunoprecipitation (MeDIP) analyses coupled with whole-genome promoter arrays confirmed that Kdm1a GT/GT and KO/KO ES cells retain normal patterns of DNA methylation at gene promoters and repetitive sequences (including major satellite, minor satellite, and IAPs) (Supplemental Fig. 1D,E,G). Methylation levels of IAP elements and major satellite sequences were also normal in Kdm1a GT/GT blastocysts (Supplemental Fig. 1F). To ensure that DNMT1 levels had not been restored by secondary compensatory changes during our ES cell isolation, we used Kdm1a FL/FL ES cells containing a 4-hydroxytamoxifen (4OHT)-inducible Cre-ERT to acutely delete Kdm1a and generate FLrec/FLrec cells. Treatment with 4OHT lead to a rapid and stable loss of KDM1A protein (Fig. 1B), but DNMT1 levels remained constant (Fig. 1 C,D). As an internal control for monitoring DNMT1 levels with immunofluorescence, we transfected Kdm1a FL/FL ES cells with Cre recombinase to generate a mixed cell population. KDM1A protein disappeared from the transfected cells within 72 h, but both KDM1A-positive and KDM1A-negative colonies expressed DNMT1 (Fig. 1E). Consistent with the maintained expression of DNMT1 in these Kdm1a mutant cells, global DNA methylation remained unchanged following the acute deletion of Kdm1a from FLrec/FLrec ES cells (Fig. 1F). Taken together, our findings indicate that KDM1A function can be dissociated from the maintenance of DNMT1 levels and global DNA methylation with our cell derivation procedure.
Figure 1.
Kdm1a mutant ES cells have normal levels of DNMT1 and DNA methylation. (A) Immunoblot analysis using the indicated antibodies was performed on Kdm1a GT/GT (lanes 1–4), +/GT (lanes 6,8), or +/+ (lanes 5,7) ES cells. (B) Immunoblot analysis of Kdm1a FL/FL Cre-ERT ES cells treated with vehicle or 4OHT and passaged once (p1), twice (p2), or six times (p6) after treatment. (C) Immunoblot analysis of ES lines containing the GT and KO alleles at the indicated passage, or a Kdm1a FL/FL Cre-ERT ES line treated with vehicle (Veh) or 4OHT after two passages. (D) Relative DNMT1 levels from C were quantified using ImageJ software. (E) Kdm1a Fl/Fl ES cells were transfected with Cre recombinase. Seventy hours after transfection, cells were immunostained with the indicated antibodies and overlaid with DAPI. (F) Bisulfite sequencing analysis of the indicated repetitive elements was performed on Kdm1a FL/FL Cre-ERT ES cells treated with vehicle or 4OHT and passaged two (p2) or six (p6) times.
LTR promoters become activated in Kdm1a mutant ES cells
To examine how KDM1A influences gene expression in ES cells, we performed microarray analysis on Kdm1a mutant ES lines. Four independently derived Kdm1a GT/GT ES lines were compared with wild-type and +/GT littermate-derived ES lines. To distinguish chronic from acute changes in gene expression, we also analyzed mRNA from three Kdm1a FL/FL ES lines transfected with either GFP or Cre-GFP after FACS enrichment. We found a total of 208 and 745 probe sets that were differentially expressed in Kdm1a GT/GT and FLrec/FLrec ES cells, respectively (Supplemental Table 2). The GT/GT data set has increased statistical power, but is also susceptible to detecting secondary (and, perhaps, compensatory) gene expression changes that arise from the prolonged lack of KDM1A. We therefore focused on the 46 genes that were also misregulated upon acute deletion of Kdm1a (Supplemental Table 3). Forty-three of the genes were up-regulated and only three were down-regulated in Kdm1a mutants, one of which was Kdm1a. Quantitative RT–PCR (qRT–PCR) was used to confirm that Tcstv1, Tcstv3, Similar to Tho4, Eif1a (Gm2022), and Zscan4 were up-regulated in all four Kdm1a GT/GT lines as well as the three FLrec/FLrec lines (Supplemental Fig. 2A,B; data not shown). To establish that the up-regulation of these genes was caused by a lack of KDM1A rather than secondary phenomena, we tested whether restoring KDM1A expression to mutant ES cells reduced the levels of the overexpressed genes. Cre-mediated rescue of the GT allele partially restored KDM1A expression (Supplemental Fig. 2D), and likewise partially repressed gene expression of the putative target genes (Supplemental Fig. 2C). The inability to completely restore high KDM1A protein levels in the GTrescue allele is likely due to the βgeo insertion remaining after cre-mediated excision of the splice acceptor (Supplemental Fig. 1A).
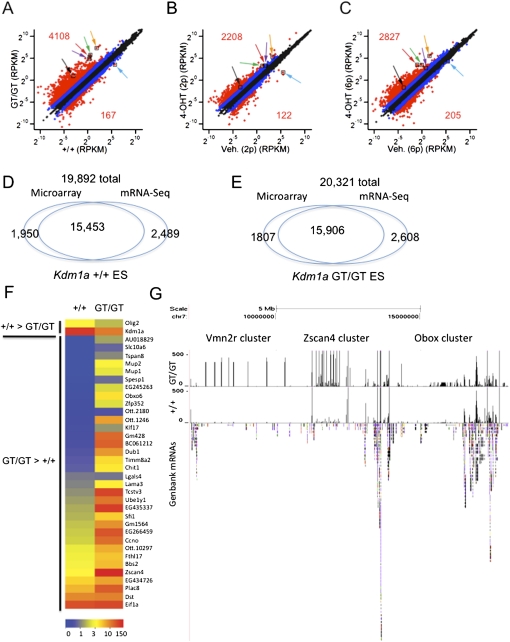
To confirm these findings and better quantify the transcriptome of Kdm1a mutant ES cells, we performed mRNA-seq analysis on polyA-enriched mRNAs from (1) Kdm1a GT/GT ES cells, (2) wild-type ES cells, and (3) Kdm1a FL/FL; Cre-ERT ES cells treated with vehicle or 4OHT for one and six passages. We achieved >16 million 43-base-pair (bp) reads per sample, with at least 86.6% of the reads aligning to the mouse genome. We plotted the expression of all UCSC (University of California at Santa Cruz) known genes in reads per kilobase of exon model per million mapped reads (RPKM) (Fig. 2A–C). The majority of genes maintained expression within twofold in cells either chronically (GT/GT) or acutely (FLrec/FLrec) lacking KDM1A, but a subset of genes were markedly up-regulated in the mutants (Fig. 2A–C). Furthermore, the fold changes in expression were larger in cells that lacked KDM1A over longer periods, consistent with progressive changes in transcription regulation. There was a 78% overlap in transcript detection between microarray and mRNA-seq methods in wild-type and Kdm1a mutant ES cells (Fig. 2D,E). Furthermore, the misexpressed transcripts identified using microarrays were also detected with mRNA-seq (Fig. 2F).
Figure 2.
