Symmetrical modification within a nucleosome is not required globally for histone lysine methylation
The findings presented here argue against the ‘methylation copying within each nucleosome' model during the inheritance of histone methylation-mediated epigenetic information.
Keywords: epigenetic inheritance, histone, methylation, symmetry
Abstract
Two copies of each core histone exist in every nucleosome; however, it is not known whether both histones within a nucleosome are required to be symmetrically methylated at the same lysine residues. We report that for most lysine methylation states, wild-type histones paired with mutant, unmethylatable histones in mononucleosomes have comparable methylation levels to bulk histones. Our results indicate that symmetrical histone methylation is not required on a global scale. However, wild-type H4 histones paired with unmethylatable H4K20R histones showed reduced levels of H4K20me2 and H4K20me3, suggesting that some fractions of these modifications might exist symmetrically, and enzymes mediating these modifications might, to some extent, favour nucleosome substrates with premethylated H4K20.
Introduction
Histone modifications have important roles in the regulation of nearly all types of chromatin-templated biological events, including replication, transcription and recombination (Allis et al, 2006; Kouzarides, 2007; Li et al, 2007; Campos & Reinberg, 2009). Shortly after the discovery of the first histone methyltransferase, Su(var)3–9 (Rea et al, 2000), methyltransferases responsible for six main histone lysine methylation sites (H3K4, H3K9, H3K27, H3K36, H3K79 and H4K20) were identified (Zhang & Reinberg, 2001; Allis et al, 2006; Shilatifard, 2006).
Histone lysine methylation is a sophisticated process. All of the six main lysine methylation sites can be mono-, di- or tri-methylated (me1, me2 or me3), the type of methylation that occurs is determined by the action of various enzymes at distinct chromatin regions (Allis et al, 2006). These discrete methylation states are then recognized by different chromatin-binding proteins for specific biological events (Allis et al, 2006). In addition, histone methylations are often regulated by other histone modifications. For example, H3K4 and H3K9 methylations are mutually exclusive (Noma et al, 2001; Boggs et al, 2002); H3K4 and H3K79 methylations are regulated by H2B ubiquitination (Briggs et al, 2002; Dover et al, 2002; Sun & Allis, 2002; Lee et al, 2007); and H3R2 methylation antagonizes H3K4 methylation (Guccione et al, 2007; Kirmizis et al, 2007). It is an interesting question whether lysine methylation can regulate the methylation status of the same lysine residue on the other identical core histone within the same nucleosome.
Mechanisms for mitotic epigenetic inheritance—or the propagation of chromatin modifications to newly deposited histones in daughter cells—remain mostly unknown. Models often involve modification-copying events from neighboring modified nucleosomes or from within the same nucleosome (Nakatani et al, 2004; Martin & Zhang, 2007; Margueron et al, 2009; Probst et al, 2009; Margueron & Reinberg, 2010; Xu & Zhu, 2010). However, it is unknown whether histone lysine methylations are required to exist symmetrically on the two copies of identical histones in the same nucleosome. The answer to this question would have interesting implications because symmetrical methylation is a prerequisite for models involving methylation-copying events within the same nucleosome. In addition, symmetrical methylation might be necessary for other chromatin mechanisms, considering that certain histone methyltransferases that methylate the same lysine residue can coexist in one stable protein complex, such as the G9a/GLP complex (Tachibana et al, 2005). Here, we report that none of the 18 main histone lysine methylation states is required to exist in a symmetrical manner on a global scale, and we argue that the general epigenetic inheritance mechanism does not necessarily involve strict methylation-copying events within the same nucleosome.
Results And Discussion
Development of a detection system
As no technique exists that can differentiate between symmetrically and asymmetrically modified nucleosomes, we developed an alternative approach to test whether symmetrical modification is a global requirement for any of the 18 main histone lysine methylation states.
We established six stable HeLa cell lines, each expressing a different Flag-tagged mutant histone (H3K4R, H3K9R, H3K27R, H3K36R, H3K79R or H4K20R). These mutants cannot be methylated at the mutated sites, because of the arginine substitution. The expression level of these mutant histones was less than 10% of their endogenous counterparts, as determined by western blot analysis (supplementary Fig S1A,B online). Mononucleosomes containing the Flag-tagged, arginine-substituted histones were selectively purified from the respective stable cell lines (Fig 1). As the expression of Flag-tagged histones in comparison to endogenous histones was low (supplementary Fig S1A,B online), most nucleosomes containing a Flag-tagged histone also included an endogenous wild-type counterpart. As determined by Coomassie blue staining and western blot analysis, affinity-purified mononucleosomes contain the Flag-H3 K–R mutants and the endogenous H3 histones at an approximate ratio of 1:1 (supplementary Fig S1C,D online). In the case of Flag-H4K20R-containing mononucleosomes, western blot analysis with H4 antibodies reproducibly showed a stronger signal on the endogenous H4 than it did for the Flag-H4K20R histones (supplementary Fig S1E online). This is probably not a result of contamination of the endogenous H4 histones, because identical mononucleosome preparation and affinity purification protocols were used for all six of the Flag-tagged K–R mutants. We subsequently realized that the H4 antibodies used in the western blot analysis react poorly with the H4K20R mutants, compared with the H4 histones (supplementary Fig S1F online).
