Abstract
We cloned the gene that encodes prothoracicotropic hormone (PTTH) in the northern house mosquito, Culex pipiens, investigated its expression profile in short-day (diapause-destined) and long-day (nondiapause-destined) individuals from the fourth instar larval stage through two-months of adulthood, as well as after a blood meal. The deduced Cupip-PTTH amino acid sequence contains seven cysteines with a specific spacing pattern. Sequence alignment suggests that Cupip-PTTH is 23% identical to Drosophila melanogaster PTTH, but is ≥ 59% identical to the PTTHs of other mosquitoes. Cupip-PTTH has structural characteristics similar to Bombyx mori PTTH and some vertebrate nerve growth factors with cysteine-knot motifs. PTTH transcripts exhibit a daily cycling profile during the final (fourth) larval instar, with peak abundance occuring late in the scotophase. The fourth larval instar is one day longer in short-day larvae than in long-day larvae, resulting in larger larvae and adults. This additional day of larval development is associated with one extra PTTH cycle. No cycling was observed in pupae, but PTTH transcripts were slightly higher in short-day pupae than in long-day pupae throughout much of the pupal stage. PTTH expression persisted at a nearly constant level in diapausing adult females for the first month but then dropped approximately 50%, while expression decreased at the beginning of adulthood in nondiapausing females and then remained at a low level as long as the females were denied a blood meal. But, when nondiapausing females were offered a blood meal, PTTH transcripts rose approximately 7-fold in 2 h and remained elevated for 24 h. A few diapausing females (~ 10%) will take a blood meal when placed in close proximity of a host, but much of the blood is ejected and such meals do not result in mature eggs. Yet, elevated PTTH mRNA expression was also observed in diapausing females that were force fed. Our results thus point to several distinctions in PTTH expression between short-day and long-day mosquitoes, but both types of females responded to a blood meal by elevating levels of PTTH mRNA.
Keywords: PTTH, gene expression, diapause, development time, body size, day length, blood meal, Culex pipiens
Introduction
The concept that the insect brain controls molting and metamorphosis was first demonstrated in pioneering experiments on the gypsy moth Lymantria dispar by Kopeć (1922). Using diapausing Hyalophora cecropia as a model, Williams (1947) described a hormone from the brain that stimulates the prothoracic glands to secrete a hormone essential for metamorphosis, a hormone later identified as ecdysone (Hanser & Karlson, 1957). This brain factor, now known as prothoracicotropic hormone (PTTH), has been well characterized and examined in Lepidoptera, especially in Bombyx mori and Manduca sexta. (Rybczynski, 2005; Marchal et al., 2009).
Sequence alignments of known and deduced PTTH amino acid sequences indicate that PTTH is not highly conserved among insect orders as its function might suggest. However, one consistent feature is the presence of seven highly conserved cysteine residues located in the mature PTTH protein. PTTH is also thought to be a homodimeric protein linked by an interchain disulfide bond formed by one of these cysteine residues, while the other six cysteine residues form three intrachain disulfide linkages (Ishibashi et al., 1994). The similarity of the disulfide-bond pattern between Bombyx PTTH and the vertebrate growth factor family with cysteine-knot motif, such as transforming growth factor-β2 (TGF-β2), β-nerve growth factor (β-NGF) and platelet-derived growth factor-BB (PDGF-BB) suggests that PTTH shares a common ancestor with these growth factors (Noguti et al., 1995). Thus, seven cysteine residues with similar spacing to Bombyx PTTH is the most highly conserved characteristic of the PTTH sequences.
Although most studies on PTTH have been conducted in Lepidoptera, the PTTH gene from Drosophila melanogaster was recently characterized, and its function was examined by ablating PTTH-producing neurons; surprisingly PTTH is not essential for Drosophila molting and metamorphosis but instead regulates developmental timing and body size (McBrayer et al., 2007). Whether this is also true for other Diptera and other orders remains unknown. Among mosquitoes, a PTTH gene, located on the X chromosome, was first noted in the genome of Anopheles gambiae (Riehle at al., 2002). Genome and EST sequences from two other mosquitoes, Aedes aegypti and Culex quiquenfasiatus, also revealed homologous PTTH genes (McBrayer et al., 2007; Marchal et al., 2009). Thus far, there are no specific reports on expression or functional analysis of PTTH in these important disease vectors.
In the present study, we cloned and characterized a PTTH gene from another mosquito, Culex pipiens, one of the infamous West Nile virus vectors. Cx. pipiens females overwinter in an adult reproductive diapause programmed by the short daylength and low temperatures of late summer and early autumn (Spielman & Wong, 1973). Nondiapausing females readily take a blood meal and produce eggs. By contrast, diapausing females rarely take a blood meal and if they do so, they eject most of the blood without converting the protein into mature eggs (Mitchell & Briegel, 1989). We investigated the expression pattern of the PTTH gene from the fourth instar until two months into the adult stage for both short- and long-day individuals, and also examined the expression of PTTH mRNA after blood feeding. Though PTTH has been linked to the regulation of pupal diapause (Denlinger et al., 2005), there are no reports examining a potential role for this hormone in other diapausing stages. We report a cyclic pattern of PTTH transcripts expression during the fourth larval instar, and higher expression levels early in the pupal stage and during early adulthood in mosquitoes programmed for diapause than in those not programmed for diapause. We also note that blood feeding elicits a rapid boost in PTTH mRNA expression in both diapausing and nondiapausing females.
Results
Cloning, characterization, and sequence analysis of a PTTH cDNA in Cx. pipiens
Using the deduced protein sequence of the PTTH gene in D. melanogaster (ID: NP_608537) as the query, a protein blast was performed against the NCBI database, and a conserved hypothetical protein (ID: XP_ 001844784) was detected in Cx. quinquefasciatus. Based on the corresponding coding sequence in its genome, gene specific primers, CPPTFP1 and CPPTRP1 (Fig. 1), were designed and synthesized for PCR to obtain a corresponding 485-bp internal fragment from a Cx. pipiens cDNA pool. This fragment encodes 161 amino acid residues in which only 2 amino acid residues (D→N, and E→A) are different between Cx. quinquefasciatus and Cx. pipiens, thus suggesting that our cloned fragment is indeed PTTH from Cx. pipiens.
Figure 1.
Structure of cDNA sequences and deduced amino acid sequences of PTTH in Culex pipiens. The start codon (ATG) and stop codon (TAA) are shaded and boxed; the putative polyadenylation signal (ATTAAA) is underlined. Sequences of the primers used for obtaining internal fragments (CPPTFP1 and CPPTRP1) and RACE products (CPPTGSP1 and CPPTGSP2) are underlined and arrowed from the 5′ to 3′-end. The deduced mature PTTH amino acid sequence is shaded.
RACE was employed to clone the full-length PTTH cDNA sequence using primers CPPTGSP1 for the 3′-end and CPPTGSP2 for the 5′-end (Fig. 1). 5′-end RACE yielded a 529-bp fragment including a 13-bp 5′-untranslated region (UTR) and a 516-bp partial open reading frame (ORF) encoding 172 amino acid residues. 3′-end RACE yielded a 478-bp fragment including a 373-bp partial ORF encoding 123 amino acid residues and a 105-bp 3′-UTR. There is a 246-bp overlap between the 5′-end RACE product and the 3′-end RACE product. The above results indicate a 760-bp PTTH cDNA with a 642-bp ORF starting at nucleotide 14 and terminating at nucleotide 655, which encodes 213 amino acid residues with a deduced molecular mass of 24 KDa and a calculated pI of 6.9. Location of the PTTH start codon (ATG) was deduced by similarities to those in D. melanogaster and Cx. quinquefasciatus (Fig. 2A). In addition, a putative polyadenylation signal (ATTAAA) was located at nucleotide 653. The cloned Cx. pipiens PTTH cDNA sequence was deposited in GenBank and assigned accession number HM246665.
Figure 2.
Sequence alignment and phylogenetic analysis of PTTH amino acid sequences indicate that Cupip-PTTH is more similar to its counterparts in other mosquitoes than to Drosophila. A: Sequence of Cupip-PTTH was compared with PTTHs from Culex quinquefasciatus (Cuqui), Aedes aegypti (Aeaeg), Anopheles gambiae (Angam) and Drosophila melanogaster (Drmel). Open diamond indicates the putative cleavage sites that release mature PTTH, and seven filled triangles indicate seven conserved cysteines. Conserved amino acid residues are indicated as white letters in a black background; similar residues are indicated as white letters in a gray background; non-conserved residues are indicated as black letters in a white background; gaps, indicated as broken lines, are inserted to optimize the alignment. The numbers of amino acid residues are designated arbitrarily for sequence alignment and are not their exact positions in the full sequences. B: Evolutionary relationship of Culex pipiens, Culex quinquefasciatus, Aedes aegypti, Anopheles gambiae, and Drosophila melanogaster based on PTTH amino acid sequences. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007). Evolutionary history is inferred using the Neighbor-Joining method and evolutionary distances were computed using the Poisson correction method. The tree is drawn to scale, with branch lengths in units of the number of amino acid substitutions per site.
