Abstract
Alternative splicing of precursor mRNA (pre-mRNA) is a strategy employed by most eukaryotes to increase transcript and proteomic diversity. Many metazoan splicing factors are members of multigene families, with each member having different functions. How these highly related proteins evolve unique properties has been unclear. Here we characterize the evolution and function of a new Drosophila splicing factor, termed LS2 (Large Subunit 2), that arose from a gene duplication event of dU2AF50, the large subunit of the highly conserved heterodimeric general splicing factor U2AF (U2-associated factor). The quickly evolving LS2 gene has diverged from the splicing-promoting, ubiquitously expressed dU2AF50 such that it binds a markedly different RNA sequence, acts as a splicing repressor, and is preferentially expressed in testes. Target transcripts of LS2 are also enriched for performing testes-related functions. We therefore propose a path for the evolution of a new splicing factor in Drosophila that regulates specific pre-mRNAs and contributes to transcript diversity in a tissue-specific manner.
Keywords: alternative splicing, splicing regulation, U2AF, splicing evolution
Alternative splicing is the complex process by which many different eukaryotic mRNAs are generated from nuclear precursor mRNAs (pre-mRNAs). The splicing of one transcript in several different ways allows the generation of vast proteomic diversity from a comparatively smaller number of genes (Nilsen and Graveley 2010). These alternatively spliced transcripts are often restricted to particular tissues and encode proteins that are critical to proper tissue function (Wang and Burge 2008). Regulation of pre-mRNA splicing is achieved through the interaction of RNA sequence elements and a variety of related RNA-binding protein factors (Black 2003; Ben-Dov et al. 2008; Wang and Burge 2008). Many different alternative splicing patterns exist (Black 2003). All of these involve the employment of one splice site over another. The efficiency with which splice sites are recognized and their ability to recruit functionally competent spliceosome components regulate splice site utilization (Nelson and Green 1988; Yu et al. 2008). These efficiencies can be modulated by the binding of factors that enhance or repress splice site use (Blanchette et al. 2005). The recognition and determination of 3′ splice sites is primarily carried out by U2-associated factor (U2AF) (Ruskin et al. 1988; Zamore and Green 1989). The essential, highly conserved U2AF general splicing factor is a heterodimer composed of large (U2AFLS) and small (U2AFSS) subunits that promotes spliceosome assembly (Ruskin et al. 1988; Singh et al. 1995). U2AF is conserved among all eukaryotic species, from Schizosaccharomyces pombe to humans. U2AFLS (dU2AF50 in Drosophila) recognizes the polypyrimidine tract at the 3′ end of the intron (Zamore and Green 1989; Kanaar et al. 1993), while its cooperating partner, U2AFSS (dU2AF38 in Drosophila), interacts with the intron-terminal AG dinucleotide (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). U2AFLS additionally cooperates with the branch point adenosine-binding SF1 through interactions in its C-terminal pseudo-RNA recognition motif (RRM) (Kent et al. 2003; Selenko et al. 2003). Following these contacts, the 3′ end of the intron is then competent for interaction with U2 snRNP. U2AF therefore functions to promote spliceosome assembly. Much work has been done concerning the evolutionary conservation of the cis-acting RNA sequence elements. Many sequence elements are widely conserved even across vast evolutionary distances and often lead to similar splicing patterns in the orthologous transcripts (Brooks et al. 2011). However, little is understood about how related family members of the RNA-binding proteins that mediate these splicing effects arise and diverge to acquire distinct and diverse functions (Baek and Green 2005; Akerman et al. 2009). These distinct functions allow evolutionarily related proteins to form regulatory networks, with each member controlling the splicing of specific transcripts through the recognition of specific sequence motifs. Here, we identified and characterized the appearance and evolutionary divergence of a Drosophila splicing factor that we termed LS2 (Large Subunit 2, also known as CG3162). LS2 arose from a retroduplicated copy of the highly conserved, positively acting dU2AF50, and has diverged sufficiently from dU2AF50 such that it is highly specialized in its specificity, function, and expression.
Results
LS2 evolved from a retroduplicated copy of dU2AF50
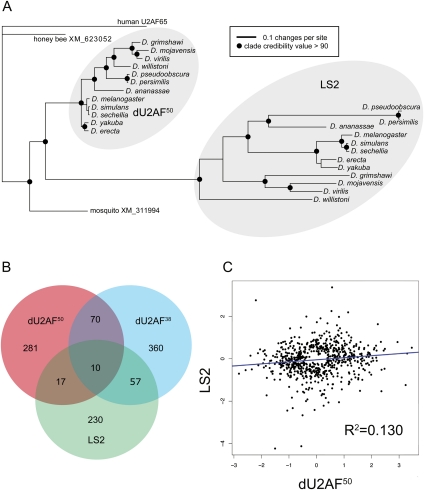
LS2 and dU2AF50 are 55% identical and 70% similar at the primary sequence level (Supplemental Fig. 1). Using the amino acid sequences of several U2AF large subunit and LS2 genes, we determined that the LS2 gene arose via a duplication event before the most recent common ancestor of all Drosophila (Fig. 1A). The LS2 orthologs are in syntenic positions in each Drosophila genome. We could not detect an LS2 ortholog in mosquitoes or honeybees. Given the estimated ages of the most recent common ancestor of Drosophila and mosquitoes, and the most recent common ancestor of the 12 Drosophila species analyzed (Tamura et al. 2004), we conclude that the duplication event that gave rise to LS2 occurred between 60 and 250 million years ago. Sequence analysis of the dU2AF50 orthologs revealed little divergence between the orthologs, consistent with the conserved function of the U2AF large subunit and its requirement for viability (Kanaar et al. 1993). However, the LS2 orthologs were comparatively highly diverged. Thus, while the dU2AF50 orthologs are under much constraint and negative selection to retain their current function, the LS2 orthologs may be free to acquire new functions and may be under positive selection. While the dU2AF50 in Drosophila melanogaster contains five introns, LS2 does not contain any introns. This implies the use of an RNA intermediate during the gene duplication process, consistent with the idea of a retroduplication event.
Figure 1.
LS2 arose and diverged in function from dU2AF50 in Drosophila. (A) Phylogenetic tree of LS2, dU2AF50, and orthologs from honey bees, mosquitoes, and humans. The non-Drosophila gene sequences are the single most similar genes to both dU2AF50 and LS2 in each of the three outgroup genomes. Clades with >90 credibility value are denoted with a black circle. (B) Venn diagram showing the overlap of splice junctions affected by dU2AF50, dU2AF38, and LS2 RNAi knockdown. (C) A scatter plot of splice junctions affected by dU2AF50 and LS2 RNAi knockdown. Axes represent the log2 change of splice junction intensity in response to RNAi of the indicated protein.