Gene expression profiling of Kdm1a mutant ES cells. (A–C) mRNA-seq was performed on polyA-enriched mRNAs from Kdm1a +/+ and GT/GT ES cells (A) or Kdm1a FL/FL Cre-ERT ES cells treated with vehicle or 4OHT and passaged two (B) or six (C) additional times. Each plot displays the expression value of UCSC known genes passing the minimal expression filter in wild-type (X-axis) and corresponding mutant ES (Y-axis) cells. Data points in red indicate a greater than fourfold difference in the mutants (numbers indicate the total number of genes with a greater than fourfold change), while those in blue indicate a greater than twofold difference. Colored arrows label the following genes: Tcstv1 (green), Tcstv3 (red), Zscan4d (orange), Zfp352 (black), Eif1a (purple), and Kdm1a (blue). (D,E) Comparison of the number of UCSC known transcripts detected by Affymetrix microarrays (using absent and present calls) or mRNA-seq (using a minimum threshold of RPKM > 1) in Kdm1a +/+ (D) or GT/GT (E) ES cells. (F) Expression levels of genes (in RPKM) in Kdm1a +/+ and GT/GT ES cells. Genes displayed were those identified as misexpressed by microarray analysis in Kdm1a mutant ES cells. (G) UCSC genome browser screenshot of mRNA-seq reads aligning to chromosome 7 in wild-type and Kdm1a GT/GT ES cells. GenBank mRNAs are displayed below to highlight gene density. Positions of gene clusters displaying increased expression in Kdm1a mutant cells are indicated. Reads aligning with the Vmn2r cluster do not align to any known or predicted genes and might correspond to regulatory RNAs.
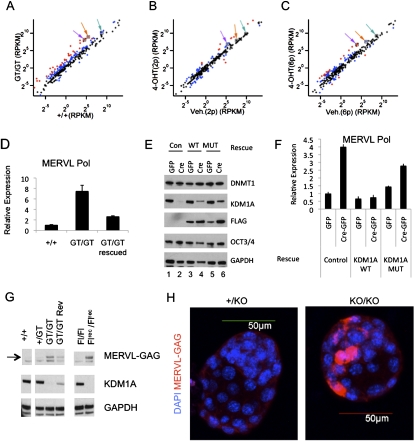
Intriguingly, when we examined the genomic organization of the derepressed genes, we found that the majority (28 of 43, 65%) overlapped segmental duplications (defined as sequences at least 1 kb in length with >90% similarity) (Supplemental Table 3). In addition, these derepressed genes were found in gene-poor regions of chromosomes and were significantly enriched with LTR-containing ERVs and LINE-1 retrotransposons (Fig. 2G; Supplemental Fig. 3A–E). To determine whether KDM1A might also regulate the expression of REs, we aligned mRNA-seq reads to the Repbase database of repetitive sequences. We found that MERVL retroviruses were up-regulated >10-fold in Kdm1a GT/GT ES cells, prompting us to further study its repression by KDM1A (Fig. 3A–C). Because there are ∼500 full-length and nearly 350 protein-coding MERVL elements (Ribet et al. 2008), we tested whether activation of these elements was restricted to a small subset or represented a broader population. We found many small nucleotide polymorphisms (SNPs) in the expressed MERVL elements that were increased in Kdm1a mutant cells, indicating that a large number of MERVL elements become activated (Supplemental Fig. 3F). The increase in MERVL expression was partially reversed in GTrescue/GTrescue cells (Fig. 3D) and completely reversed by rescuing with human wild-type KDM1A, but not a catalytic point mutant (Fig. 3E,F). Importantly we also detected high levels of MERVL Gag proteins in Kdm1a mutant ES cells, but only trace levels in wild-type ES and GTrescue/GTrescue cells (Fig. 3G). To exclude the possibility that this an epiphenomenon restricted to ES cells, we examined whether MERVL misregulation also occurred in mutant embryos. Using immunofluorescence microscopy, we found a marked increase in MERVL Gag expression in Kdm1a mutant blastocysts (Fig. 3H). Thus, Kdm1a is critical for silencing of MERVL retroviruses in preimplantation development. The inappropriate activation of latent MERVL viruses precedes the earliest known phenotypic abnormalities in Kdm1a mutants.
Figure 3.
KDM1A represses MERVL retroviruses in ES cells and preimplantation development. (A–C) mRNA-seq was performed on polyA-enriched mRNAs from Kdm1a +/+ and GT/GT ES cells (A), or Kdm1a FL/FL Cre-ERT ES cells treated with vehicle or 4OHT and passaged two (B) or six (C) additional times. Each plot displays the expression value (in RPKM) of repetitive sequences in wild-type (x-axis) and mutant (Y-axis) ES cells. Data points in red indicate a greater than fourfold difference in the mutants, while those in blue indicate a greater than twofold difference. Colored arrows label the following: MERVL (purple), L1- (orange), and SINE-B2 (green). (D) qRT–PCR analysis was performed on Kdm1a +/+, GT/GT, and GT/GT rescued clones with MERVL Pol-specific primers and plotted relative to Gapdh. (E,F) Kdm1a FL/FL ES cells stably expressing a neo resistance control plasmid (Con; lanes 1,2), Flag-hKDM1A (WT; lanes 3,4), or catalytically inactive Flag-hKDM1A (MUT; lanes 5,6) were transfected with either GFP alone or Cre-GFP as indicated to remove the endogenous mouse Kdm1a alleles. GFP+ cells were collected 24 h later and plated for an additional 48 h before being subjected to immunoblot analysis with the indicated antibody (E) or qRT–PCR with MERVL primers and plotted relative to Gapdh (F). Error bars represent SD. (G) Whole-cell extracts from ES cells of the indicated genotype were subjected to immunoblots with the indicated antibodies. (H) Immunofluorescence microscopy using anti-MERVL Gag antibodies (red) was performed on Kdm1a +/KO or KO/KO blastocysts and overlaid with DAPI (blue).
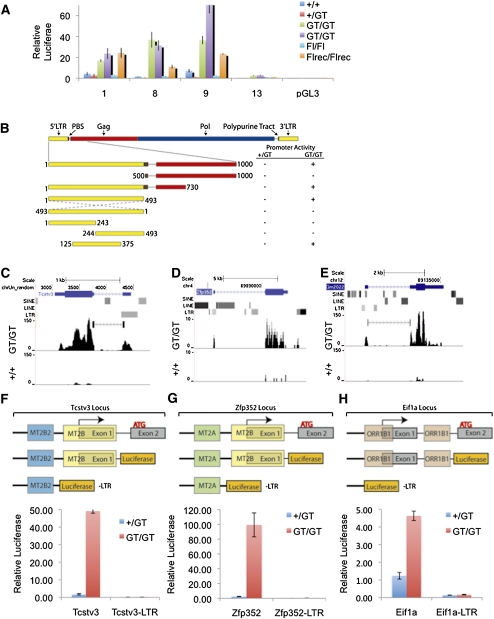
To test whether MERVL derepression in Kdm1a mutant ES cells was due to improper activation of its retroviral LTR, we cloned ∼1 kb of sequence encompassing the 5′ LTR and a portion of the Gag sequence from four unique MERVL retroviruses (aligned in Supplemental Fig. 4A). These sequences were inserted upstream of a promoterless luciferase reporter and transfected into Kdm1a wild-type and mutant ES lines. Although activity was very low in wild-type ES cells, all four MERVL reporters were highly up-regulated in Kdm1a mutant ES cells (Fig. 4A). Importantly, we found that neither the Gag- nor the primer-binding site (PBS) of the MERVL virus was required for this activity (Fig. 4B, Supplemental Fig. 4B). In contrast, the central portion of the 5′ LTR containing a direct repeat and a TATA box was essential for activity in Kdm1a mutant ES cells (Fig. 4B; Supplemental Fig. 4B). As expected, inversion of the LTR completely blocked activity of the reporter (Fig. 4B;Supplemental Fig. 4B). In sum, these data suggest that the MERVL LTR is serving as a bona fide promoter that is repressed by KDM1A in wild-type ES cells.