The deduced amino acid sequence of Cupip-PTTH was aligned with homologs from three other mosquitoes including Cx. quinquefasciatus (ID: XP_001844784), Aedes aegypti (ID: DV370510), Anopheles gambiae(ID: XP_555854), and D. melanogaster, and the results indicate that the three mosquito PTTHs are quite similar, especially for the deduced mature PTTH, but very low homology is observed between Cx. pipiens and D. melanogaster as shown in Fig. 2A. Not unexpectedly, the most identical homolog of Cupip-PTTH is Cuqui-PTTH, in which only 3 amino acid residues out of 213 differ (aa7: S →A, aa38: D→N, and aa92: A→E for Cupip-PTTH, see Fig. 2A); the mature Cupip-PTTH is completely identical to Cuqui-PTTH. At the amino acid level, deduced whole Cupip-PTTH is 70% identical to Aeaeg-PTTH, 59% identical to Angam-PTTH, but only 23% identical to Drmel-PTTH. The deduced mature Cupip-PTTH is 86% identical to Aeaeg-PTTH, 84% identical to Angam-PTTH, but only 39% identical to Drmel-PTTH.
These five PTTHs were subjected to a simple phylogenetic analysis by MEGA4 (Fig. 2B), and the results comply well with the known taxonomy. Four mosquitoes are well segregated from D. melanogaster; in the four mosquitoes, based on the evolutionary distance, Cx. quinquefasciatus is the closest species to Cx. pipiens, and Ae. aegypti is closer to Cx. pipiens than An. gambiae. These results indicate that PTTH may be a good candidate gene for discerning phylogeny within the Diptera.
Structural prediction of Cupip-PTTH
Both online programs used here produced similar stereo-structures of Cupip-PTTH, based on a similar structure model from the Protein Data Bank (PDB) database. The structure with the highest accuracy score and full length sequence produced by I-TASSER is shown in Fig. 3. There are four major antiparallel β-strands forming two β-sheets, with 6 cysteines forming three intra-chain disulfate bonds that reside mainly in the coil regions and cluster on one side of the β-sheets. The deduced structure of Cupip-PTTH is consistent with the predicted structure of Bombyx mori PTTH and other growth factors with cysteine-knot motifs in vertebrates as reported previously (Noguti et al., 1995).
Figure 3.
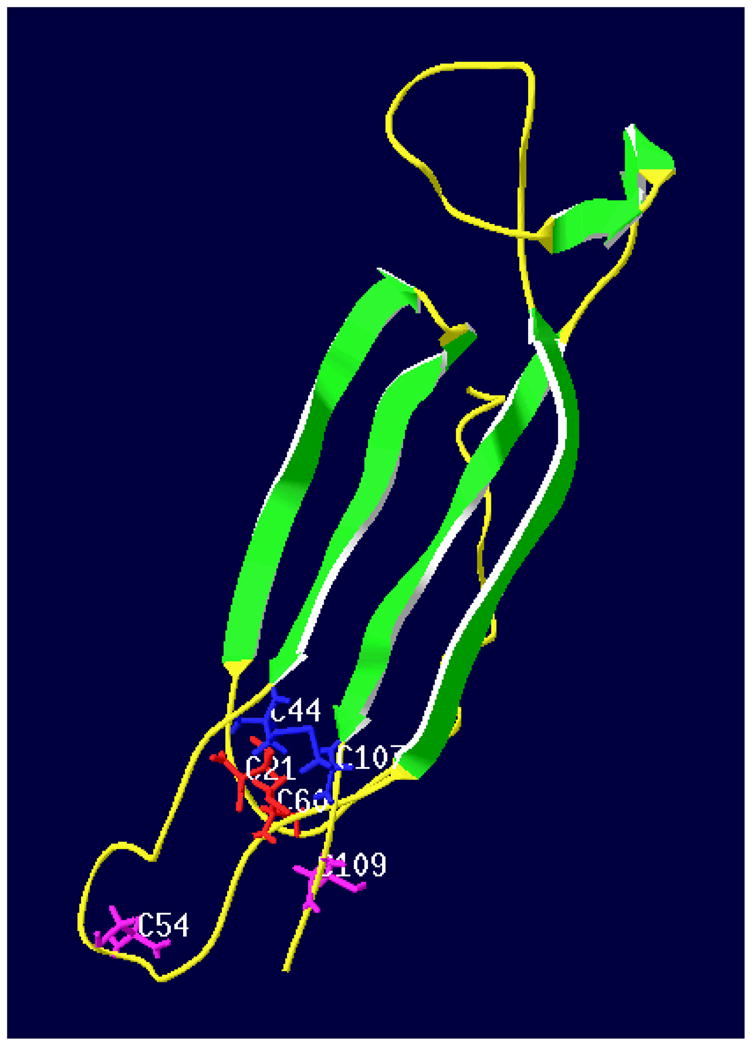
Three-dimensional structure of Cupip-PTTH. Green ribbons represent 6 β-strands including two β-sheets, each containing two antiparallel β-strands. The β-strands are arrowed from N-terminal to C-terminal. Yellow threads indicate coil areas. Six cysteines form three intra-chain disulfate bonds shown as C21, C44, C54, C60, C107, and C109. Groups including side chains of cysteines are colored in red (C21 and C60), blue (C44 and C107), and purple (C54 and C109) to show the formation of the three disulfate bonds. Predicted by I-TASSER program and viewed by SWISS-Pdb Viewer Deep View v4.0.1.
Distribution of PTTH mRNA using Northern blots
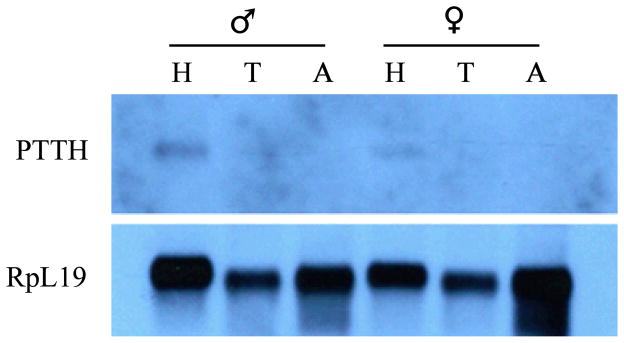
Northern blots indicate that PTTH mRNAs are present in both female and male adults as shown in Fig. 4. The PTTH mRNA was mainly present in the head, presumably within the brain, but can not be detected in thoracic or abdomen segments of either male or female adults by Northern blot.
Figure 4.
Segment distribution of PTTH mRNA in both sexes including head (H), thorax (T), and abdomen (A). Total RNA was isolated from above segments and 1~1.5 μg mRNA was further purified from total RNA and subjected to Northern blot analysis. PTTH mRNA was detected only in the heads of males and females and not in the thorax or abdomen. RpL19 mRNA was used as an internal control.