LS2 controls splicing of a transcript pool that is distinct from that of dU2AF50
To determine whether LS2 was simply a redundant form of dU2AF50, we used Drosophila splice junction microarrays to determine the splicing events sensitive to dU2AF50, dU2AF38, and LS2 after RNAi knockdown in Drosophila S2 cells (Blanchette et al. 2005). We verified that LS2 expression was efficiently knocked down (Supplemental Fig. S2A). Analysis of the microarray results revealed that dU2AF50, dU2AF38, and LS2 affected the splicing of 378, 497, and 311 splice junctions in 206, 276, and 168 genes, respectively (Supplemental Fig. 2B; Supplemental Table 1). dU2AF50 is a core splicing factor, and as such may not be expected to specifically regulate distinct transcripts. Nevertheless, our data are consistent with previous studies in which core spliceosomal factors did have such specificity (Park et al. 2004; Sridharan et al. 2011). Although the collection of splice junctions sensitive to dU2AF50 and LS2 depletion overlapped to a small extent, the majority of them were unique to either protein (Fig. 1B). To more precisely characterize the relationship between the splice junction targets of each protein, we incorporated the magnitude and direction of splice junction changes upon knockdown. These plots showed little correlation between the responses to dU2AF50 and LS2 knockdown (Fig. 1C). Thus, these two proteins have distinct splice junction specificities and functions. In contrast, there was a strong correlation of the responses to dU2AF50 and dU2AF38 knockdown (Supplemental Fig. 2C), consistent with their known physical and functional interactions (Rudner et al. 1998b). We also observed an intermediate correlation of the responses to LS2 and dU2AF38 knockdown, implying a possible functional interaction. Finally, we validated several of the predicted splicing changes predicted by the microarray using semiquantitative RT–PCR (Supplemental Fig. 2D).
LS2 has diverged from dU2AF50 in RNA sequence recognition specificity
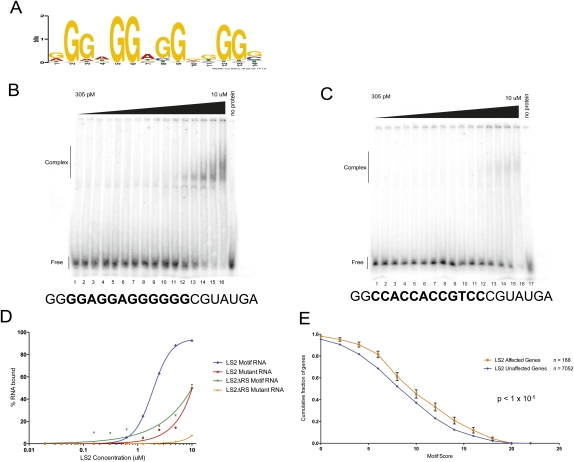
To directly determine whether dU2AF50 and LS2 recognize similar or different RNA-binding sites, we used in vitro binding site selection (SELEX) to determine an optimized RNA-binding sequence motif for LS2. Similar analyses with the large subunit of U2AF showed that U2AFLS preferentially recognizes pyrimidine-rich sequences, consistent with its role in spliceosome assembly through recognition of the polypyrimidine tract (Singh et al. 2000; Sickmier et al. 2006). In contrast, although the LS2 and dU2AF50 proteins are highly related in primary sequence throughout their RRMs (Supplemental Fig. 1), the purified LS2 protein preferentially binds to a G-rich RNA motif with much less degeneracy at specific positions (Fig. 2A). This RNA-binding specificity was confirmed using quantitative electrophoretic mobility shift RNA-binding assays using an RNA probe containing the SELEX-derived motif (Fig. 2B) and a mutant probe that much more closely resembled a polypyrimidine-rich RNA (Fig. 2C). Similar to the measured equilibrium dissociation constant (Kd) of 2.2 μM for purified dU2AF50 binding to a polypyrimidine RNA, the apparent Kd of the LS2 protein for its RNA SELEX motif was 1.9 μM (Fig. 2D). The LS2 protein showed a much lower affinity for the mutant probe. Additionally, as was also the case for dU2AF50, the highly positively charged N-terminal arginine and serine-rich (RS) domain was required for high-affinity RNA binding but did not play a role in sequence specificity (Fig. 2D; Rudner et al. 1998a). Finally, the purified recombinant LS2/dU2AF38 heterodimer bound RNA much more tightly than the LS2 monomer alone (Supplemental Fig. 3). The apparent equilibrium Kd of the heterodimer for a G-rich RNA was 150 nM, similar to the affinity of the U2AF heterodimer for a polypyrimidine RNA (Rudner et al. 1998a). The heterodimer also showed greater nonspecificity in RNA binding that may be due to the presence of an additional RS domain provided by dU2AF38. However, the LS2–dU2AF38 heterodimer still bound preferentially to a G-rich RNA. The increased nonspecificity for G-rich versus pyrimidine-rich DNA of the LS2–dU2AF38 heterodimer compared with the LS2 monomer is also consistent with the previously documented RNA-binding properties of human and Drosophila U2AF (Rudner et al. 1998a).
Figure 2.
LS2 has diverged in RNA-binding sequence specificity from dU2AF50, and its binding motif is enriched in its target transcripts. (A) SELEX-derived PSSM (position-specific scoring matrix) of the RNA sequence recognized by the LS2 protein. (B,C) Electrophoretic gel mobility shift assays using purified recombinant GST-tagged LS2 protein and a synthetic RNA containing the SELEX-derived G-rich recognition motif (B) or a mutant RNA in which all of the important guanosine residues (see motif) had been mutated to cytosine (C). Protein concentrations ranged from 305 pM (lane 1) to 10 μM (lane 16) in twofold increments. (Lane 17) No protein control. (D) PhosphorImager quantification of the results in A and B. Similar experiments were done using GST-tagged truncated versions of LS2 lacking the N-terminal RS domain (data not shown). (E) Enrichment of the LS2 recognition motif in its affected target transcripts. Each point represents the fraction of genes that contain an LS2 recognition motif scoring at the X-axis value or higher.
If the derived SELEX motif for LS2 binding is correct and the target transcript pool from the LS2 RNAi knockdown splice junction microarray data are direct targets of the LS2 protein, we reasoned that the G-rich LS2-binding motif should be enriched in the LS2-affected genes over all other unaffected transcripts. Similar patterns of RNA-binding motif enrichment have been observed previously with known splicing factors with well-defined RNA-binding motifs and from in vivo transcript-binding data (Blanchette et al. 2009). We detected such an enrichment (P-value < 1 × 10−5) of preferred LS2 RNA-binding motifs in the 168 LS2-affected genes (Fig. 2E).