Figure 4.
Endogenous retroviral LTRs are activated in Kdm1a mutant ES cells. (A) Four unique MERVL LTR-Gag reporter luciferase plasmids (1, 8, 9, and 13) were cotransfected with CMV βgal into Kdm1a wild-type or mutant ES cells as indicated, and relative luciferase levels were measured 24 h later. Error bars represent SEM. (B) Schematic of MERVL retrovirus and truncation mutants generated and tested for promoter activity in Kdm1a wild-type and mutant cells. Luciferase activity (at least 20-fold over background) is indicated with a plus sign (+). (C–E) Raw mRNA-seq reads from wild-type (+/+) and Kdm1a GT/GT ES cells were aligned to the mouse genome and displayed on the UCSC genome browser at the Tcstv3 (C), Zfp352 (D), and Eif1a (Gm2022) (E) genes with RepeatMasker function enabled to visualize the position of LTR elements. (C,E) Detected junctions are also displayed for Tcstv3 and Gm2022. (F–H) The promoter-proximal sequences of Tcstv3 (F), Zfp352 (G), and Eif1a (Gm2022) (H), as well as deletion constructs (−LTR), as schematized above, were cloned upstream of a promoterless luciferase reporter gene and cotransfected with a CMV βgal reporter into Kdm1a +/GT or GT/GT ES cells. Relative luciferase activity was measured 24 h later and plotted, with error bars representing SEM.
We next examined the promoters of the KDM1A-repressed genes more closely and found that more than half (23 of 43) contained remnants of retroviral LTRs within 2 kb of their transcription start sites (Supplemental Table 3). Strikingly, several of the most highly up-regulated genes in Kdm1a mutant ES cells had transcripts initiating from within the cryptic LTRs—including Tcstv1, Tcstv3, BC099439, AU018829, AF067061, LOC639910, Ubtfl1, Chit1, Eif1a (Gm2022), and Zfp352—indicating that the LTRs were serving as promoters (Fig. 4C–E; data not shown). To determine if the promoter-proximal sequences were sufficient to confer Kdm1a-dependent repression on reporters, we cloned ∼1 kb of the promoter sequences of Tcstv3, Zfp352, and Eif1a (Gm2022), containing retroviral LTRs of the MERVL-related MT2B/C and ORR1B1 elements, respectively. These promoters were linked to luciferase and transfected into Kdm1a wild-type and mutant ES cells. We found that the reporter genes had low basal activity in wild-type ES cells, but were highly up-regulated in Kdm1a mutant ES cells (Fig. 4F–H). Importantly, deletion of regions encompassing the LTRs in these promoter fragments completely blocked activation of the reporters in Kdm1a mutant ES cells (Fig. 4F–H). Thus, retroviral LTRs that have been co-opted by cellular genes as promoters are repressed in a KDM1A-dependent manner in ES cells.
Kdm1a mutant ES cells display abnormal histone H3 modifications at LTR sequences
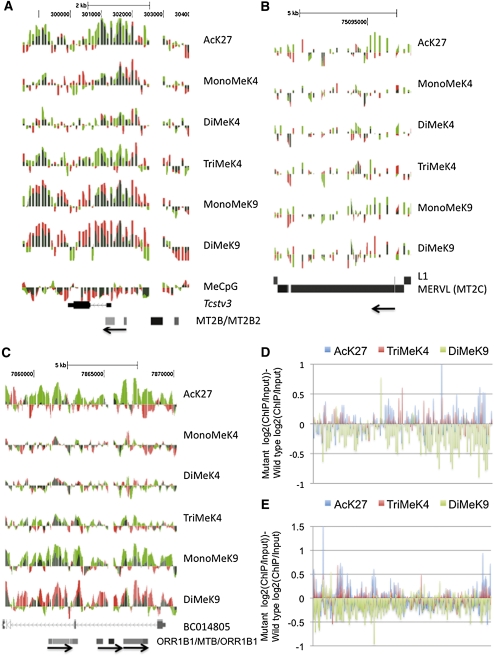
Despite using antibodies that immunoprecipitate KDM1A protein with high affinity (Fig. 6B, below), we were unable to successfully perform chromatin immunoprecipitation (ChIP) to identify KDM1A target sequences genome-wide. Since KDM1A is a demethyalting enzyme, its residence at target genes may be short-lived. To indirectly test whether KDM1A functions as a histone demethylase at its putative target genes in ES cells, we performed genome-wide ChIP and MeDIP analysis. We used a high-density tiling array encompassing portions of chromosomes 18, 19, X, and Y for ChIP, and a whole-genome promoter array for MeDIP. In Kdm1a GT/GT ES cells, we found an increase in active histone marks, including methylation of histone H3K4 and acetylation of H3K27 near the promoter/LTR sites of the Tcstv3, AF067061, and AF067063 genes (Fig. 5A; Supplemental Fig. 5A,B), as well as a cluster of highly up-regulated genes on chromosome 18 (Supplemental Fig. 6). Increased acetylation and methylation of histone H3K27 and H3K4 were also observed on MERVL retroviruses and ORR1B1 elements (Fig. 5B–E; Supplemental Fig. 5C,D). These retroviral-derived sequences also displayed reduced dimethylation of histone H3K9 (Fig. 5D,E), but had no detectable change in DNA methylation (Supplemental Fig. 7A–D). These data indicate that KDM1A is required to maintain chromatin modifications associated with gene repression, including hypomethylation of histone H3K4, hypoacetylation of histone H3K27, and dimethylation of histone H3K9 at specific genes and ERVs in ES cells.
Figure 6.