Genomic organization and Southern blot analysis of the PTTH gene
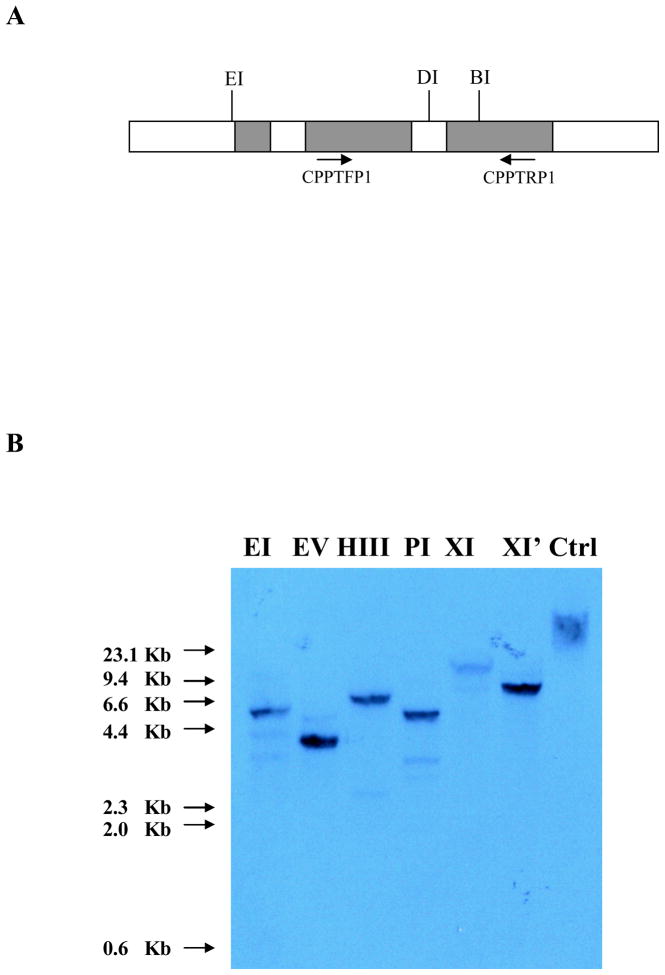
The cloned Cupip-PTTH gene contains 3 exons, 2 introns, and small pieces of the 5′-end and 3′-end sequences. The sequence was deposited in GenBank and assigned accession number HM246666. Aschematic diagram of the PTTH gene is presented in Fig. 5A, with 3 restriction enzyme sites indicated. Primers CPPTFP1 and CPPTRP1 (see Fig. 5A) were employed to obtain the probe for Southern blot analysis. The known genomic sequence of PTTH was subjected to restriction enzyme site analysis, and none of the six restriction enzyme sites, including EcoRI, EcoRV, HindIII, PstI, XbaI, and XhoI, were present in the exon or intron sequences. Thus, these enzymes were employed to digest genomic DNA, and only one major band appeared in each lane (Fig. 5B). The results indicate that only one copy of the PTTH gene is present in the Cx. pipiens haploid genome.
Figure 5.
Schematic diagram showing the structure of Cupip-PTTH gene and Southern blot analysis. A: Cupip-PTTH gene structure and restriction map. Three filled boxes indicate exons, and the open boxes indicate introns. Three restriction enzyme sites are shown as EcoRI (EI), DraI (DI), and BamHI (BI). CPPTFP1 and CPPTRP1 were the primers used for obtaining hybridization probes. B: Southern blot analysis. Ten μg of genomic DNA was digested by 50 units’ restriction endonucleases EcoRI (EI), EcoRV (EV), HindIII (HIII), PstI (PI), XbaI (XI), and XhoI (XI′) respectively; only one major band was observed in each lane. Non-digested DNA was used as a control (Ctrl); sizes of DNA molecular weight markers are shown at left.
Extended larval development in short-day fourth instar larvae
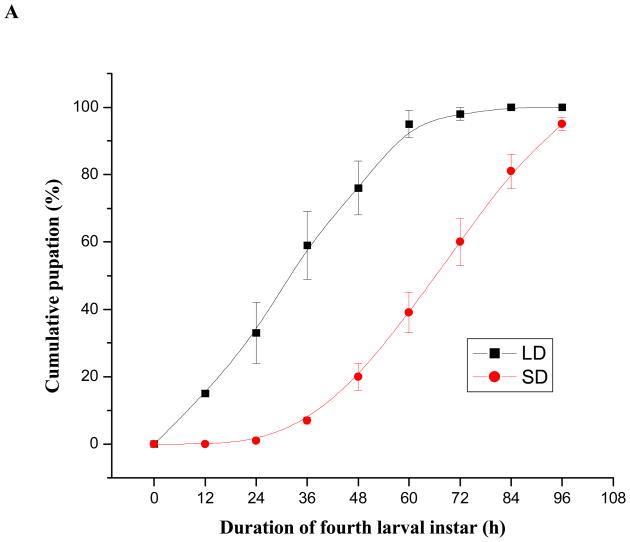
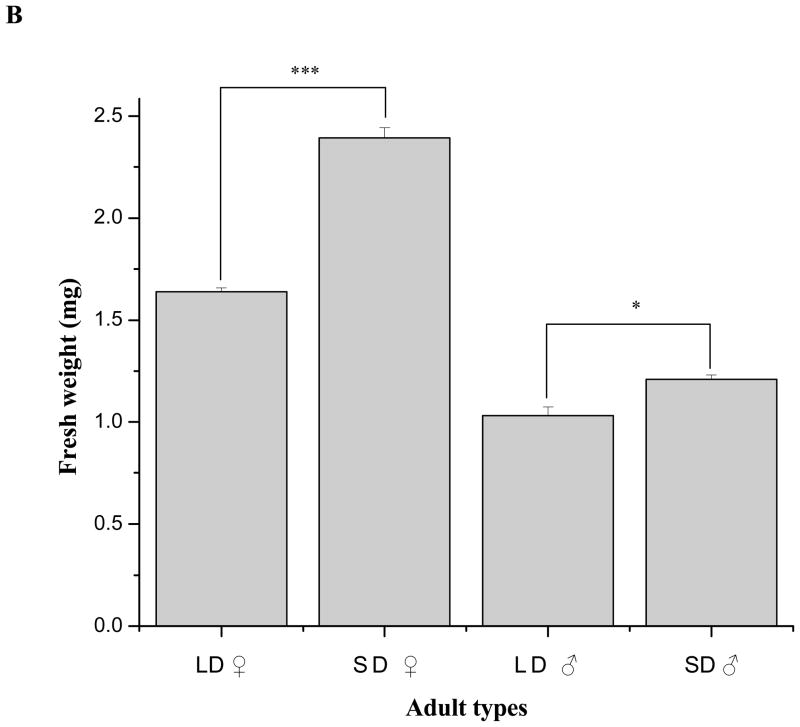
Short-day fourth instar larvae required approximately 24 h longer to complete the final larval instar than long-day larvae (Fig. 6A). Correlated with this increased duration of the final larval instar fresh body mass of newly-eclosed diapausing adults was significantly greater than that of corresponding nondiapausing adults (Student’s t-test, P<0.05); this was especially evident in females, with mean masses of 2.39 mg for diapausing females and 1.64 mg for nondiapausing females (Fig. 6B) (Student’s t-test, P<0.01). These very significant differences were also reflected in dry masses: 0.62 mg for diapausing females and 0.41 mg for nondiapausing females (Student’s t-test, P<0.01). In addition, statistically significant differences in fresh mass were observed in short- and long-day males as shown in Fig. 6B (Student’s t-test, P<0.05). Significant differences were also reflected in male dry masses: 0.33 mg for diapausing males and 0.26 mg for nondiapausing males (Student’s t-test, P<0.05).
Figure 6.
Comparisons of (A) larval development duration at 12h intervals in short-day (diapause-destined) and long-day (nondiapause-destined) individuals during the final fourth larval instar and (B) newly-molted adult wet weight. These results show that duration of the final larval instar of short-day larvae was approximately 24 h longer than that of long-day larvae and resulted in heavier diapausing adults. A: Pupation time of short-day and long-day fourth instar larvae. LD and SD indicate long-day and short-day groups, respectively. Error bars represent standard deviations of three replicates; 40–42 individuals were measured in each replicate. B: Weights of newly molted male and female adults reared under SD and LD. Error bars represent standard deviations of three replicates; 15–17 individuals were counted in each replicate. Single asterisk and triple asterisks indicate significant differences and very significant differences between the two tested group, respectively (*P<0.05, ***P<0.001, one-way ANOVA).