LS2 interacts with dU2AF38 in an RNA-independent manner
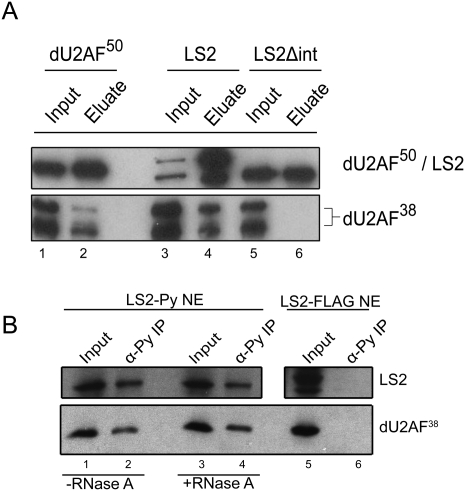
U2AFLS functions in spliceosome assembly in conjunction with the small U2AF subunit U2AFSS. U2AFSS functions as a core splicing factor whose role is to recognize the intron-terminal AG dinucleotide (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). U2AFLS and U2AFSS in humans and Drosophila interact through a hydrophobic interface (Zamore and Green 1989; Rudner et al. 1998b; Kielkopf et al. 2001) that, in both LS2 and dU2AF50, is located in between the RS domain and the first RRM. Both dU2AF50 and LS2 contain the critical hydrophobic residues necessary for this interaction (Supplemental Fig. 1). To test whether LS2 can interact physically with dU2AF38, we performed GST pull-down interaction assays with recombinant LS2 and dU2AF38 proteins. GST-tagged dU2AF50 and LS2 bound recombinant dU2AF38 (Fig. 3A, lanes 3,4), and this interaction was dependent on the presence of the putative U2AFSS interaction domain (Fig. 3A, lanes 5,6). Additionally, recombinant LS2 and dU2AF38 coeluted from an ion exchange column at 900 mM KCl, consistent with a hydrophobic interaction between the two proteins (data not shown). To test whether LS2 and dU2AF38 interact in Drosophila cells, we used a stably transfected S2 cell line that expressed an epitope-tagged LS2. Endogenous dU2AF38 could be coimmunoprecipitated with polyoma (also known as Py or Glu–Glu)-tagged LS2 from these S2 cell nuclear extracts (Fig. 3B, lanes 1,2). This interaction was resistant to RNase treatment (Fig. 3B, lanes 3,4), indicating that these two proteins were interacting physically, not simply bound to the same RNA. However, dU2AF38 could not be immunoprecipitated using a polyoma antibody from S2 cell extract containing a Flag-tagged version of LS2, indicating the specificity of the interaction (Fig. 3B, lanes 5,6). Additionally, there is likely to be a functional interaction between LS2 and dU2AF38 in vivo based on the moderate correlation and overlap of splice junction population changes in response to LS2 and dU2AF38 knockdown (Fig. 1B; Supplemental Fig. 2C). We therefore propose that LS2 has co-opted a fraction of the cellular dU2AF38 population for use on its distinct transcript pools in extraspliceosomal functions.
Figure 3.
LS2 interacts physically with dU2AF38. (A) Immunoblots from GST pull-down experiments using purified recombinant GST-tagged dU2AF50 (lanes 1,2), LS2 (lanes 3,4), or an LS2 truncation lacking the putative dU2AF38 interaction domain (lanes 5,6), and Escherichia coli lysates containing His-tagged dU2AF38 (see Supplemental Fig. 1). Eluates from the pull-downs were then immunoblotted using anti-dU2AF50 and anti-LS2 (top panel) or anti-dU2AF38 (bottom panel) antibodies. (B) Immunoblot analysis from coimmunoprecipitation experiments performed with epitope-tagged LS2 expressed in S2 cells. Polyoma (GLU–GLU) epitope-tagged LS2 was immunoprecipitated from S2 extracts in the presence (+; lanes 1,2) or absence (−; lanes 3,4) of RNase A, and the precipitates were immunoblotted using anti-LS2 (top panel) or dU2AF38 (bottom panel) antibodies. (Lanes 1,3,5) In all cases, the immunopurified proteins were compared with input lysate lanes (Input). (Lanes 5,6) To show specificity, similar experiments were done using Flag-tagged LS2 (negative control).
Expression of LS2 is highly enriched in testes
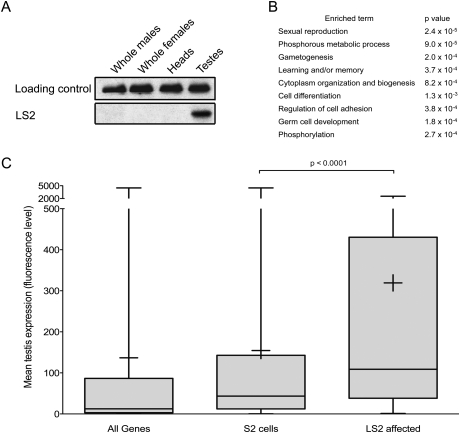
Many alternative splicing events are specific to a particular cell or tissue type. A common mechanism for achieving this specificity is to restrict expression of the necessary splicing factors to the appropriate tissues, as is the case for the mammalian nervous system-specific factors nPTB (Kikuchi et al. 2000; Markovtsov et al. 2000) and Nova (Buckanovich et al. 1993). Although dU2AF50 expression is ubiquitous, consistent with its function as a general splicing factor, FlyAtlas expression microarray data indicated that LS2 mRNA was preferentially expressed in the testes (Chintapalli et al. 2007). To confirm that this is also true for LS2 protein, we performed immunoblot analysis for LS2 using whole males, whole females, heads, and testes. While expression of LS2 in whole flies and in heads compared with the loading control was negligible, we detected significant expression in testes, consistent with the mRNA expression array results (Fig. 4A). Moreover, gene ontology (GO) analysis on the LS2-affected transcripts revealed several GO terms, consistent with a role in testes function, gamete production, and cellular regulation through phosphorylation (Fig. 4B; Supplemental Fig. 4A; Al-Shahrour et al. 2006). Fewer GO term enrichments were seen for genes affected by dU2AF50 and dU2AF38 (Supplemental Fig. 4B,C), consistent with their ubiquitous expression and function as general, spliceosome-associated splicing factors. If expression of LS2 was highly enriched in testes, we hypothesized that expression of the LS2 target transcripts should also be testes-enriched. Using FlyAtlas tissue expression data, we found that 87.4% of all genes expressed in S2 cells are also expressed in testes (Chintapalli et al. 2007). However, 97.5% of LS2 targets identified from S2 cells are expressed in testes, representing a significant enrichment (P < 0.0001, χ2 test). Furthermore, when the magnitude of expression is taken into account, the LS2 mRNA targets tend to be much more highly expressed in testes than either all Drosophila genes or those present in S2 cells (Fig. 4C).
Figure 4.
LS2 and its target transcripts are enriched in testes and regulate testes-related functions. (A) Immunoblot analysis of lysates from whole flies, heads, and testes probed with anti-PSI (loading control) and anti-LS2 antibodies. (B) Enriched GO terms for the LS2-affected genes (Al-Shahrour et al. 2006). P-values were calculated using a two-tailed Fisher's exact test. (C) Testes expression levels of all genes, genes expressed in S2 cells, and genes affected by LS2. The Y-axis is the mean expression level from four Affymetrix Dros2 expression arrays (Chintapalli et al. 2007). Whiskers represent the maximum and minimum values, boxes represent the 25th and 75th percentiles, crosses represent the mean value, and lines represent the median value. P-values were calculated using a two-tailed t-test.
LS2 acts as a splicing repressor in vitro and in vivo
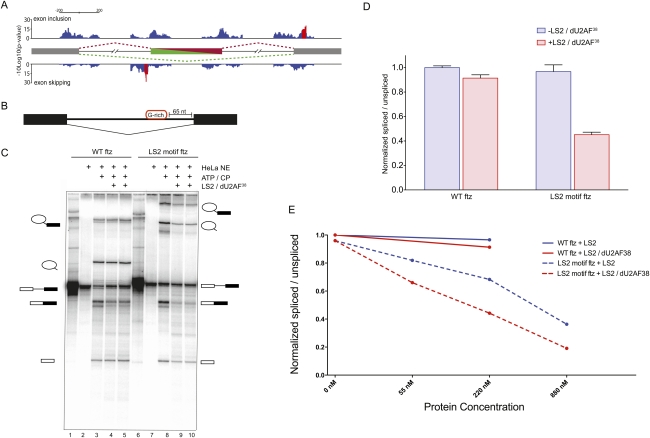
We then asked where positional enrichments of LS2 recognition motifs were located in the endogenous target transcripts of LS2 that were identified by the RNAi splice junction microarrays. Analysis of the location of the LS2 recognition motifs near affected cassette exon junctions showed an enrichment of motifs associated with exon skipping just upstream of the cassette exon (Fig. 5A). This peak was ∼60 nucleotides (nt) upstream of the 3′ splice site.