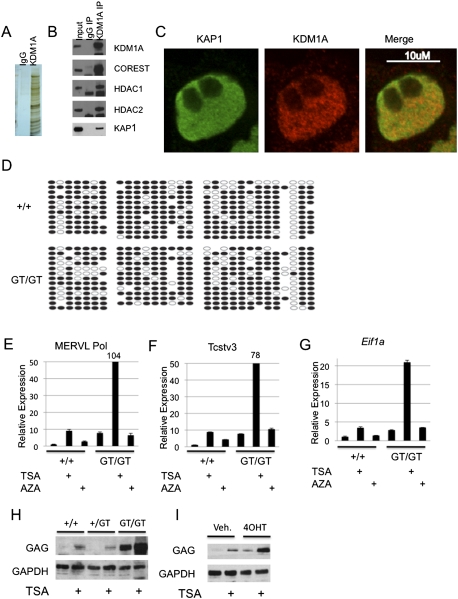
KDM1A, HDACs, and KAP1 cooperate to repress endogenous retroviral sequences in ES cells. (A,B) KDM1A complexes were affinity-purified from ES cells and subjected to LDS-PAGE and silver staining (A) or immunoblotting (B) with the indicated antibodies. (C) Wild-type ES cells were immunostained with KDM1A and KAP1 antibodies and subjected to confocal microscopy. (D) Bisulfite sequencing was performed on Kdm1a +/+ or GT/GT ES cells with primers amplifying the MERVL retrovirus. (E–G) Kdm1a +/+ or GT/GT ES cells were treated with TSA or 5Aza for 24 h. qRT–PCR was performed using primers specific for MERVL Pol (E), Tcstv3 (F), or Eif1a (Gm2022) (G) and normalized to Gapdh. The fold change was then plotted relative to untreated Kdm1a +/+ ES cells, with error bars representing SD. (H) Kdm1a +/+, +/GT, or GT/GT ES cells were treated with TSA for 24 h, and cells were subjected to immunoblotting with the indicated antibodies. (I) Kdm1a FL/FL Cre-ERT ES cells were treated with vehicle or 4OHT for 48 h, followed by treatment with TSA for 24 h. Immunoblotting was then performed with the indicated antibodies.
Figure 5.
Kdm1a mutant ES cells display histone modifications associated with transcription activation at target genes. (A–C) ChIP-on-chip analysis was performed on Kdm1a +/+ or Kdm1a GT/GT ES cells with histone modification-specific antibodies or methylated CpG antibodies (shown on the right). The normalized log2 intensity ratios were displayed as individual tracks on the UCSC genome browser for Kdm1a +/+ (red) and Kdm1a GT/GT (green) ES cells, and the tracks were overlaid. Red and green overlapping tracks appear dark green. Tracks from the Tcstv3 gene (A), a MERVL retrovirus (B), and a cluster of ORR1B1 and MTB LTRs (C) are shown. The direction of the LTRs (5′–3′) is indicated with an arrow. (D,E) The normalized log2 ChIP/Input intensity values (for AcK27, TriMeK4, and DiMeK9) from Kdm1a +/+ ES cells were subtracted from the corresponding log2 ChIP/Input values from Kdm1a GT/GT ES cells. The subtracted values for 108 MERVL elements (D) and 307 ORR1B1 elements (E) tiled on chromosome 19 are displayed.
Endogenous retroviral activation is constrained by KDM1A, HDACs, and KAP1 independently of DNA methylation
To provide more mechanistic insight into the Kdm1a-dependent repression of retroviral LTRs, we immunopurified KDM1A complexes and identified interacting proteins by mass spectrometry. We detected numerous proteins that coprecipitated specifically with KDM1A (Fig. 6A), including members of the CoREST complex (RCOR1/2, HDAC1, HDAC2, ZMYM2, PHF21a, HMG20B, and ZNF217), as well as the COREST-associated proteins CTBP1/2 (Fig. 6B; data not shown). An additional protein found at substoichiometric levels in successive affinity purifications was the tripartite motif-containing protein Trim28/KAP1, recently identified as a critical corepressor of ERVs (Rowe et al. 2010). The interaction of KAP1 and KDM1A was confirmed by immunoblotting with KAP1 antibodies (Fig. 6B). Immunofluorescence microscopy demonstrated that the proteins colocalized in nuclei, with a mixture of overlapping and nonoverlapping domains (Fig. 6C). Thus, it appears that KDM1A and KAP1 complexes are heterogeneous.
It has been proposed recently that silencing of ERVs in preimplantation development and ES cells involves a DNA methylation-independent pathway mediated by the KAP1 corepressor and the HMT ESET (Matsui et al. 2010; Rowe et al. 2010). Using bisulfite sequencing and combined bisulfite and restriction analysis (COBRA) of Kdm1a mutant ES cells, we confirmed that activation of MERVL retroviruses and Zfp352 occurs despite high levels of methylated DNA (Fig. 6D; Supplemental Fig. 8A). We next tested the effect of the HDAC inhibitor trichostatin A (TSA) and the DNA demethylating agent 5-Azacytidine (5Aza) on MERVL expression. TSA treatment caused a significant increase in MERVL mRNA and protein expression in wild-type ES cells and was synergistic with KDM1A loss of function, suggesting HDACs function cooperatively with KDM1A (Fig. 6E,H,I). In contrast, 5Aza had little effect on MERVL expression, whereas the positive control, IAP, was activated (Fig. 6E; Supplemental Fig. 8B). This suggests that DNA methylation is not a critical negative regulator of MERVL expression in ES cells. The Kdm1a-regulated genes Tcstv3, Eif1a-like, and Similar to tho4 were also sensitive to TSA but not 5Aza treatment, and TSA was synergistic with Kdm1a loss of function (Fig. 6F,G; Supplemental Fig. 8C). Thus, KDM1A and HDACs cooperate to repress LTR-regulated cellular genes and MERVL retroviruses in ES cells, whereas DNA methylation is dispensable.