Daily cylces of PTTH transcripts during the fourth instar
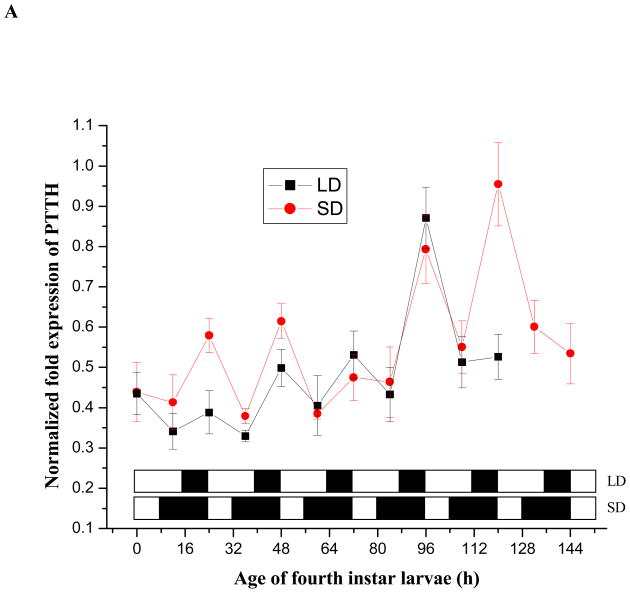
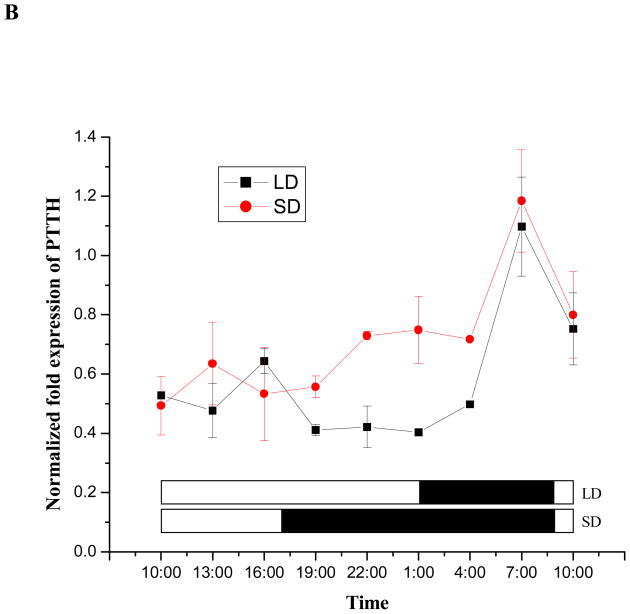
Results shown in Fig. 7A suggest a daily cycling pattern of PTTH mRNA in both short- and long-day larvae, with peak abundance occurring late in the scotophase. To further evaluate this response we also monitored PTTH mRNA expression levels at 3 h intervals from day 3 to day 4 in 4th instar larvae (Fig. 7B). In both short- and long-day larvae, the peak PTTH transcripts appeared at 7:00 AM, near the end of the scotophase. Then, mRNA levels decreased rapidly at the outset of the photophase and remained low throughout the remainder of the photophase and early scotophase (Fig. 7A and 7B). PTTH transcripts in short-day larvae began increasing slightly in the early scotophase, while expression remained low in long-day larvae that were still in the photophase (Fig. 7B). PTTH mRNA levels between short- and long-day larvae at this stage (19:00 PM-1:00 AM) were significantly different (Student’s t-test, P<0.05), but expression in both types of larvae peaked at the same time, near the end of the scotophase. Short-day fourth instar larvae remained as larvae one additional day and exhibited one additional PTTH mRNA cycle. In both types of larvae, PTTH mRNA content increased gradually throughout the fourth instar but dropped at pupation.
Figure 7.
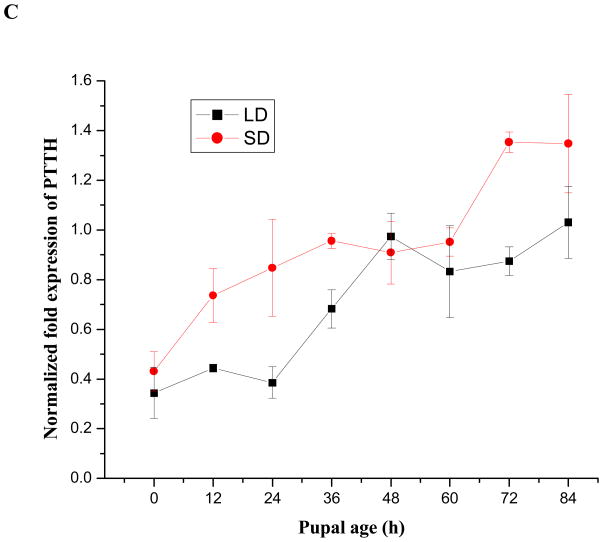
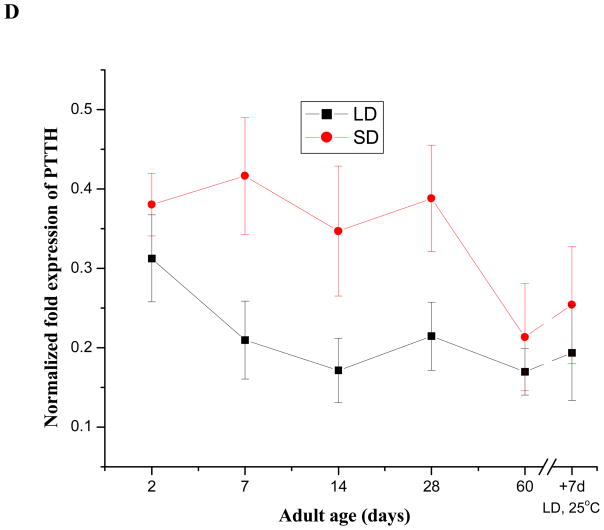
Expression patterns of PTTH mRNA in short-day(diapause-destined) and long-day (nondiapause-destined) fourth instar larvae (A), 24 h time course from day 3–10:00 AM to day 4–10: 00 AM in the fourth instar larvae at 3 h intervals (B), pupae (C) and adults (D) evaluated by qPCR. LD and SD indicate long-day and short-day groups respectively. Error bars represent standard deviations of three replicates; 15 individuals were sampled in each replicate. The lower panel in (A) and (B) indicates the corresponding photoperiods used to generate diapausing (short-day, 8 L: 16 D) and nondiapausing (long-day, 16 L: 8 D) adult females. The open and filled boxes represent photophase and scotophase, respectively. Male adults fail to enter diapause regardless of photoperiod and die when the sugar solution is removed, thus only females were monitored during the adult stage. Female adults were sampled from short-day and long-day groups until day 60. Then, day 60 females were transferred to 25°C and long days (16 L: 8 D) for 7 days to break diapause as indicated in (C).
Expression patterns of PTTH in pupae and adults
Expression patterns of PTTH mRNA in short- and long-day pupae were investigated at 12 h intervals: PTTH levels increased 3–3.5 fold throughout the 84 h pupal stage (Fig. 7C), and there was no evidence for a daily cycling pattern as observed in the fourth instar larvae. PTTH mRNA content was higher in short-day pupae than in long-day pupae during the early pupal stage (Student’s t-test, P<0.05), but expression levels were nearly the same late in the pupal stage.
Male adults of Cx. pipiens die without entering diapause, thus PTTH levels were investigated only in female adults. No differences were noted at the beginning of the adult stage (Day 2), but differences were evident in older adults (Fig. 7D) (Student’s t-test, P<0.05). The PTTH mRNA content persisted at a similar level in diapausing adults for approximately 4 weeks, and then dropped by approximately 50%. By contrast, PTTH levels decreased immediately at the onset of the adult stage in nondiapausing individuals and remained at a relatively low level as long as the females were denied a blood meal. When the females were fed only honey and water, PTTH mRNA content in both types of adults reached the lowest level after 2 months and increased only marginally when the females were transferred from 18°C to 25°C and a photoperiod of 16 L: 8 D (long days).
Upregulation of PTTH expression after blood feeding in both nondiapausing and diapausing females
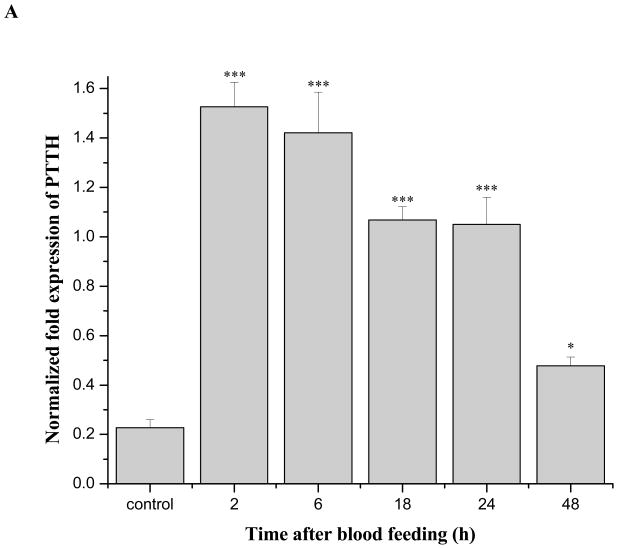
One month-old nondiapausing female adults that were offered a blood meal quickly elevated PTTH transcripts following a blood meal (Fig. 8A). A7-fold elevation was noted 2 h after the blood meal. PTTH mRNA content then decreased gradually but remained elevated 24 h after blood feeding. After 48 h, the PTTH transcript level was only one fold higher than that prior to feeding, but the difference was still statistically significant (Student’s t-test, P<0.01).
Figure 8.