Figure 5.
LS2 inhibits splicing of the ftz intron in vitro. (A) Motif location and clustering in LS2-affected cassette exons. LS2-affected cassette exons were searched for LS2 recognition motifs in 50-nt overlapping windows. Motif-containing windows were called as significant (red bars) if they contained a significant motif enrichment (P-value < 0.05) and were part of a stretch of at least five consecutive significant windows. LS2 recognition motifs associated with cassette exon inclusion are displayed on top, while motifs associated with cassette exon skipping are on the bottom. (B) Diagram of the modified ftz intron in vitro splicing substrate generated with and without a G-rich LS2 recognition motif inserted 65 nt upstream of the 3′ splice site. (C) In vitro splicing reaction of the wild-type (lanes 1–5) and LS2 SELEX motif-containing (lanes 6–10) ftz substrates using HeLa cell nuclear extract in the presence or absence of purified recombinant GST-tagged LS2/dU2AF38 heterodimer. The identity of each RNA species is shown schematically at the right and left of the panel. (CP) Creatine phosphate. Reactions were carried out without HeLa nuclear extract (lanes 1,6); with nuclear extract but without ATP and CP (lanes 2,7); with nuclear extract, ATP, and CP (lanes 3,8); and with nuclear extract, ATP, CP, and 500 ng of recombinant LS2/dU2AF38 heterodimer protein (lanes 4,5,9,10). (D) PhosphorImager quantification of the results in B. The Y-axis represents the ratio of all splicing intermediate species to the unspliced pre-mRNA, with intensities for each species normalized to their length and all ratios normalized such that the value for wild-type (WT) ftz without added LS2/dU2AF38 heterodimer is 1.0. Error bars represent standard deviations of four to six experiments. (E) In vitro splicing efficiency of wild-type (WT) ftz and LS2 motif ftz in the presence of varying amounts of purified recombinant LS2 and LS2/dU2AF38 heterodimer. Quantification was performed as in D.
In order to investigate the molecular mechanism by which LS2 affects splicing of specific transcripts, we modified the efficiently spliced Drosophila ftz intron by adding a G-rich LS2-binding site motif 65 nt upstream of the 3′ splice site (Fig. 5B). This placed the LS2-binding site upstream of both the polypyrimidine tract and the branch point adenosine. We then monitored splicing of this modified pre-mRNA in HeLa cell nuclear splicing extract in the presence or absence of purified recombinant LS2/dU2AF38 heterodimer protein or LS2 protein alone. In these in vitro splicing assays, the splicing efficiency of the LS2-binding motif-containing pre-mRNA was significantly decreased in the presence of purified recombinant LS2/dU2AF38 heterodimer (Fig. 5C [lanes 9,10], D), as well as in the presence of the uncomplexed LS2 protein (Fig. 5E; Supplemental Fig. 5), indicating that LS2 has repressive activity even in human splicing extracts. Additionally, both LS2 alone and the LS2/dU2AF38 heterodimer repressed splicing of the G-rich motif-containing RNA in a concentration-dependent manner (Fig. 5E; Supplemental Fig. 6A). However, splicing of the substrate lacking the G-rich LS2-binding motif was unaffected by the addition of the LS2/dU2AF38 heterodimer or uncomplexed LS2 protein (Fig. 5C [lanes 4,5], D,E), indicating that the effect of LS2 is specific and dependent on its ability to bind RNA through its specific recognition motif. The ability of LS2 to repress splicing without the need for the dU2AF38 small subunit is consistent with the ability of dU2AF50 and human U2AF65 to activate splicing without dU2AF38 or U2AF35, respectively (Zamore et al. 1992; Kanaar et al. 1993).
LS2 was not able to substitute for the 3′ splice site definition activity of dU2AF50; that is, LS2/dU2AF38 could not activate the splicing of substrates in which the polypyrimidine tract had been replaced by the G-rich LS2 recognition motif (Supplemental Fig. 6B).
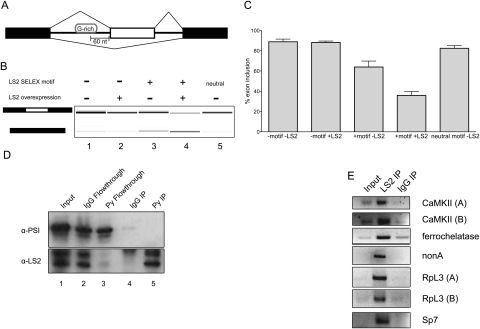
Next, we asked whether LS2 also displayed similar activities in vivo. A minigene construct made from the Drosophila PEP gene containing a cassette exon was used to test the effect of LS2 in S2 cells (Fig. 6A). Here, we inserted a G-rich LS2 recognition motif in the first intron 60 nt upstream of the 3′ splice site. This motif was again upstream of both the polypyrimidine tract and the branch point adenosine. In this assay, splicing repression would be manifested near the cassette exon, leading to increased skipping of the internal exon. We monitored the exon inclusion levels of both the wild-type and motif-inserted minigenes by RT–PCR in response to the overexpression of LS2.The basal level of inclusion of the cassette exon in this minigene was ∼90% (Fig. 6B,C). Overexpression of LS2 had very little effect on the splicing of the wild-type construct. Similarly, the insertion of a neutral, unrelated sequence motif 60 nt upstream of the 3′ splice site had a very modest effect. However, insertion of an LS2 recognition motif significantly shifted the splicing toward exclusion of the cassette exon, likely due to the action of endogenous LS2. Moreover, unlike the wild-type construct, the splicing of the motif-containing construct was sensitive to the level of LS2 because overexpression of LS2 further shifted the splicing toward exon exclusion (Fig. 6B,C). These results are consistent with the repressive activities of LS2 detected in vitro, its role as a potent splicing repressor, and the bioinformatically predicted positional enrichments of LS2 RNA-binding motifs. We also detected another LS2-binding motif enrichment, associated with exon inclusion, located ∼120 nt 3′ of the downstream splice site (Fig. 5A). Repression at the downstream splice site may kinetically allow splicing to occur at the cassette exon, causing its inclusion. Both enrichments are therefore consistent with the proposed function of LS2 as a splicing repressor. Although previous studies had identified G-runs as important splicing regulatory motifs in mammals (Xiao et al. 2009), these runs were associated mainly with 5′ splice sites and are bound by hnRNP H. The LS2 recognition sequence is not a G-run, but rather a motif with guanosines enriched at specific positions. Additionally, the motif's action as a splicing repressor is greatly increased by overexpression of LS2.
Figure 6.