Kdm1a mutant ES cells generate high levels of extraembryonic tissues
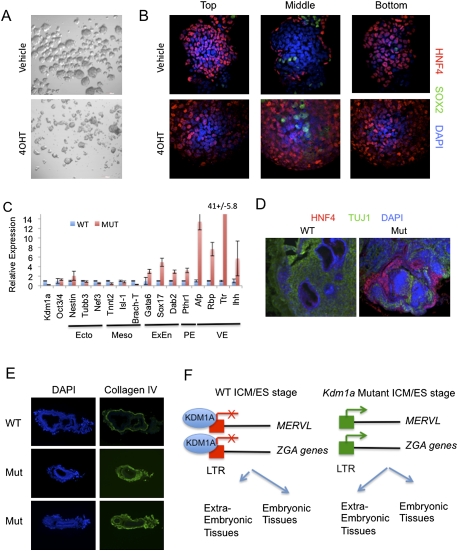
Intriguingly, a number of the KDM1A-repressed genes that we identified are transiently expressed during zygotic genome activation (ZGA) shortly after egg fertilization. These include Zscan4, Tcstv1/3, Eif1a (Gm2022), Similar to Tho4, and Zfp352 (Kigami et al. 2003; Liu et al. 2003; Svoboda et al. 2004; Zhang et al. 2006; Ribet et al. 2008). Each of these genes either uses an LTR as a promoter or contains a MERVL element within 5 kb of its transcriptional start site. We thus refer to this subset of cellular developmental genes as LTR-linked ZGA genes. At the two/four-cell stage of development, each blastomere is capable of generating both embryonic and extraembryonic derivatives, unlike the Oct4+ cells of the inner cell mass, which are restricted to generating only fetal tissues. ES cells are derived from the ICM; consequently, their fate is likewise restricted to the embryonic lineages. Since Kdm1a mutant ES cells expressed ZGA genes indicative of an earlier blastomere fate, we tested if Kdm1a mutant ES cells had expanded fate potential. We found that both wild-type and Kdm1a mutant ES cells formed embryoid bodies when cultured without leukemia inhibitory factor (LIF) (Fig. 7A). Both wild-type and Kdm1a mutant ES cells lost expression of the pluripotency marker Oct3/4 after several days of differentiation (Supplemental Fig. 9A) and showed a similar increase in the embryonic mesendoderm marker Brach-T, which differs from a report from Foster et al. (2010) (Supplemental Fig. 9B). Strikingly, Kdm1a mutant embryoid bodies expressed significantly increased levels of genes characteristic of extraembryonic endoderm (Gata4, Gata6, and Dab2), visceral endoderm (Hnf4, Afp, Ttr, Ihh, Rbp, ApoE, Foxa2, and Msg), and parietal endoderm (Sparc, Sna, and Pthr1), and reduced expression of the neural ectoderm markers Sox1, Nestin, Pax6, and Tubb3 (Supplemental Fig. 9A–K; data not shown). In addition, Kdm1a-deficient ES cells expressed higher levels of the trophoblast stem cell markers Cdx2 and Hand1 (Supplemental Fig. 9L). After 8 d in culture, Kdm1a mutants had significantly increased numbers of HNF4+ visceral endoderm-like cells that were unusually located throughout the embryoid body (Fig. 7B). Despite the increased potential to generate extraembryonic tissues, Kdm1a mutant ES cells retained the ability to generate beating cardiomyocytes and HB9/TUJ1-positive motor neurons (Supplemental Fig. 9M,N). Thus Kdm1a mutant ES cells acquire an expanded fate potential, rather than become restricted to producing a particular cell lineage.
Figure 7.
Kdm1a mutant ES cells have increased potency to generate extraembryonic lineages. (A,B) Kdm1a FL/FL Cre-ERT ES cells were treated with 4OHT and maintained for 48 h before growing in bacterial-grade dishes in the absence of LIF to induce differentiation. (A) After 48 h, embryoid bodies were imaged by phase-contrast microscopy. After 8 d of differentiation, embryoid bodies were immunostained with HNF4 and SOX2 antibodies and overlaid with DAPI. Optical confocal sections were then taken through the entire embryoid body. (C) qRT–PCR analysis with the indicated primers was performed on tumors derived from Kmd1a Fl/FL CreERT ES cells treated with vehicle (WT) or 4OHT (Mut). Error bars represent SD. (Ecto) Ectoderm; (Meso) mesoderm; (ExEn) extraembryonic endoderm; (PE) parietal endoderm; (VE) visceral endoderm. (D) Tumors derived from Kdm1a wild-type (WT) and mutant (Mut) ES cells were immunostained with the VE marker HNF4 and the neural marker TUJ1 and counterstained with DAPI. (E) Cryosections from Kdm1a wild-type (WT) and KO/KO (Mut) embryos at embryonic day 6.5 were immunostained with anti-collagen IV antibodies and counterstained with DAPI. (F) Model of KDM1A function in the coordinated repression of MERVL and extraembryonic cell fate potential. In ES cells derived from the ICM, MERVL retroviruses and a subset of ZGA genes that are LTR-linked are repressed by KDM1A, and cell fate is restricted to embryonic lineages. In Kdm1a mutants, MERVL and LTR-linked ZGA genes are improperly activated, resulting in an expanded fate potential to generate extraembryonic tissues.
To test the differentiation potential of Kdm1a mutant ES cells in vivo, we generated teratomas in nude mice. Kdm1a mutant teratomas expressed significantly higher levels of extraembryonic/parietal/visceral endoderm markers, including Gata6, Sox17, Dab2, Pthr1, Afp, Rbp, Ttr, and Ihh (Fig. 7C). Furthermore, in mutant teratomas, we detected an epithelial layer that was positive for the visceral endoderm marker HNF4 that was not detected in wild-type teratomas (Fig. 7D). To demonstrate that this expanded fate potential was not only a phenomenon unique to ES cells, we also examined Kdm1a mutant embryos at gastrulation stages. We found that Kdm1a mutant embryos produced significantly greater levels of basement membrane that stained positive for collagen IV (Fig. 7E). This marker is produced by the parietal endoderm (an extraembryonic endodermal tissue), suggesting an expansion of this lineage in Kdm1a mutants. Taken together, these data indicate that the misregulation of LTR-linked ZGA genes in Kdm1a mutants has important functional consequences for the allocation of cells to particular lineages.
Discussion
In this study, we explored the function of KDM1A in early development by isolating ES cell lines containing conditionally deletable and rescuable alleles of Kdm1a. We discovered that KDM1A is required to silence ERVs and regulate histone methylation and acetylation at LTR sequences. Interestingly, we found that a number of cellular genes expressed during ZGA, but then silenced by the blastocyst stage, have promoters derived from cryptic LTRs. These LTR-linked ZGA genes become derepressed in Kdm1A mutant cells, which coincides with an expanded cell fate potential. We propose a model whereby KDM1A recruitment to LTRs alters the chromatin around these sites, helps to guard the genome against excessive retroviral activity, and prevents embryonic cell lineages from inappropriately developing into extraembryonic tissues (Fig. 7F).
KDM1A regulation of transcription correlates with histone modifications
We performed a series of microarray and mRNA-seq analyses on wild-type and Kdm1a mutant ES lines isolated from blastocyst embryos and identified a set of genes that consistently became derepressed. Others have reported that KDM1A stabilizes DNMT1 and maintains global DNA methylation using mutant ES cells that had undergone double targeting, prompting us to test if this was the underlying basis for the changes in gene expression that we detected (Wang et al. 2009). However, we were unable to find a correlation between gene derepression and DNA hypomethylation. First, we detected DNMT1 in multiple Kdm1a mutant ES lines. Second, DNMT1 expression was maintained in cells chronically lacking KDM1A as well as cells following acute deletion of Kdm1a. Third, MeDIP, bisulfite sequencing, and COBRA failed to identify changes in DNA methylation in Kdm1a mutants. Fourth, we detected reproducible chromatin modifications at LTR sequences. Fifth, Dnmt1 mutant mice develop beyond gastrulation, when Kdm1a mutants die (Li et al. 1992; Wang et al. 2007), suggesting that the loss of DNMT1 is not the critical defect in Kdm1a mutant embryos. Although Kdm1a mutant ES culture conditions appear to favor DNMT1 down-regulation over extended passages (Wang et al. 2009; Foster et al. 2010), our studies find a strong correlation between KDM1A-dependent gene regulation and histone modifications rather than DNA methylation.