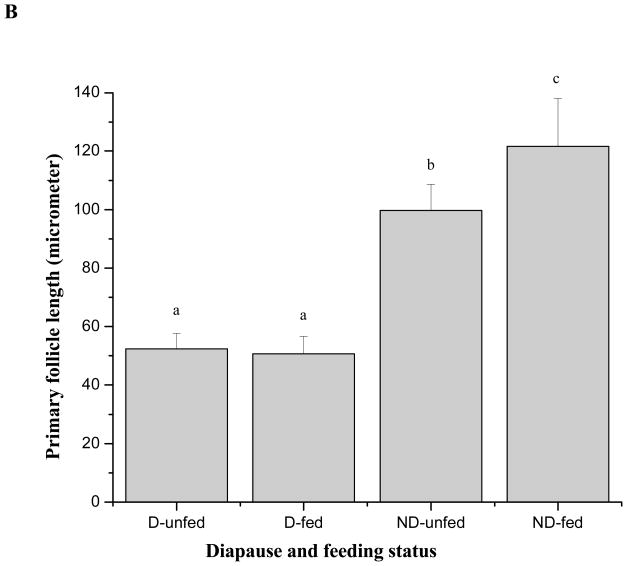
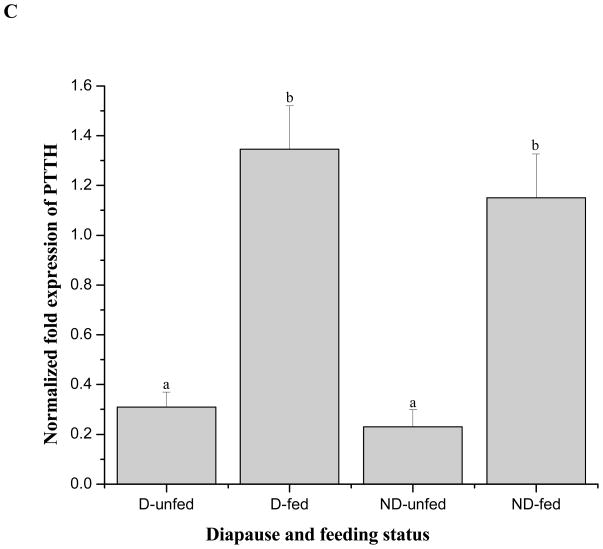
Expression of PTTH mRNA after blood feeding in nondiapausing and diapausing females. A: Time-course of PTTH mRNA expression after a blood meal in nondiapausing females. PTTH mRNA increased 7 fold within 2 h and the elevation persisted for at least 24 h. Nondiapausing females used in the experiment were 4-weeks old when offered a blood meal. Nondiapausing females fed a 10% sugar solution were used as controls. Error bars represent standard deviations of three replicates; 15 individuals were sampled in each replicate. Double asterisk and triple asterisks indicate significant differences and very significant differences between the tested group and control group, respectively (*P<0.05, ***P<0.001, one-way ANOVA). B: Length of primary follicles measured in diapausing unfed (D-unfed), diapausing fed (D-fed), nondiapausing unfed (ND-unfed), and nondiapausing fed (ND-fed) groups 5 h after the fed groups received a blood meal. Three follicles were selected randomly for measurement in each female, and 16 females were employed in each group. Bars with different letters are significantly different at P = 0.05, one-way ANOVA. C: Expression of PTTH mRNA in D-unfed, D-fed, ND-unfed, and ND-fed groups 5 h after the fed groups received a blood meal. Error bars represent standard deviations of three replicates; 10 individuals were sampled in each replicate. PTTH mRNA was elevated 4–5 fold after blood feeding in both diapausing and nondiapausing group. All mosquitoes were held at 18°C. Bars with different letters are significantly different at P = 0.05, one-way ANOVA.
Diapausing females were also offered a blood meal. Within 45 min, 64% of nondiapausing females (n=340) took a blood meal from host chickens, while only 10% of the diapausing females (n=850) did so within 3 h. Diapause status was verified by measuring and comparing follicle length in the various groups (Fig. 8B). Diapausing females that accepted a blood meal had the same length follicles as those that were unfed, and the size was significantly smaller than those in the nondiapausing groups (P<0.05, one-way ANOVA). Furthermore, much of the blood imbibed by the diapausing females was ejected (many spots of blood on the transparent plastic cage wall) within the next day after feeding, but no blood ejection was observed from nondiapausing females. All of this evidence indicates that the few short-day females that accepted a blood meal were indeed in diapause.
In spite of the fact that diapausing females did not utilize the blood meal to mature eggs, PTTH mRNA also increased approximately 4.4 fold in the blood-fed diapausing females 5 h after blood feeding, similar to the increase in blood-fed nondiapausing females (5 fold) as shown in Fig. 8C. These results suggest that PTTH is significantly elevated shortly after blood feeding regardless of the diapause status of the females (P<0.05, one-way ANOVA).
Discussion
Sequence alignment indicates huge variation in PTTH at the primary structure level, suggesting that the PTTH molecule evolved rapidly from a common ancestral gene. Following the first report on the cDNA structure of PTTH in B. mori (Kawakami et al., 1990), the structures of quite a few PTTH cDNAs have been described, including Samia cynthia (Ishizaki & Suzuki, 1994), Antheraea pernyi (Sauman & Reppert, 1996), Hyalophora cecropia (Sehnal et al., 2002), Manduca sexta (Shionoya et al., 2003), and several noctuids including Heliothis virescens (Xu & Denlinger, 2003), Helicoverpa zea (Xu et al., 2003), Helicoverpa armigera (Wei et al., 2005), and Spodoptera exigua (Xu et al., 2007). These PTTHs are exclusively from Lepidoptera. However, with the rapid development of genome sequencing projects, putative PTTHs were also noted from Diptera including D. melanogaster, An. gambiae (Riehle at al., 2002), and Cx. quinquefasciatus (Marchal et al., 2009). The homology drops dramatically when PTTH from Drosophila is compared with that from Cx. pipiens, though both are Diptera. But, PTTH sequences still are well conserved at the family level. For example, among the four PTTHs known from Saturniidae, the lowest identity of mature PTTH is 71% between Samia cynthia and Antheraea pernyi; among the five PTTHs known from Noctuidae, the lowest identity of mature PTTH is 68%, between H. zea and S. exigua (Xu et al., 2007). In the four Culicidae compared in the present study the lowest identity of mature PTTH is 84%, between Cx. pipiens and An. gambiae.
An essential structural characteristic of PTTH is the 7 cysteines with a specific spacing pattern in the amino acid sequence. Although PTTH amino acid sequences are quite different between Lepidoptera and Diptera, the 7 cysteines and their positions are strikingly conserved as shown by Marchal et al. (2009). By molecular modeling it was suggested that Bomor-PTTH adopts a similar stereo-structure to vertebrate growth factors with the cysteine-knot motif such as TGF-β2, β-NGF, or PDGF-BB, including two β-sheets consisting of four antiparallel β-strands (Noguti et al., 1995). Our results indicate that 7 cysteines with a similar spacing pattern are also present in Cupip-PTTH. Furthermore, molecular modeling by prevailing protein 3-D structure programs indicates a stereo-structure similar to Bomor-PTTH and vertebrate growth factors containing the cysteine-knot motif. Thus, the PTTH sequence we obtained here is quite similar to those known PTTHs at the tertiary structure level, likely indicating that PTTHs adopt a similar structure for ligand-receptor binding despite differing primary structures.
The genomic organization of PTTH and the location of PTTH mRNA were also investigated by Northern-blot. Our Southern blot results indicated only a single copy of the PTTH gene in the Cx. pipiens haploid genome, a conclusion consistent with B. mori PTTH (Adachi-Yamada et al., 1994). Northern blot analysis showed that PTTH mRNA was expressed primarily in the adult head and not in the thorax or abdomen of Cx. pipiens. PTTH mRNA and protein were localized specifically in two pairs of lateral neurosecretory cells in larval and pupal brains of Lepidoptera including B. mori (Kawakami et al., 1990), M. sexta (Shionoya et al., 2003) and H. armigera (Wei et al., 2005). PTTH mRNA was also detected in these specific neurons within D. melanogaster embryos and larval brains (McBrayer et al., 2007). Thus, PTTH appears to be consistently expressed in corresponding neurosecretory cells of the brain in different species, a feature more highly conserved than the amino acid sequences.
What function PTTH may serve in the adult is not at all clear. The prothoracic gland is the target of PTTH in larvae and pupae, but this gland undergoes apoptosis during metamorphosis, and adults lack this structure. Yet, PTTH was first purified from adult heads of B. mori, and a recent report also demonstrated the presence of PTTH in adult heads of Manduca sexta (Rybczynski et al., 2009). Ecdysone is, of course, present in adult insects, originating in the reproductive organs (Gilbert et al., 2002). The neuropeptide ovary ecdysteroidogenic hormone (OEH) is known to stimulate ecdysteroid secretion in mosquito ovaries (Brown et al., 1998; Riehle et al., 2002), but the presence of PTTH in adult mosquitoes does raise the question whether it too could be a player in stimulating adult ecdysteriod synthesis.