LS2 acts as a splicing repressor in vivo, is enriched at specific positions in its target transcripts, and binds its predicted targets. (A) The minigene splicing reporter used with or without an LS2 G-rich binding motif inserted 60 nt upstream of the cassette exon (white). (B) Effect on exon inclusion in S2 cells as measured by RT–PCR of RNA expression from the minigenes carrying the LS2 recognition motif and LS2 overexpression. (Lane 1) Splicing pattern without LS2 motif or LS2 overexpression. (Lane 2) Splicing pattern without LS2 motif, but with LS2 overexpression. (Lane 3) Splicing pattern with LS2 motif, but without LS2 overexpression. (Lane 4) Splicing pattern with LS2 motif and LS2 overexpression. (Lane 5) Splicing pattern with neutral motif (see the Supplemental Material) and without LS2 overexpression. The left schematic denotes inclusion (top) or exclusion (bottom) product. (C) Quantification of results in B. Error bars represent standard deviations from three independent biological replicates. (D) Drosophila S2 cells stably expressing epitope-tagged (Glu–Glu, also called Py) LS2 protein were lysed and LS2 was immunoprecipitated using either anti-Py antibodies (lane 5) or nonimmune IgG (lane 4), and was detected using anti-PSI antibodies (top panel) or anti-LS2 antibodies (bottom panel). Input protein is shown in lane 1 (5% of input). PSI protein was detected and used as a negative control for immunoprecipitation. Both immunoprecipitation pellets and flowthrough material for IgG (lanes 2,4) or anti-Py antibody (lanes 3,5) are shown. (E) Immunoprecipitation of LS2 nuclear RNP complexes followed by RT–PCR of predicted affected transcripts using equal amounts of immunoprecipitated or input RNA. These included two CaMKII isoforms, ferrochelatase, nonA, two RpL3 isoforms, and Sp7. cDNA amplification products specific for each gene were compared between input RNA, LS2-immunopurified, and nonimmune IgG-immunopurified RNA samples.
LS2 interacts with its predicted targets in Drosophila S2 cells and has functionally diverged from dU2AF50
To determine whether LS2 interacts with its targets as predicted by the splice junction microarray, we performed immunoprecipitations of LS2 protein from stably transfected cells expressing polyoma epitope-tagged LS2. Using PSI protein as a negative control, we determined that the immunoprecipitation was specific for LS2 (Fig. 6D). We then used RT–PCR of anti-LS2-immunoprecipitated or nonimmune IgG-immunoprecipitated RNA with gene-specific primers to assay for specific LS2 protein–RNA interactions. We detected a significant enrichment of several predicted target transcripts in the LS2 immunoprecipitates over both the input and negative control nonimmune IgG immunoprecipitates (Fig. 6E). The action of LS2 as a splicing repressor rather than an activator demonstrates its functional divergence from dU2AF50. Consistent with this divergence, activation of 3′ splice sites could not be detected in constructs where the normal polypyrimidine tract was replaced by the LS2 G-rich sequence motifs (Supplemental Fig. 6B). If LS2 could serve as a surrogate, albeit opposite, form of dU2AF50, we hypothesized that the polypyrimidine tracts of LS2-affected splice junctions would be weaker than expected; that is, several of the pyrimidines in the polypyrimidine tract would be replaced by guanosines to allow LS2 binding. Toward this end, we analyzed the polypyrimidine tracts and 3′ splice sites of targets of several alternative splicing factors, including LS2, as well as those of all 48,550 introns interrogated by the splice junction microarray, using MaxEntScore (Yeo and Burge 2004). This analysis takes into account only the last 22 nt of the intron and therefore would not be expected to detect the motif enrichment that was detected 60 nt upstream of the 3′ splice site (Fig. 5A). Using these data, we conclude that the polypyrimidine tracts of LS2 target splice junctions are not any stronger or weaker than expected and do not contain anything resembling the G-rich LS2 SELEX motif (Supplemental Fig. 7A), indicating that LS2 has diverged from the 3′ splice site-centric role of dU2AF50. Interestingly, we also noted that the introns affected by LS2 are significantly longer than those affected by other characterized alternative splicing factors (Supplemental Fig. 7B). While the median lengths of all Drosophila introns and those affected by dU2AF50 knockdown were 85 nt and 121 nt, respectively, the median length of introns affected by LS2 knockdown was 422 nt (P-value < 0.0001, Student's t-test).
Although it is curious that knockdown of a core splicing factor like dU2AF50 resulted in splicing changes at specific junctions and not a global down-regulation in splicing, this was consistent with previous studies that had shown similar effects with S. pombe U2AF temperature-sensitive mutants and RNAi depletion of Drosophila core spliceosome proteins (Park et al. 2004; Sridharan et al. 2011).
LS2 is specifically expressed in differentiated cells in the Drosophila testes
Recent mRNA-seq studies have shown that cells in the Drosophila testes undergo extensive changes in alternative splicing patterns upon differentiation, and testes, like the brain and nervous system, are known hot spots of alternative splicing in mammals (Venables and Eperon 1999; Elliott and Grellscheid 2006; Gan et al. 2010). In general, the overall complexity of alternative splicing events decreases upon differentiation in the Drosophila testis, when the testes stem cell population adopts more restricted cell fates as the spermatocytes develop and mature. Consistent with this decrease in the overall complexity of alternative splicing patterns, there is a concomitant decrease in expression of a majority of splicing factors during testes differentiation (Gan et al. 2010). In contrast, this recent mRNA-seq study reported that LS2 is one of the few splicing factors whose expression increases dramatically upon testes differentiation (Gan et al. 2010). Consistent with this mRNA profiling data, immunofluorescence localization studies using flies expressing GFP-tagged Histone-2Av and affinity-purified anti-LS2 antibody indicated that, while LS2 protein was expressed in differentiated spermatocytes, it was not expressed in the undifferentiated stem cells at the testis tip (Supplemental Fig. 8). These testis tips were phenotypically normal, however, as evidenced by the ample GFP fluorescence from the tagged histone in the tip.
Discussion
Although the evolutionary patterns of splice sites and splice signals have been well documented (Brooks et al. 2011), little is known about how the proteins that recognize these sites and signals acquired their distinct functions and specificities. Many of these factors belong to large multigene families, with the SR proteins and hnRNP proteins being two notable examples (Dreyfuss et al. 1993; Shepard and Hertel 2009). It has been difficult, though, to determine how and when these family members diverged. Our findings indicate that the Drosophila genome acquired a new gene encoding a novel splicing factor, LS2, >60 million years ago through a retrotransposition gene duplication event. The quickly evolving LS2 gene subsequently diverged from its progenitor in its RNA-binding sequence specificity, expression pattern, and function to become an independent factor with a vastly different regulatory capacity and influence. We believe this to be the clearest example yet described of how gene duplication and divergence can result in the many related, yet distinct, splicing factors found in mammalian genomes. Furthermore, these results give an example of how these processes can transform a duplicated copy of a ubiquitously expressed and generally acting splicing factor into a tissue-specifically expressed and highly specialized component of a dedicated biological system.