The intrinsic catalytic activity of KDM1A is specific for demethylating mono- and dimethyl lysine (Shi et al. 2004), yet we detected a wide variety of histone modification changes in Kdm1a mutant ES cells. For example, we found elevated levels of trimethyl H3K4 in Kdm1a mutants. Perhaps this occurs indirectly due to increased recruitment of RNA polymerase and its associated HMTs. Likewise, hyperacetylation of H3K27 is also detected in Kdm1a mutants. Since HDAC proteins copurify in KDM1A complexes, it is likely that an additional way KDM1A contributes to gene repression is recruitment of HDACs to LTR sites. This is consistent with our finding that HDAC inhibitors activate KDM1A target genes in ES cells, and with the findings that CoREST complexes lacking KDM1A or carrying catalytic mutants of KDM1A have reduced HDAC activity (Lee et al. 2006; Foster et al. 2010). Finally, it is unclear how H3K9 becomes demethylated in Kdm1a mutants, although it is likely due to the altered activity of repressive HMTs. Intriguingly, conditional deletion of the histone H3K9 methyltransferase G9A leads to up-regulation of genes that overlap with Kdm1a mutants (Yokochi et al. 2009). Thus, KDM1A and G9A activity may be linked.
Epigenetic regulation of ERVs
ERVs are expressed and epigenetically silenced in complex patterns during embryonic development. For example, MERVL expression peaks at the two/four-cell stage, whereas IAP expression is high in blastocyst embryos before being silenced during gastrulation (Svoboda et al. 2004). It is widely accepted that the long-term silencing of ERVs and retrotransposons is controlled by DNA methylation (Reik 2007). However it is also becoming clear that DNA methylation is not sufficient for silencing of ERVs in preimplantation development and mouse ES cells. Indeed IAPs are highly activated in ES cells lacking the HMT ESET and the corepressor KAP1. Intriguingly, this activation occurs despite the continued presence of DNA methylation, suggesting the ESET-mediated histone modifications are dominant (Matsui et al. 2010; Rowe et al. 2010). Furthermore, a recent genome-wide methylation analysis in human ES cells failed to detect a correlation between CpG methylation and gene expression (Lister et al. 2009). Likewise, we now demonstrate that MERVL is activated in Kdm1a mutants despite high levels of DNA methylation. These data suggest that DNA methylation is not sufficient for ERV silencing in ES cells, and that additional repressive machinery is required to establish silent chromatin domains. However, this machinery appears to be combinatorial in nature, as different classes of ERVs (like MERVL, IAP, and MusD) harbor distinct histone modifications and are differentially affected by KAP1, ESET, and KDM1A loss of function (Mikkelsen et al. 2007; Matsui et al. 2010; Rowe et al. 2010). Therefore mammalian embryos appear to have evolved a mixture of epigenetic pathways to protect their genomes from damage caused by overexpression or prolonged expression of these mutagenic agents.
Although we cannot rule out the possibility that KDM1A indirectly represses MERVL elements and LTR-linked ZGA genes via regulation of the expression or activity of another factor, we favor a model in which KDM1A is recruited along with KAP1 to their targets in ES cells to maintain chromatin in a repressed state via active histone H3K4 demethylation. This model best explains the altered histone modifications, the increased activity of LTR promoters, and the progressive changes in gene expression that occur upon Kdm1a deletion. Since KAP1 is a general corepressor for KRAB (Kruppel-associated box) zinc finger proteins and is recruited by the KRAB-containing Zfp809 to repress the murine leukemia virus in ES cells (Wolf and Goff 2007, 2009; Wolf et al. 2008), it is likely to be recruited to its target sites by yet-to-be identified KRAB-ZFPs that have undergone strong positive selection during evolution (Emerson and Thomas 2009). Interestingly, we observed ZNF217 and several novel ZFPs in KDM1A immunoprecipitates (TS Macfarlan and SL Pfaff, unpubl.). Thus, KAP1 and KDM1A's activity may be regulated in a developmental and gene-specific manner by the coordinated expression of zinc finger DNA-binding partners.
ERVs and mammalian extraembryonic tissue development
Early studies of genome structure quickly recognized that repetitive DNA is abundant, but it was unclear whether this represented parasitic nucleic acid or provided selective advantages for animals. It is now thought that ERVs helped to drive genome evolution in a variety of ways, such as facilitating recombination that causes gene deletions, duplications, and rearrangements, and providing regulatory sequences for cellular gene transcription (Goodier and Kazazian 2008; Cohen et al. 2009). In addition, the gag and envelope proteins encoded by ERVs appear to benefit the host by interfering with new viral infections as well as mediating cell fusions that establish the multinucleated syncytiotrophoblasts of the placenta (Benit et al. 1997; Jern and Coffin 2008).
ERVs are expressed at high levels in placental tissues, and for decades it has been considered likely that they contributed to the evolution of viviparity (live birth) (Black et al. 2010). In addition to co-opting envelope proteins for extraembryonic cell fusion, the placental gene CYP19A1 derived its promoter from the MER21A LTR, presumably because it is highly active in this tissue (Cohen et al. 2009). The cellular interactions that distinguish extraembryonic from embryonic cell fate occur very early in development. Shortly after the totipotent two/four-cell stage, the trophectoderm and embryonic lineages become specified. By the late blastocyst stage, the cells allocated to the inner cell mass are committed to produce the embryonic tissues but not extraembryonic endoderm and placenta. This distinction is preserved in vitro, as ES cells derived from the inner cell mass have a low capacity for generating extraembryonic tissue and normally suppress the expression of endogenous (and exogenous) retroviruses.
Our studies identified a novel function for MERVL LTRs in the specification of the totipotent two/four-cell fate that serves as the precursor for both embryonic and extraembryonic tissue types. In the absence of Kdm1a, MERVL expression is up-regulated in both ES cells and blastocyst stage embryos. It is unclear why MERVL retroviruses are normally repressed in the embryonic lineages, although this might help to protect the germline from acquiring excessive mutations, and therefore could provide a selective advantage. Interestingly, we also discovered that a subset of early zygotic genes expressed exclusively in two/four-cell embryos (during ZGA) became up-regulated in ES cells lacking Kdm1a. This derepression was associated with a remarkable change in ES cell developmental potential. Rather than becoming more restricted in cell fate, ES cells lacking KDM1A function have an expanded fate potential. Namely, Kdm1a mutant ES cells acquire the ability to generate both extraembryonic and embryonic cell types. This finding suggests that inhibitors of KDM1A activity or ectopic expression of one or more of the downstream target genes of KDM1A may prove to be beneficial for cell reprogramming. We found that the concomitant up-regulation of MERVL elements and derepression of early zygotic gene expression occur prior to gastrulation. Therefore, one or both of these genetic changes could account for the early lethality of Kdm1A mutant embryos, and suggest that reprogramming strategies should incorporate on–off control over this developmental pathway.