Our observations with Cx. pipiens demonstrate that larvae programmed by short days to enter diapause require one more day to attain the pupal stage than their long-day counterparts reared at the same temperature (18°C). This results in a heavier adult weight, a feature commonly observed in diapause-destined insects (Hahn & Denlinger, 2007, 2011). Based on observations with Drosophila melanogaster suggesting that PTTH influences the timing of development and hence larval size (McBrayer et al., 2007), we tested the hypothesis that there may be different PTTH mRNA profiles in short-day and long-day larvae that could provide an endocrine basis for the delay in pupation in short-day larvae. We found no evidence to suggest major differences in the PTTH mRNA profiles during the final larval instar. The only obvious distinction was that the short-day larvae required one additional day to pupate, and they thus displayed one additional pulse of PTTH mRNA.
Our preliminary examination of PTTH mRNA abundance throughout the final larval instar suggested considerable daily variation in the abundance of PTTH mRNA. When we examined this more thoroughly at 3 h intervals throughout a one-day period, the periodicity became even more apparent. PTTH mRNA levels remained low throughout the photophase and reached a peak late in the scotophase. This result is consistent with the scotophase peak observed in the blood-feeding bug, Rhodnius prolixus (Steel & Vafopoulou, 2006), as well as in B. mori (Rybczynski, 2005). An 8-h periodicity was noted for PTTH transcript expression in larvae of D. melanogaster (McBrayer et al., 2007). However, in pupae of Cx. pipiens, we saw no evidence to suggest cycling of PTTH expression. Instead, PTTH mRNA levels progressively increased from pupation to adult eclosion in both short- and long-day pupae. Levels in short-day pupae were significantly higher than in long-day pupae in both early pupae and shortly before adult eclosion, but these differences were modest.
In adults, no differences in PTTH mRNA expression levels were noted in the two types of females at eclosion, but shortly thereafter, expression declined rapidly in nondiapausing females and remained low as long as the females were denied a blood meal. By contrast, PTTH mRNA expression persisted at an elevated level in diapausing females for the first four weeks and then declined. When a blood meal was not provided expression levels were low in both types of females after 2 months, and transfer from 18 to 25°C also had no impact on the expression level.
But, when offered a blood meal, PTTH transcript levels in nondiapausing females rose rapidly, a 7-fold increase within 2 h. This increase in Cx. pipiens persisted for 24 h and then declined. Transcript AGAP000859, the PTTH homolog in An. gambiae, shows a similar profile following a blood meal, as indicated by microarray analysis (Marinotti et al., 2005). In both Cx. pipiens (Baldridge & Feyereisen, 1986) and An. gambiae (Dana et al., 2005) ecdysteroid titers also rise following the blood meal, reflecting a pattern similar to that observed with PTTH mRNA. This correlation suggests a possible role for PTTH in mediating ecdysteroid production or some other critical activity dependent on the blood meal. Interestingly, expression of OEH reported in the above microarray analysis declined continuously after the blood meal, thus indicating that PTTH and OEH mRNA expression patterns are distinctly different.
Though diapausing females of Cx. pipiens do not normally take a blood meal until diapause is terminated, a small portion of the females will do so when they are placed in close proximity of a host (Mitchell & Briegel, 1989). We too observed this; in our experiments approximately 10% of the diapausing females accepted a blood meal when offered a 3 h opportunity. But, as reported by Mitchell & Briegel (1989), the blood was not utilized and, in fact, much of it was ejected during a 24-h interval after feeding. Even though these blood-fed, diapausing females were not physiologically-programmed for taking and digesting blood, blood feeding still caused a boost in PTTH mRNA equivalent to that observed in nondiapausing females. These results suggest that PTTH mRNA elevation is a direct response to feeding, possibly elicited by gut or abdomen expansion, rather than to some downstream event such as blood digestion or ovarian development. Our results indicate that a blood meal effectively elicits robust expression of PTTH mRNA even in a diapausing female, but this alone is not sufficient to bring about the full termination of diapause.
Several key endocrine components of the adult diapause in Cx. pipiens are well established. As with most adult diapauses (Denlinger et al., 2005), the diapause of Cx. pipiens can be characterized as a juvenile hormone (JH) deficiency syndrome. The titer of JH is low in diapausing adults (Readio et al., 1999), and the diapause can be readily terminated by application of JH or a JH analog (Spielman, 1974; Sim & Denlinger, 2008). Insulin signaling appears to play a regulatory role upstream of JH (Sim & Denlinger, 2008, 2009a), but these two endocrine regulators are likely not to be the only hormones impacting the decision to enter, maintain or terminate diapause. The distinctly different PTTH mRNA profiles observed in this study during early adulthood by short- and long-day females likely reflect differences in the physiological status of the female as she prepares to enter diapause.
Whether the PTTH mRNA patterns of expression we noted here for Cx. pipiens also translate into similar patterns of neuropeptide synthesis and/or release is not yet known. While the dynamics of PTTH mRNA expression does correlate well with pupal diapause induction and termination in Heliothis virescens (Xu & Denlinger, 2003), monitoring of the actual peptide is required before fully understanding the meaning of the PTTH mRNA levels observed in this study. But, from the results we have observed in Cx. pipiens, there are major developmental changes in PTTH mRNA levels, especially at the time of diapause entry and in response to blood feeding, and such changes likely reflect a significant contribution of this neuropeptide to physiological functions in the adult female.
Experimental procedures
Insects
Our colony of Culex pipiens was established from mosquitoes collected in Columbus, Ohio in 2000 and maintained as described (Robich & Denlinger, 2005). Approximately 250 newly hatched larvae were reared in 5 × 18 × 28 cm plastic containers at 25°C with a photoperiod of 16 L: 8 D. Larvae were fed ground fish food (TetraMin) until they pupated. For diapause induction, third instar larvae were transferred to 18°C and a photoperiod of 8 L: 16 D. Pupae and adults were maintained under the same environmental conditions. Adults were placed in 30.5× 30.5 × 30.5 cm screened cages and supplied honey sponges for 10–13 days after eclosion. Nondiapause larvae, pupae and adults were reared at the same temperature (18°C) with a photoperiod of 16 L: 8 D; honey sponges were continuously supplied to the adults. Chicken was supplied as a host for 45 min for nondiapausing females and 3 h for diapausing females, respectively. Diapause status was confirmed by checking development of the primary follicles in the ovaries as described (Sim & Denlinger, 2008).
To compare developmental durations and PTTH transcripts levels of the diapause-and nondiapause-destined fourth instar larvae, late third instar larvae were collected based on the size of the larvae and the appearance of dark head capsules; newly molted fourth instar larvae were collected the next morning. Larvae were reared as described above, using the same population density and feeding frequency for the diapause- and nondiapause-destined groups, and the incidence of pupation was recorded every 12 h. Larvae used for monitoring PTTH transcript levels were sampled every 12 h or every 3 h for 24 h time course and homogenized in Trizol® reagent (Invitrogen, CA) immediately according to the manufacturer’s instructions. After centrifugation the homogenate was stored at −80°C until used.
1–2 day old diapausing and nondiapausing adults that had not been fed were collected and weighed. Fresh weights were determined by briefly chilling the adults at −20°C, separating the sexes, and weighing on a microbalance (Mettler-Toledo, OH). The mosquitoes were then heated at 60°C until they obtained a constant weight, which was recorded as the dry weight.
Amplification of the PTTH cDNA sequences
Total RNA was isolated from the whole body of 2–3 day old mosquito adults using Trizol® reagent as above. The quality and quantity of the RNA were examined by a spectrometer and agarose gel electrophoresis. Total RNA (1.5 μg) was reverse transcribed using SuperScript III™ Reverse Transcriptase (Invitrogen, CA) and Oligo dT20 according to the manufacturer’s instructions. The synthesized first strand cDNA (1 μl) was used for PCR.
Based on the sequence of a PTTH-like gene in Culex quinquefasciatus, primers CPPTFP1 (5′-GCTAGT TAC GTG CCAAACACAG-3′) and CPPTRP1 (5′-TTG TTAGCACTC GGACCTTGTA -3′) were designed and synthesized. Using these two primers PCR was performed under the following conditions: 3 min at 94°C; 35 cycles of 30 sec at 94°C, 30 sec at 60°C, 30 sec at 72°C; 7 min at 72°C. The PCR product was analyzed on 1.0% agarose gel and a band of approximately 485 bp was excised from the gel and centrifuged in an Ultrafree®- DA tube (Millipore, MA). The purified PCR fragment was ligated into pCR®2.1-TOPO® vector and then transformed into TOP10 competent cell according to the manufacturer’s instructions (Invitrogen, CA). Positive colonies were picked and sequenced at the Plant-Microbe Genomic Facility, Ohio State University.