Generally, new genes in Drosophila that are formed by retrotransposition events show a propensity to leave the X chromosome for the autosomes (Betran et al. 2002). More specifically, the phenomenon of acquisition of male-specific expression and function following gene duplication of an X-linked parental copy has been described for a multitude of genes in the Drosophila genome (Parisi et al. 2003). This may be due to the possibly disadvantageous overexpression of X-linked genes in males due to dosage compensation (Baker et al. 1994) or the increased risk of uncomplemented deleterious mutations due to X chromosome hemizygosity in males (Oliver 2002). The autosomal LS2 gene appears to be a very old instance of this phenomenon. dU2AF50 is X-linked, and the LS2 ortholog is found on chromosome 2R in the same syntenic context in all 12 sequenced Drosophila genomes. The burst of protein sequence evolution common to all of the Drosophila LS2 orthologs, combined with the maintenance of an intact but fast-evolving ORF, may allow for identification of specific LS2 amino acid residues that have undergone positive selection—a common fate for such genes (Proschel et al. 2006) during establishment and evolution.
In addition to showing a general male bias in expression and function, it has also been observed that many duplications of X-linked genes in Drosophila end up with a large testes-specific bias in expression (Bai et al. 2008), as is the case for the LS2 gene. Many of these genes, including LS2, have specifically identified motifs in their promoter regions that may contribute to a testes-biased expression pattern (Bai et al. 2009). Consistent with this, chromatin immunoprecipitation with sequencing (ChIP-seq) data from the modEncode consortium show a large enrichment of acetylation at histone H3 Lys 9 (H3K9), usually associated with transcriptionally active regions, in the promoter region of LS2 in males but not in females (Liang et al. 2004; Celniker et al. 2009). LS2 gene expression increases significantly upon testis cell differentiation, and, through its action as a splicing repressor, may serve to suppress the many possible alternative splicing events typical of an undifferentiated stem cell in order to funnel the population of mature spliced mRNA isoforms toward a simpler, cell type-specific pattern. Although we cannot distinguish whether LS2 expression and function is a cause or consequence of testes differentiation, we suggest that LS2 acts to promote testes differentiation through its action on testes-important target pre-mRNA transcripts. Because, in mammals, many RNA-binding proteins are members of multigene families (Martinez-Contreras et al. 2007), similar evolutionary associations among related RNA-binding protein family members are likely to exist in other organisms. Many of these factors and their functions have yet to be characterized. The relationship between LS2 and dU2AF50 investigated here may provide a conceptual framework for future studies of the appearance and evolution of other splicing factors, including those of the multigene families commonly found in all mammalian genomes.
Materials and methods
dU2AF50/LS2 sequence alignments
Sequence alignments of dU2AF50 and LS2 were performed using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Visualizations were done using Jalview.
dU2AF50/LS2 phylogenetic analysis
Annotated orthologs of dU2AF50 (CG9998) and LS2 (CG3162) were extracted from the 11 other Drosophila genus sequenced genomes. For each, the genomic context (neighboring genes) was manually inspected to confirm orthology. The closest homologs in mosquitoes (Anophele gambiae), honey bees (Apsis mellifera), and humans were identified by blastp search. For each of these outgroups, the most similar protein sequence to both CG9948 and CG3162 was a single gene: XP_311994.3 for mosquitoes, XP_623055.1 for honey bees, and NP_001012496.1 for humans. The genomic context in mosquito, honey bee, and human genomes was inspected and did not support a closer relationship to dU2AF50 or LS2—none of the neighboring genes was shared with any of these species. Of note, however, is that in no case was the mosquito, honey bee, or human homolog X-linked. This set of protein sequences was aligned using muscle version 3.7 using default parameters. The protein sequence alignment was then used as a fixed guide to align the corresponding codons of each genes’ coding sequence. Phylogeny of these sequences was inferred using MrBayes (version 3.1.2) under a three-partition model in which the first and second codon positions evolve in a two-state model with γ rate variation. The third codon position was modeled to evolve under a six-state model with a separate γ rate. MCMC sampling was allowed to proceed for 900,000 generations, of which 100,000 were discarded as burn-in. The resulting consensus tree and its clade credibility values are shown in Figure 1A.
Splice junction microarray analysis
For each microarray hybridization, 1 μg of total RNA from the LS2 knockdown and 1 μg of total RNA from a nonspecific knockdown were amplified and converted to aRNA using the MessageAmp II aRNA Amplification kit following the manufacturer recommendations (Ambion) and labeled with Cy5 and Cy3 monoreactive dye, respectively (GE Healthcare). The custom splicing-sensitive microarray used was based on FlyBase version 5.15 and interrogates 49,364 annotated splicing events from 13,344 different genes with three overlapping 36-nt oliognucleotide probes: one centered at the splice junction, and two probes offset by 3 nt on each side of the splice junction. In addition, one fully exonic probe per 100 nt of each mRNA, on average, were added. The 348,650 different probes were distributed randomly onto two custom Agilent 220K arrays and used for hybridization of each cDNA sample. The microarrays were then processed and scanned following the manufacturer's recommendation (Agilent Technologies). The Feature Extraction reports were loaded into R (http://www.r-project.org) and Lowess-normalized using the marray package (Gentleman et al. 2004; Smyth 2004). The genes with affected alternative splicing were first identified using ANOVA, comparing the group of exonic probes common to all transcripts with the different groups of splice junction probes corresponding to every splicing event of a given gene. The genes with Q-values <0.001 (adjusted using Benjamini-Hocheberg correction) were then subjected to t-tests to identify the group of junction probes significantly affected with a P-value of ≤0.001.
RNA SELEX
RNA SELEX was performed as described previously (Amarasinghe et al. 2001), with minor modifications described in the Supplemental Material.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays were performed using purified recombinant GST-tagged LS2 protein and in vitro transcribed RNA probes. For detailed methods, see the Supplemental Material.
LS2/dU2AF38 interaction assays
GST pull-downs were performed using GST-tagged dU2AF50 protein or GST-tagged LS2 protein, purified as described above. Recombinant dU2AF38 protein was also expressed as described above. To 1 mL of dU2AF38-expressing Escherichia coli lysate, 50 μg of purified dU2AF50 large subunit was added. The final concentrations of the GST-LS2 and dU2AF38 proteins were ∼650 nM and 400 nM, respectively. The reaction mixture was then rotated for 1 h at 4°C. Fifty microliters of glutathione agarose beads, washed in buffer A (see GST-LS2 protein purification, above) was then added and the reaction was rotated for another hour at 4°C. The beads were then pelleted and washed four times with 1 mL of buffer A. The beads were then boiled in 50 μL of protein sample buffer. Samples were then run on an SDS-PAGE gel and analyzed by Coomassie staining (data not shown) or immunoblotting. For the coimmunoprecipitation of LS2 and dU2AF38 proteins, RNP-enriched nuclear extracts from S2 cells stably expressing polyoma-tagged LS2 protein were used (Pinol-Roma et al. 1990). RNP-enriched extract was stored in HNEB2 (10 mM HEPES at pH 7.6, 100 mM KCl, 2.5 mM MgCl2, 0.2% NP-40, 0.2 mM PMSF). Twenty-five microliters of protein G beads (GE Healthcare) containing cross-linked anti-polyoma (GLU–GLU) antibodies was washed three times with 1 mL of HNEB2. Fifty microliters of RNP-enriched extract was then added and the reaction was incubated for 1 h at 4°C with rotation. For RNase-treated extracts, the extracts were pretreated with 20 μg/mL RNase A for 20 min at room temperature and then allowed to continue digestion during incubation with the anti-polyoma beads (1 h at 4°C). The beads were then washed four times with 1 mL of wash buffer (20 mM HEPES-KOH at pH 7.6, 400 mM LiCl, 2.5 mM MgCl2, 0.2% NP-40, 0.2 mM PMSF, 0.5 mM DTT). The beads were then boiled in 25 μL of SDS protein sample buffer and analyzed by SDS-PAGE and immunoblotting.