The coregulation of MERVL elements and cellular genes by KDM1A prompted us to examine the regulatory sequences to identify shared features. Genome-wide chromatin studies revealed that the same pattern of histone modifications associated with MERVL up-regulation in Kdm1a mutant cells also occurred at the promoters of the cellular genes. Remarkably, we found that a large number of the cellular genes had promoters derived from ancient retroviral insertions. These observations suggest that the regulatory elements within the MERVL LTRs that confer KDM1A-mediated repression in embryonic cell lineages may have been useful regulatory modules for ensuring that inappropriate cellular genes are suppressed in embryonic cells. Although our studies focused on the subset of cellular genes containing well-preserved LTRs directly within their promoters, our model proposes that retroviral insertions influenced neighboring gene expression by helping to recruit the chromatin-modifying enzyme KDM1A. Consequently, some of the KDM1A-regulated genes lacking obvious LTR promoters have had viral insertions at more distant sites (like Zscan4) or within introns (like Scml2), such that their transcription may be influenced by chromatin spreading, although this needs further examination. Interestingly, the integration of viruses can often cause genomic rearrangements because of the recombinogenic nature of their repetitive sequences. Depending on the orientation of the repetitive sequences, the recombination events can lead to gene duplications and reciprocal deletions, and can also lead to the appearance of solo LTRs that predominate the repetitive landscape. We noted that many of the genes that became up-regulated in Kdm1A mutants were regulated by solo LTRs or were part of segmental duplications that were highly variable in copy number across species (and, in some cases, were absent in closely related species). Some have even been documented to be copy number variant within the mouse lineage, indicating that they are rapidly evolving (Church et al. 2009). In summary, our findings indicate that chromatin regulation mediated by KDM1A is part of the host's defense against excessive endogenous retroviral activity. This repressive strategy appears to have been co-opted to suppress a subset of cellular genes in early embryonic development.
Materials and methods
Mouse lines
Kdm1a GT mice were generated by injection of RRK075 ES cells (Bay Genomics, MMRRC) into C57Bl/6J blastocysts. The resulting chimeric mice were then mated with B6 mice to obtain Kdm1a +/GT mice. Sox2::Cre and Cre-ERT mice were from Jackson Laboratories (strain #004783 and #004847). Kdm1a FL and KO mice were described previously (Wang et al. 2007).
ES cell derivation and culture
Mouse ES cell lines were derived as described (Bryja et al. 2006). Where indicated, ES cells were transfected with pEGFP C1 (Clontech) or pCAGS Cre-GFP (Addgene) using Lipofectamine 2000 using a 1:4 ratio of DNA to Lipofectamine. GFP+ cells were then collected using a FACSDiVA. Kdm1a FL/FL ES cells containing the Cre-ERT transgene were treated with 1 μM 4OHT or vehicle for 24 h to recombine the FL allele, which was routinely evaluated by genotyping PCR. For stable rescue clones, Kdm1a FL/FL ES cells were transfected as above with pCMV Flag-KDM1A wild type or KDM1A MUT (K661A) and treated with G418 (400 μg/mL) for 7 d before picking colonies and screening for KDM1A expression using anti-Flag antibodies (Sigma). For differentiation of ES cells, ∼106 cells were grown in 10 mL of mDiff medium (1:1 Knockout DMEM:DMEM/F12 [Invitrogen], 5% KOSR [Invitrogen], 1× NEAA [Mediatech], 2 mM L-glutamine, 14.3 mM 2-mercaptoethanol) in bacterial-grade 10-cm2 dishes, and medium was changed every 2 d. Retinoic acid (1 μM) and smoothened agonist (1 μM; Calbiochem) were added to induce motor neuron differentiation after 2 d of embryoid body formation.
Immunofluorescence microscopy
Cells were fixed with 4% PFA for 10 min, followed by washing with PBS-T (0.05% Tween). Cells were then blocked in PBS-T containing 3% BSA for 10 min and stained with primary antibody for 1 h at room temperature. After washing three times for 10 min with PBS-T, cells were stained with secondary antibody (1:1000 anti-mouse, anti-rat, or anti-rabbit IgG Alexa Fluor 488, 555, or 647) for 1 h at room temperature and washed again three times with PBS-T. Coverslips were stained with DAPI in PBS for 5 min before being inverted onto slides in mounting medium. Cells were then imaged using an Olympus IX81 confocal, an Olympus FV1000 confocal, or a Zeiss Axioskop 2 epifluorescence microscope.
Immunoblotting
Cell pellets were extracted in 1% NP40 containing 10 mM Tris, 150 mM NaCl, and 1× protease inhibitors. Total protein (10–50 μg) in LDS sample buffer was then loaded onto a 4%–12% NuPage gel (Invitrogen), electrophoresed, and transferred to nitrocellulose membranes. Membranes were blocked in PBS-T containing 5% nonfat dry milk. Primary antibodies were incubated overnight at 4°C. After washing extensively with PBS-T, secondary antibodies (anti-rabbit or anti-mouse IgG HRP conjugate, 1:10,000 dilution) were incubated for 1 h at room temperature. After washing extensively with PBS-T and water, blots were developed using the ECL plus detection system (Amersham).
Bisulfite sequencing/COBRA analysis
Bisulfite treatment of genomic DNA was carried out using the Epitect Bisulfite kit (Qiagen). Bisulfite-converted DNA was then PCR-amplified using Accuprime Taq polymerase (Invitrogen), followed by TOPO TA cloning (Invitrogen) and sequencing (Eton Bio). For COBRA, bisulfite-converted DNA was PCR-amplified using Accuprime Taq, phenol chloroform-extracted, and ethanol-precipitated before digestion with either HpyCH4IV or Taqa1. Digested and undigested DNA was then run on a 2% agarose gel and stained with ethidium bromide.
Affymetrix arrays, qRT–PCR, and mRNA-seq
RNA was prepared from mouse ES cells using the RNEasy kit (Qiagen) with on-column DNase digestion. dscDNA was generated from 100 ng–1 μg of total RNA using the GeneChip3′ IVT Express kit (Affymetrix), fragmented, and hybridized to Affymetrix Mouse Genome 430 2.0 expression arrays. Differentially expressed genes were determined using Vampire and the default settings (http://genome.ucsd.edu/microarray).
For qRT–PCR analysis, cDNA was generated using SuperScript III (Invitrogen) and polydT priming. qPCR was performed using SYBRGreen master mix (Applied Biosystems). All reactions were performed in triplicate. Standard curves were generated for each primer pair (sequences are available on request), and expression levels were performed relative to Gapdh (in arbitrary units). Error bars represent SD.
For mRNA-seq, sample libraries were prepared using the mRNA-seq sample prep kit (Illumina). Library samples were amplified on flow cells using the mRNA-seq cluster generation kit (Illumina), and then sequenced using the 36-cycle sequencing kit on the Genome Analyzer (Illumina). Raw sequence data were then aligned to the mouse genome using the short read aligner Bowtie and the default setting (two mismatches per 25 bp and up to 40 genomic alignments) (http://bowtie-bio.sourceforge.net/index.shtml). RPKM values were also determined by Bowtie. For repetitive sequences, we aligned sequencing reads to the Repbase database using Bowite (http://www.girinst.org/repbase/index.html). Heat maps of RPKM values were generated using CiMminer (http://discover.nci.nih.gov/cimminer).