The cDNAs used to obtain 5′ and 3′-end sequences of Cupip-PTTH were synthesized using a SMART™ RACE cDNA Amplification Kit (Clontech, CA) according to the manufacturer’s instructions. Gene specific primers, CPPTGSP1 (5′-AGC GAG GAA TCT GAC GAG AAC CAA AT-3′) for 5′-RACE, CPPTGSP2 (5′-TAG CAC TCG GAC CTT GTACGG GATTT-3′) for 3′-RACE, were designed and synthesized accordingly. Using a universal primer and gene-specific primer, PCR conditions for RACE were as follows: 94°C for 3 min; 35 cycles of 94°C for 30 sec; 68°C for 30 sec; 72°C for 1 min; extra extension at 72°C for 7 min. Distinct bands were excised from the agarose gel and the cDNAs were purified, cloned and sequenced as above.
Primers designation, DNA and amino acid sequences analysis and alignment, and phylogenetic analysis were similar to procedures described previously (Zhang & Denlinger, 2010).
Mature PTTH three-dimensional structure modeling
The putatively mature Cupip-PTTH amino acid sequence was subjected to web-based programs, I-TASSER (Zhang, 2008) and Protein Homology/anologY Recognition Engine (PHYRE) (Kelly & Sternberg, 2009), to predict secondary and three-dimensional structures. The predicted model with highest accuracy scores was adopted and visualized by Swiss-Pdb Viewer DeepView version 4.0.1 (Guex & Peitsch, 1997).
Northern blots
Since the abundance of PTTH transcripts was known to be very low in other species and in our experiments, we used an mRNA purification kit, Mini Oligotex® mRNA kit (Qiagen, Valencia, CA), to further purify mRNA from total RNA. 1~1.5 μg of purified mRNA from each sample was loaded onto the gels with ribosome protein L19 (RpL19) mRNA (GenBank No. FJ266017) serving as an internal control. Procedures for probe labeling and Northern blots were as described (Zhang & Denlinger, 2010).
Genomic organization and Southern blots of Cupip-PTTH
High molecular weight genomic DNA was purified from 2–3 day old male adult mosquitoes using a Gentra Puregene tissue kit (Qiagen, Valencia, CA). Size and quality of the isolated DNA were evaluated by agarose gel electrophoresis, and concentration was determined by measuring absorbance values at wavelengths of 230, 260 and 280 nm with a UV spectrometer. Using the reported C. quinquefasciatus genomic sequence and the sequenced Cupip-PTTH cDNA as references, primers were designed to obtain full gene sequence of Cupip-PTTH. Approximately 100 ng genomic DNA was used as a template to amplify sequences of introns in the PTTH gene, as well as upstream and downstream sequences of the PTTH gene. Ten micrograms of genomic DNA was incubated with restriction endonucleases EcoRI, EcoRV, HindIII, XbaI and XhoI overnight at 37°C, respectively. The digested DNAs and no digestion control were separated on a 0.8% agarose gel as described (Zhang & Denlinger, 2010).
Quantitative reverse-transcription PCR
Total RNA isolation and the examination of quality and quantity were the same as indicated above. To eliminate possible genomic DNA contamination, 1.5 μg RNA was treated by amplification-grade DNase I (Invitrogen, CA) for 15 min at room temperature according to the manufacturer’s instructions. The treated RNAs were reverse transcribed using the SuperScript™ III First-Strand synthesis system (Invitrogen, CA), and the synthesized cDNAs were used for quantitative PCR analysis. RPL19 was consistently expressed during the larval and pupal stages in both diapause- and nondiapause-destined groups and therefore was used as an internal control gene for qPCR. During the adult stage, two genes, RpL19 and 28S (GenBank No. DQ401446) ribosomal RNA, were employed as internal controls as previously used for comparison of gene expression in diapausing and nondiapausing Cx. pipiens adults (Sim & Denlinger, 2009b). Based on preliminary results, couch potato (unpublished data) is a more stable gene than 28S ribosomal RNA or RpL19 for the PTTH mRNA assay after blood feeding. Therefore, couch potato was used as an internal control for PTTH expression after blood feeding. Primers used for qPCR and sizes of amplicons are presented in Table 1.
Table 1.
Primers used for qPCR and the size of amplicons.
| Gene name | Forward primer | Reverse primer | Size of amplicon | Efficiency (%) |
|---|---|---|---|---|
| ptth | 5′-cgaactatggcaaccagaaact-3′ | 5′-atccatttggttctcgtcagat-3′ | 191 bp | 98.4 |
| RpL19 | 5′-gctttgtttgatcgtgtgtga-3′ | 5′-aacatatctcccccaatccag-3′ | 105 bp | 101.4 |
| 28 S rRNA | 5′-acgtgaaactgcctagggctc-3′ | 5′-tgggacaagcaaccagatg-3′ | 200 bp | 97.8 |
| couch potato | 5′-ttcgtcacgttcagcacacggt-3′ | 5′-gtgttactcttggcgaattcc-3′ | 119 bp | 102.4 |
Quantitative PCR was performed in a 20 μl volume (final primer concentration = 400 nM) using iQ™ SYBR® Green Supermix on an iQ™ 5 qPCR machine (Bio-Rad, Hercules, CA) under the following conditions: 3 min at 94°C; 40 cycles of 10 sec at 95°C, 30 sec at 60°C, 30 sec at 72°C. Target fragments of each gene were cloned and sequenced as above to verify sequences. Plasmids were also serially diluted 7 times using 10-fold dilutions to construct a standard curve for each gene. qPCR efficiencies generated by these primers are presented in Table 1. Based on standard curves, only those primers with efficiencies between 95% and 105%, R2 >0.995 and showing only a single peak in the melt curve were employed for the next qPCR experiments. After qPCR, the products were loaded in a 1% agarose gel to verify the presence of only a single product.
Three biological replicates were prepared for each sample, and 10–15 individuals were collected for each replicate. Two or three technical replicates were performed for each biological replicate on the qPCR plates. The geNorm program was employed to compare expression stability of reference genes, and normalization factors were calculated for the target gene (Vandesompele et al., 2002). Expression levels of PTTH were depicted as the arithmetic means of three biological replicates with standard deviations. Student’s t-test or one-way ANOVA was used to examine the statistical difference between different samples. P<0.05 and P<0.01 were considered significant and very significant differences respectively.
Acknowledgments
This work was supported by National Institutes of Health-National Institute of Allergy and Infectious Diseases Grant R01 AI058279 and the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, Grant Number 2006-35607-16582. We also thank Ms. Megan Meuti and Dr. Julie Reynolds for help with blood-feeding experiments.