Motif enrichment in affected transcripts
The SELEX data were analyzed using MEME (Bailey and Elkan 1994), and the position-specific scoring matrix (PSSM) of the preferred LS2-binding sites was used to search the LS2-affected transcripts identified from the splice junction microarray, compared with the rest of the expressed transcriptome not affected by LS2 RNAi knockdown. The relative fraction of transcripts containing at least one LS2-binding site with different motif scores was calculated and plotted. The error bars in Figure 2E correspond to the bootstrapped standard deviation of the population of transcripts at the different motif score.
Motif placement in affected transcripts
In order to analyze the enrichment of the LS2-binding motif(s) in genes with LS2 RNAi knockdown-affected alternative splicing, only simple alternative splicing patterns corresponding to alternative cassette exons, competing donor sites, competing acceptor sites, and intron retention events were considered for modeling purposes. The affected alternative splicing events from every simple splice pattern type were further divided into two groups, either positively or negatively affecting exon inclusion of the longer isoform. A 400-nt region surrounding each affected splice site of the corresponding splicing events was used to identify the best motif score within a window of 50 nt. For each window, a t-test was performed to compare the population of motif scores from the affected events with the population of the best motif scores of the corresponding window in all of the other known Drosophila alternative splicing events of a given type but not affected in the LS2 RNAi knockdown samples. The P-values from these tests were plotted below every splice type analyzed. The red bars in Figure 5A correspond to regions of at least five consecutive best score windows with P-values of ≤0.05 separated by at most three windows above the P-value cutoff.
Testis enrichment of LS2 protein and its pre-mRNA targets
Whole-cell lysates containing approximately half of a whole fly, one head, and one pair of testes were separated by SDS-PAGE and immunoblotted for the PSI and LS2 proteins. GO analysis of the LS2-affected transcripts was done using Babelomics version 3.2 (http://babelomics3.bioinfo.cipf.es; Al-Shahrour et al. 2006). The testis mRNA expression levels of LS2 RNAi knockdown-affected transcripts were calculated using expression microarray data from FlyAtlas (http://www.flyatlas.org; Chintapalli et al. 2007). The mean fluorescence level from four independent Affymetrix Dros2.0 expression arrays was used as the testes expression level of that particular gene. For the S2 cell data sets, only genes identified as present in S2 cells in at least one out of four microarray experiments were used. For the LS2 RNAi knockdown-affected data set, only the 168 genes whose splicing was changed upon knockdown of LS2 were used.
In vivo splicing assays
Cassette exon constructs containing exons 1–3 of the PEP (CG6143) gene either without or with two LS2 recognition motifs (GGCGGCGGTGGGGGGTGGTGGCGGG) or a neutral motif (TGCACCCTCTGATGCACCCTCTGA) inserted 60 nt upstream of the cassette exon were created using overlap PCR. These constructs were cloned into pMT-V5-His (Invitrogen) and their expression was under the control of the metallothionein promoter. To overexpress LS2, we used an LS2-cDNA cloned into pUC-hyg-MT such that expression of LS2 was also under control of the metallothionein promoter. Twenty-four hours before transfection, 2 mL of 1 × 106 cells per milliliter were seeded in a six-well plate. The cells were then transfected with 0.5 μg each of PEP − motif, PEP + motif, or PEP + neutral motif-containing plasmid DNA, and pUC-hyg-MT-LS2 or blank pUC-hyg-MT plasmids. Transfections were done using Effectene (Qiagen). One day later, Cu2SO4 was added to 50 μM. Two days after copper addition, the cells were harvested and total RNA was isolated. Reverse transcription was done using random hexamers, and PCR was done using specific primers that amplify the exogenous PEP and not the endogenous PEP. The quantities and sizes of the RT–PCR products were then analyzed using an Agilent 2100 Bioanalyzer.
In vitro splicing assays
In vitro splicing assays were performed as described previously (Padgett et al. 1983).
Similar to the in vivo splicing assays, an LS2 recognition motif was inserted into the ftz intron 65 nt upstream of the 3′ splice site. The ftz intron was transcribed in vitro using T7 RNA polymerase and α32P-UTP. It was then gel-purified using a 5% polyacrylamide denaturing gel. In vitro splicing reactions were then set up in 20-μL final volumes with 8 μL of HeLa nuclear extract, 8 μL of 2.5× SP mix, 3 μL of LS2/dU2AF38 heterodimer (∼500 ng) or blank buffer, and 1 μL of RNA (20 fmol, ∼20,000 counts per minute [cpm]). SP mix (2.5×) contained the following: 5 mM ATP, 50 mM creatine phosphate, 25% glycerol, 50 mM HEPES (pH 7.6), 7.5% PEG 8000, 62.5 mM potassium glutamate, and 10 mM MgCl2. The KCl concentrations of the HeLa nuclear extract and heterodimer fractions were both 100 mM. The final concentrations of glutamate, chloride, and potassium were therefore 25 mM, 55 mM, and 80 mM, respectively. The reactions were incubated for 3 h at 30°C, then phenol/chloroform-extracted, ethanol-precipitated, and washed once with 70% ethanol. They were then resuspended in 10 μL of urea/bromophenol blue/xylene cyanol and subjected to denaturing gel electrophoresis on a prerun 0.4-mm-thick 12% polyacrylamide-urea gel for 7 h at 25 W. The gel was then fixed for 20 min in 10% methanol and 10% acetic acid. It was then dried and exposed using a PhosphorImager. Quantitation was done using a Typhoon PhosphorImager with ImageQuant software (GE Healthcare). Quantitation was done by first normalizing the intensity of each band according to its length in nucleotides. The spliced ratio was then calculated by adding up the intensities of all splicing intermediates and products and dividing by the unspliced pre-mRNA. Those results were then normalized by setting the splicing efficiency of wild-type ftz pre-mRNA in the absence of heterodimer to 1.0.
RNA immunoprecipitation (RIP) of predicted LS2 target pre-mRNAs
Polyoma (Glu–Glu)-tagged LS2 protein was immunoprecipitated from S2 nuclear RNP-enriched extracts (Pinol-Roma et al. 1990). The extracts were stored in HNEB2 (see above). Nine-hundred microliters of extract was incubated with 100 μL of beads containing anti-polyoma antibodies that had been washed four times with 1 mL HNEB2. As a control, an immunoprecipitation using IgG was also done. The reaction was incubated for 4 h at 4°C. The resin was then washed four times with 1 mL of wash buffer (20 mM HEPES at pH 7.6, 100 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.2 mM PMSF, 50 U/mL RNasin [Promega]). The beads were then resuspended in 100 μL of 1× RQ1 DNase buffer (Promega). Five units of RQ1 DNase (Promega) was then added and the reaction was incubated for 1 h at 37°C. RNA was then eluted by phenol/chloroform-extracting the beads and ethanol-precipitating. The pellet was washed twice with 1 mL of 70% ethanol. The pellet was then resuspended in 15 μL of H2O. RNA concentration was measured using a Nanodrop spectrophotometer. Equal amounts of polyoma or IgG-immunoprecipitated RNA and RNA isolated from the starting RNP-enriched extracts were then used for RT–PCR using random hexamers. Individual bound transcripts were then assayed using HotStart PCR (Qiagen) and gene-specific primers.