KDM1A affinity purification/mass spectrometry
A KDM1A affinity matrix was generated by mixing 25 μg of polyclonal KDM1A antibody (Abcam, ab17721) with 250 μL of protein A/G Dynabeads for 10 min at room temperature, followed by covalent cross-linking using 20 mM DMP (Pierce, no. 21666) in 0.2 M triethanolamine (pH 8.2), for 30 min at 20°C. The cross-linked Dynabeads were washed with PBS, followed by one wash with 0.1 M citrate buffer (pH 2.5). The antibody/beads were washed with PBS before overnight incubation with 350 μL of 10 mg/mL ES cell extract as described above. The antibody/bead mixture was washed seven times with 1% NP40 buffer, followed by elution of bound proteins with 50 μL of 0.1 M glycine (pH 2.5). The eluate was neutralized with 5 μL of 1 M Tris (pH 8.0). Samples were then subject to LDS-PAGE and silver-staining (Invitrogen). Individual lanes of the gel were cut into 10–15 slices and digested with trypsin prior to mass spectrometry analysis using ESI-MS/MS as described (Herrera et al. 2009; Schubert et al. 2009). Data were analyzed using the Mascot algorithm (Matrix Science).
Genome-wide MeDIP and ChIP analysis
MeDIP and ChIP-on-chip analysis were performed on Kdm1a +/GT and GT/GT as described previously (Li et al. 2003; Weber et al. 2005). For hybridization to arrays, 3 μg each of ChIP (or MeDIP) and input DNA were labeled with Cy5 and Cy3, hybridized, washed, and scanned according to Nimblegen's ChIP–chip protocol. For ChIP analysis, samples were hybridized to Array 10 (Roche Nimblegen, catalog no. 05542600001) of mouse whole-genome 10 array set, carrying 2.1 million features tiling chromosome 18, 19, X, and Y at a median probe spacing of 100 bp. For MeDIP, samples were hybridized to Nimblegen 2.1M Deluxe promoter arrays. The two-color images were scanned and aligned using Nimblescan software. The relative intensities were normalized by Nimblescan to generate a *.PAIR file. The genomic coordinates were loaded, and then log2 ratio of relative intensities were calculated to give a *.GFF file. The chromosome, coordinates, and log2 ratio from the *.GFF files were then converted to *.WIG using Perl scripts and uploaded onto UCSC genome (mm8) browser for visualization. Tracks from wild-type and Kdm1a GT/GT ES cells were colored red and green, respectively, and overlaid using Photoshop. Each displayed track is from a representative biological sample. For MeDIP, MonoMeK4, DiMeK4, TriMeK4, and AcK27, subtraction plots were generated by averaging the log2 intensities from two biological replicates.
Luciferase reporter assays
Genomic promoter fragments from the Tcstv1, Zfp352, and Eif1a genes or MERVL LTR-Gag sequences were amplified by PCR using Accuprime Taq (Invitrogen) and cloned into pGL3 basic vector using NheI and either XhoI or Nco1 sites. Luciferase clones were then cotransfected with CMV β-gal into ES lines in 96-well dishes in triplicate. Luciferase and β-gal activity were measured 24 h later. Error bars represent SEM.
Teratoma formation assay
Approximately 106 ES cells were injected into the rear flanks of athymic nude mice (Foxn1nu, Jackson Laboratories, 002019). After 4–5 wk, mice were euthanized and teratomas were isolated and flash-frozen on dry ice for qRT–PCR or fixed in 4% PFA for immunostaining.
Acknowledgments
We thank Yelena Dayn, David Chambers, Ryan Lister, Joseph Nery, Jonna Barrie, Ling Ouyang, James Nguyen, Wolfgang Fischer, Jessica Read, Gerard Manning, Eric Scheeff, Henry Juguilon, Mathias LeBlanc, and Benjamin Spike for technical assistance. We thank Thierry Heidmann for MERVL Gag antibodies. We thank Gordon Gill and members of the Pfaff laboratory for discussions and comments on the manuscript. T.S.M and W.D.G. were supported by CIRM. S.L.P. was supported by grants from the National Institute of Neurological Disorders and Stroke and CIRM, and is an investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2008511.
Supplemental material is available for this article.
References
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G 1999. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci 96: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benit L, De Parseval N, Casella JF, Callebaut I, Cordonnier A, Heidmann T 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol 71: 5652–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Black SG, Arnaud F, Palmarini M, Spencer TE 2010. Endogenous retroviruses in trophoblast differentiation and placental development. Am J Reprod Immunol 64: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryja V, Bonilla S, Cajanek L, Parish CL, Schwartz CM, Luo Y, Rao MS, Arenas E 2006. An efficient method for the derivation of mouse embryonic stem cells. Stem Cells 24: 844–849 [DOI] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112 doi: 10.1371/journal.pbio.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448: 105–114 [DOI] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet 5: e1000325 doi: 10.1371/journal.pgen.1000325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CT, Dovey OM, Lezina L, Luo JL, Gant TW, Barlev N, Bradley A, Cowley SM 2010. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol Cell Biol 30: 4851–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, et al. 2007. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128: 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding MC, Zhang L, Mann MR 2010. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell 6: 457–467 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH Jr 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera F, Chen Q, Fischer WH, Maher P, Schubert DR 2009. Synaptojanin-1 plays a key role in astrogliogenesis: possible relevance for Down's syndrome. Cell Death Differ 16: 910–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. 2008. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci 105: 19199–19204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Coffin JM 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42: 709–732 [DOI] [PubMed] [Google Scholar]
- Kigami D, Minami N, Takayama H, Imai H 2003. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod 68: 651–654 [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R 2006. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol 26: 6395–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci 100: 8164–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TY, Chen HH, Lee KH, Choo KB 2003. Display of different modes of transcription by the promoters of an early embryonic gene, Zfp352, in preimplantation embryos and in somatic cells. Mol Reprod Dev 64: 52–60 [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437: 436–439 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W 2005. Epigenetic reprogramming in mammals. Hum Mol Genet 14: R47–R58 doi: 10.1093/hmg/ddi114 [DOI] [PubMed] [Google Scholar]
- Nottke A, Colaiacovo MP, Shi Y 2009. Developmental roles of the histone lysine demethylases. Development 136: 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463: 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Ribet D, Louvet-Vallee S, Harper F, de Parseval N, Dewannieux M, Heidmann O, Pierron G, Maro B, Heidmann T 2008. Murine endogenous retrovirus MuERV-L is the progenitor of the ‘orphan’ ε viruslike particles of the early mouse embryo. J Virol 82: 1622–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Schubert D, Herrera F, Cumming R, Read J, Low W, Maher P, Fischer WH 2009. Neural cells secrete a unique repertoire of proteins. J Neurochem 109: 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM 2004. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol 269: 276–285 [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. 2007. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446: 882–887 [DOI] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. 2009. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41: 125–129 [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Cammas F, Losson R, Goff SP 2008. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J Virol 82: 4675–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM 2009. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci 106: 19363–19368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL 2001. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc Natl Acad Sci 98: 1454–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Walker E, Tamplin OJ, Rossant J, Stanford WL, Hughes TR 2006. Zfp206 regulates ES cell gene expression and differentiation. Nucleic Acids Res 34: 4780–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]