References
- Adachi-Yamada T, Iwami M, Kataoka H, Suzuki A, Ishizaki H. Structure and expression of the gene for the prothoracicotropic hormone of the silkmoth, Bombyx mori. Eur J Biochem. 1994;220:633–643. doi: 10.1111/j.1432-1033.1994.tb18665.x. [DOI] [PubMed] [Google Scholar]
- Baldridge GD, Feyereisen R. Ecdysteroid titer and oocyte growth in the northern house mosquito, Culex pipiens L. Comp Biochem Physiol A Comp Physiol. 1986;83:325–9. doi: 10.1016/0300-9629(86)90583-9. [DOI] [PubMed] [Google Scholar]
- Brown MR, Graf R, Swiderek KM, Fendley D, Stracker TH, Champagne DE, Lea AO. Identification of a steroidogenic neurohormone in female mosquitoes. J Biol Chem. 1998;273:3967–3971. doi: 10.1074/jbc.273.7.3967. [DOI] [PubMed] [Google Scholar]
- Dana AN, Hong YS, Kern MK, Hillenmeyer ME, Harker BW, Lobo NF, Hogan JR, Romans P, Collins FH. Gene expression patterns associated with blood-feeding in the malaria mosquito Anopheles gambiae. BMC Genomics. 2005;6:5. doi: 10.1186/1471-2164-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denlinger DL, Yocum GD, Rinehart JP. Hormonal control of diapause. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Insect Molecular Science. Elsevier; Amsterdam: 2005. pp. 615–650. [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Gilbert LI, Rybczynski R, Warren JT. Control and biochemical nature of the ecdysteroidogenic pathway. Annu Rev Entomol. 2002;47:883–916. doi: 10.1146/annurev.ento.47.091201.145302. [DOI] [PubMed] [Google Scholar]
- Hahn DA, Denlinger DL. Meeting the energetic demands of insect diapause: nutrient storage and utilization. J Insect Physiol. 2007;53:760–773. doi: 10.1016/j.jinsphys.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Hahn DA, Denlinger DL. Energetics of insect diapause. Annu Rev Entomol. 2011;56:103–121. doi: 10.1146/annurev-ento-112408-085436. [DOI] [PubMed] [Google Scholar]
- Hanser G, Karlson P. Über die Wirkung des Metamorphosehormons auf die Epidermis von Ephestia Dauerraupen. Biol Zentbl. 1957;76:129–141. [Google Scholar]
- Ishibashi J, Kataoka H, Isogai A, Kawakami A, Saegusa H, Yagi Y, Mizoguchi A, Ishizaki H, Suzuki A. Assignment of disulfide bond location in prothoracicotropic hormone of the silkworm, Bombyx mori: A homodimeric protein. Biochemistry. 1994;33:5912–5919. doi: 10.1021/bi00185a031. [DOI] [PubMed] [Google Scholar]
- Ishizaki H, Suzuki A. The brain secretory peptides that control moulting and metamorphosis of the silkmoth, Bombyx mori. Int J of Dev Biol. 1994;38:301–310. [PubMed] [Google Scholar]
- Kawakami A, Kataoka H, Oka T, Mizoguchi A, Kimura-Kawakami M, Adachi T, Iwami M, Nagasawa H, Suzuki A, Ishizaki H. Molecular cloning of the Bombyx mori prothoracicotropic hormone. Science. 1990;247:1333–1335. doi: 10.1126/science.2315701. [DOI] [PubMed] [Google Scholar]
- Kelly LA, Sternberg MJE. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kopeć S. Studies on the necessity of the brain for the inception of insect metamorphosis. Biol Bull. 1922;42:323–342. [Google Scholar]
- Marchal E, Vandersmissen HP, Badisco L, Van de Velde S, Verlinden H, Iga M, Van Wielendaele P, Huybrechts R, Simonet G, Smagghe G, Vander Broeck J. Control of ecdysteroidogenesis in prothoracic glands of insects: A review. Peptides. 2010;31:506–519. doi: 10.1016/j.peptides.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JM. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Mol Biol. 2005;14:365–373. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- McBrayer Z, Ono H, Shimell M, Parvy JP, Beckstead RB, Warren JT, Thummel CS, Dauphin-Villemant C, Gilbert LI, O’Connor MB. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell. 2007;13:857–871. doi: 10.1016/j.devcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CJ, Briegel H. Fate of the blood meal in force-fed, diapausing Culex pipiens (Diptera: Culicidae) J Med Entomol. 1989;26:332–341. doi: 10.1093/jmedent/26.4.332. [DOI] [PubMed] [Google Scholar]
- Noguti T, Adachi-Yamada T, Katagiri T, Kawakami A, Iwami M, Ishibashi J, Kataoka H, Suzuki A, Go M, Ishizaki H. Insect prothoracicotropic hormone: A new member of the vertebrate growth factor superfamily. FEBS Lett. 1995;376:251–256. doi: 10.1016/0014-5793(95)01296-8. [DOI] [PubMed] [Google Scholar]
- Readio J, Chen MH, Meola R. Juvenile hormone biosynthesis in diapausing and nondiapausing Culex pipiens (Diptera: Culicidae) J Med Entomol. 1999;36:355–360. doi: 10.1093/jmedent/36.3.355. [DOI] [PubMed] [Google Scholar]
- Riehle MA, Garczynski F, Crim JW, Hill CA, Brown MR. Neuropeptides and peptide hormones in Anopheles gambiae. Science. 2002;298:172–175. doi: 10.1126/science.1076827. [DOI] [PubMed] [Google Scholar]
- Robich RM, Denlinger DL. Diapause in the mosquito Culex pipiens evokes a metabolic switch from blood feeding to sugar gluttony. Proc Natl Acad Sci USA. 2005;102:15912–15917. doi: 10.1073/pnas.0507958102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybczynski R. Prothoracicotropic hormone. In: Gilbert LI, Iatrou K, Gill S, editors. Comprehensive Insect Molecular Science. Elsevier; Amsterdam: 2005. pp. 61–123. [Google Scholar]
- Rybczynski R, Snyder CA, Hartmann J, Gilbert LI, Sakurai S. Manduca sexta prothoracicotropic hormone: evidence for a role beyond steroidogenesis. Arch Insect Biochem Physiol. 2009;70:217–229. doi: 10.1002/arch.20295. [DOI] [PubMed] [Google Scholar]
- Sauman I, Reppert SM. Molecular characterization of prothoracicotropic hormone (PTTH) from the giant silkmoth Antheraea pernyi: developmental appearance of PTTH-expressing cells and relationship to circadian clock in central brain. Dev Biol. 1996;178:418–429. doi: 10.1006/dbio.1996.0228. [DOI] [PubMed] [Google Scholar]
- Sehnal F, Hansen I, Scheller K. The cDNA-structure of the prothoracicotropic hormone (PTTH) of the silkmoth, Hyalophora cecropia. Insect Biochem Mol Biol. 2002;32:233–237. doi: 10.1016/s0965-1748(01)00107-2. [DOI] [PubMed] [Google Scholar]
- Spielman A, Wong J. Environmental control of ovarian diapause in Culex pipiens. Ann Entomo. Soc Am. 1973;66:905–907. [Google Scholar]
- Spielman A. Effect of synthetic juvenile hormone on ovarian diapause of Culex pipiens mosquitoes. J Med Entomol. 1974;11:223–225. doi: 10.1093/jmedent/11.2.223. [DOI] [PubMed] [Google Scholar]
- Shionoya M, Matsubayashi H, Asahina M, Kuniyoshi H, Nagata S, Riddiford LM, Kataoka H. Molecular cloning of the prothoracicotropic hormone from the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. 2003;33:795–801. doi: 10.1016/s0965-1748(03)00078-x. [DOI] [PubMed] [Google Scholar]
- Sim C, Denlinger DL. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc Natl Acad Sci USA. 2008;105:6777–6781. doi: 10.1073/pnas.0802067105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim C, Denlinger DL. A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquito Culex pipiens. Insect Mol Biol. 2009a;18:325–332. doi: 10.1111/j.1365-2583.2009.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim C, Denlinger DL. Transcription profiling and regulation of fat metabolism genes in diapausing adults of the mosquito Culex pipiens. Physiol Genomics. 2009b;39:202–209. doi: 10.1152/physiolgenomics.00095.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel CG, Vafopoulou X. Circadian orchestration of developmental hormones in the insect, Rhodnius prolixus. Comp Biochem Physiol A Mol Integr Physiol. 2006;144:351–364. doi: 10.1016/j.cbpa.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:7. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei ZJ, Zhang QR, Kang L, Xu WH, Denlinger DL. Molecular characterization and expression of prothoracicotropic hormone during development and pupal diapause in the cotton bollworm, Helicoverpa armigera. J Insect Physiol. 2005;51:691–700. doi: 10.1016/j.jinsphys.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Williams CM. Physiology of insect diapause. II. Interaction between the pupal brain and prothoracic glands in the metamorphosis of the giant silkworm, Platysamia cecropia. Biol Bull. 1947;93:89–98. [PubMed] [Google Scholar]
- Xu J, Su J, Shen J, Xu W. Molecular characterization and developmental expression of the gene encoding the prothoracicotropic hormone in the beet armyworm, Spodoptera exigua. Sci China C Life Sci. 2007;50:466–472. doi: 10.1007/s11427-007-0060-y. [DOI] [PubMed] [Google Scholar]
- Xu WH, Denlinger DL. Molecular characterization of prothoracicotropic hormone and diapause hormone in Heliothis virescens during diapause, and a new role for diapause hormone. Insect Mol Biol. 2003;12:509–516. doi: 10.1046/j.1365-2583.2003.00437.x. [DOI] [PubMed] [Google Scholar]
- Xu WH, Rinehart JP, Denlinger DL. Structural characterization and expression analysis of prothoracicotropic hormone in the corn earworm, Helicoverpa zea. Peptides. 2003;24:1319–1325. doi: 10.1016/j.peptides.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Denlinger DL. Molecular characterization of heat shock protein 90, 70 and 70 cognate cDNAs and their expression patterns during thermal stress and pupal diapause in the corn earworm. J Insect Physiol. 2010;56:138–150. doi: 10.1016/j.jinsphys.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]