Testes immunofluorescence
Testes from 1- to 3-d-old males expressing GFP-tagged His2Av were dissected into cold Ringers solution (0.35 g NaCl in 50 mL of water). Approximately 10 pairs of testes were then fixed using 1× PBX (PBS supplemented with 0.1% Triton X-100 and 0.5% BSA) with 4% formaldehyde for 15 min at room temperature. The testes were then washed three times with 1× PBX for 2 min each. Blocking was done for 1 h at room temperature in 1 mL of 2% normal goat serum. Fixed testes were then incubated with primary antibody diluted 1:500 in 1× PBX overnight at 4°C. They were then washed three times with 1 mL of 1× PBX for 15 min each. Secondary (donkey anti-rabbit AlexaFluor 568) was then added at 1:400 dilution for 2 h at room temperature. Testes were then washed three times with 1 mL of 1× PBX for 15 min each. Testes were then mounted on a slide and imaged using an Axioimager 373 microscope.
Acknowledgments
We thank J. Aspden, M. Francis, S. Majumdar, and M. Harrison for helpful comments and discussion; A. Brooks for discussion and assistance concerning bioinformatics; B. Shapiro for assistance with phylogenetic analysis; and L. Jones for the Histone-GFP fly stock. We also thank Lisa Isailovic, Nik Chmiel, and Jennifer Doudna for some early contributions to this work. This work was supported by NIH grant R01GM61987. J.M.T., M.B., and D.C.R. conceived the experiments. J.M.T. purified the proteins, generated antibodies and cell lines, did the biochemical and molecular biological experiments, and performed part of the bioinformatic analyses at University of California at Berkeley. N.A. and M.B. performed bioinformatics motif-finding analysis, RNAi knockdowns, and splice junction microarray analysis at the Stowers Institute. R.E.G. performed the phylogenic analysis on the U2AF and LS2 genes. J.M.T., N.A., R.E.G., M.B., and D.C.R. wrote the paper.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2009011.
Supplemental material is available for this article.
References
- Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y 2009. A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol 10: R30 doi: 10.1186/gb-2009-10-3-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J 2006. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res 34: W472–W476 doi: 10.1093/nar/gkl172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe AK, MacDiarmid R, Adams MD, Rio DC 2001. An in vitro-selected RNA-binding site for the KH domain protein PSI acts as a splicing inhibitor element. RNA 7: 1239–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Green P 2005. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc Natl Acad Sci 102: 12813–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Casola C, Betran E 2008. Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics 9: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Casola C, Betran E 2009. Quality of regulatory elements in Drosophila retrogenes. Genomics 93: 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Baker BS, Gorman M, Marin I 1994. Dosage compensation in Drosophila. Annu Rev Genet 28: 491–521 [DOI] [PubMed] [Google Scholar]
- Ben-Dov C, Hartmann B, Lundgren J, Valcarcel J 2008. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem 283: 1229–1233 [DOI] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M 2002. Retroposed new genes out of the X in Drosophila. Genome Res 12: 1854–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Brenner SE, Rio DC 2005. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev 19: 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Green RE, MacArthur S, Brooks AN, Brenner SE, Eisen MB, Rio DC 2009. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Mol Cell 33: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, Graveley BR 2011. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res 21: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Posner JB, Darnell RB 1993. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron 11: 657–672 [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG 1993. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 62: 289–321 [DOI] [PubMed] [Google Scholar]
- Elliott DJ, Grellscheid SN 2006. Alternative RNA splicing regulation in the testis. Reproduction 132: 811–819 [DOI] [PubMed] [Google Scholar]
- Gan Q, Chepelev I, Wei G, Tarayrah L, Cui K, Zhao K, Chen X 2010. Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Res 20: 763–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Roche SE, Beall EL, Green MR, Rio DC 1993. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science 262: 569–573 [DOI] [PubMed] [Google Scholar]
- Kent OA, Reayi A, Foong L, Chilibeck KA, MacMillan AM 2003. Structuring of the 3′ splice site by U2AF65. J Biol Chem 278: 50572–50577 [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK 2001. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106: 595–605 [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Ichikawa M, Arai J, Tateiwa H, Fu L, Higuchi K, Yoshimura N 2000. Molecular cloning and characterization of a new neuron-specific homologue of rat polypyrimidine tract binding protein. J Biochem 128: 811–821 [DOI] [PubMed] [Google Scholar]
- Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, et al. 2004. Distinct localization of histone H3 acetylation and H3–K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci 101: 7357–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol 20: 7463–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B 2007. hnRNP proteins and splicing control. Adv Exp Med Biol 623: 123–147 [DOI] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841 [DOI] [PubMed] [Google Scholar]
- Nelson KK, Green MR 1988. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev 2: 319–329 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver B 2002. Genetic control of germline sexual dimorphism in Drosophila. Int Rev Cytol 219: 1–60 [DOI] [PubMed] [Google Scholar]
- Padgett RA, Hardy SF, Sharp PA 1983. Splicing of adenovirus RNA in a cell free transcription system. Proc Natl Acad Sci 80: 5230–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299: 697–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR 2004. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci 101: 15974–15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S, Swanson MS, Matunis MJ, Dreyfuss G 1990. Purification and characterization of proteins of heterogeneous nuclear ribonucleoprotein complexes by affinity chromatography. Methods Enzymol 181: 326–331 [DOI] [PubMed] [Google Scholar]
- Proschel M, Zhang Z, Parsch J 2006. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics 174: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Breger KS, Kanaar R, Adams MD, Rio DC 1998a. RNA binding activity of heterodimeric splicing factor U2AF: at least one RS domain is required for high-affinity binding. Mol Cell Biol 18: 4004–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Kanaar R, Breger KS, Rio DC 1998b. Interaction between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol Cell Biol 18: 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B, Zamore PD, Green MR 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52: 207–219 [DOI] [PubMed] [Google Scholar]
- Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Kramer A, Sattler M 2003. Structural basis for the molecular recognition between human splicing factors U2AF(65) and SF1/mBBP. Mol Cell 11: 965–976 [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ 2009. The SR protein family. Genome Biol 10: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL 2006. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell 23: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176 [DOI] [PubMed] [Google Scholar]
- Singh R, Banerjee H, Green MR 2000. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor sex-lethal. RNA 6: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Sridharan V, Heimiller J, Singh R 2011. Genomic mRNA profiling reveals compensatory mechanisms for the requirement of the essential splicing factor U2AF. Mol Cell Biol 31: 652–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol 21: 36–44 [DOI] [PubMed] [Google Scholar]
- Venables JP, Eperon I 1999. The roles of RNA-binding proteins in spermatogenesis and male infertility. Curr Opin Genet Dev 9: 346–354 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835 [DOI] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Nutiu R, Wang ET, Burge CB 2009. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat Struct Mol Biol 16: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW 2008. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell 135: 1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Green MR 1989. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci 86: 9243–9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR 1992. Cloning and domain analysis of the mammalian splicing factor U2AF. Nature 355: 609–614 [DOI] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838 [DOI] [PubMed] [Google Scholar]