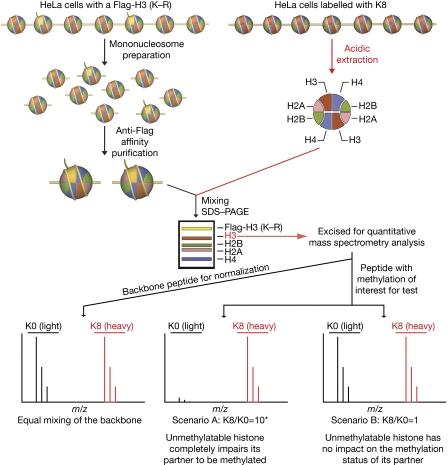
Figure 1.
Experimental scheme. HeLa cells labelled with K8 ([13C6, 15N2] heavy isotope-labelled L-lysine) were cultured in K8 medium for 1 month. If any methylation is required to exist in a symmetrical manner, unmethylatable histones should have a negative impact on the methylation status of its partner histone (Scenario A). Asterisk: The expected K8/K0 ratio for scenario A is approximately 10, because the labelling efficiency of the HeLa cells used in this study is approximately 90% (supplementary Fig S4 online). PAGE, polyacrylamide gel electrophoresis.
To test whether these mutant histones are only incorporated into distinct areas of the genome—which might introduce bias into this study—we designed 12 primer pairs amplifying random genomic sequences on chromosome 20 with 5 Mb intervals and one randomly chosen extra pair of primers at 7 Mb of chromosome 20 for normalization (supplementary Fig S2A,B online). Quantitative PCR was performed using HeLa mononucleosomes and the six affinity-purified Flag-histone-containing mononucleosomes as templates. Although certain fluctuations were observed, all six Flag-tagged, arginine-substituted histones were readily detected in all tested genomic regions—including gene-coding regions and intergenic regions—without marked abundance variation (supplementary Fig S2C online). This suggests that these affinity-purified mononucleosomes were representative of the genome.
To quantitatively assess the histone methylation levels of the endogenous H3 (or H4) copurified with the Flag-tagged H3 (or H4), stable isotope labelling-based quantitative mass spectrometry (Vermeulen et al, 2007) was performed. Histone samples prepared from HeLa cells cultured in media supplemented with Lys 8 (K8, L-lysine labelled with 13C6 and 15N2) were used as a reference for relative quantification (Fig 1; explanatory illustration in supplementary Fig S3 online). These HeLa cells showed more than 90% labelling efficiency (supplementary Fig S4 online). The K8-labelled histone samples were mixed with Flag-containing mononucleosomes purified from stable cells labelled with regular lysine (K0). Endogenous H3 (or H4) histones were well separated from Flag-H3 (or Flag-H4) on SDS–polyacrylamide gel electrophoresis, allowing the specific excision of the endogenous H3 (or H4) and subsequent quantitative mass spectrometry (Fig 1; supplementary Fig S1C,E online). The endogenous H3 (or H4) histones were mixed in an approximate ratio of 1:1. The exact mixing ratio was determined by quantification results of the backbone peptides, and the ratio of each modification was normalized against the mixing ratio of the backbone peptides.
Symmetrical methylation is not required for H3K9
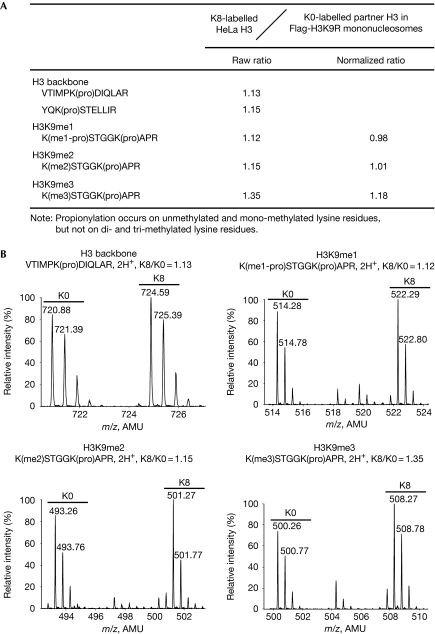
As one of the H3K9-specific methyltransferase complexes contains two homologous enzymes—G9a and GLP (Tachibana et al, 2005)—we first evaluated the H3 histones copurified with Flag-H3K9R histones. Following the analysis of two backbone peptides, YQKSTELLIR and VTIMPKDIQLAR, the K8/K0 ratio of histone H3 was determined to be 1.14 (Fig 2). All three H3K9 methylation states (me1, me2 and me3) were successfully detected by mass spectrometry and quantified using MSQuant software (Mortensen et al, 2010). For all three H3K9 methylation states, the normalized K8/K0 ratio was close to 1 (Fig 2). These results indicate that the endogenous H3 histones copurified with unmethylatable Flag-H3K9R histones had similar levels of H3K9 methylation to the global H3K9 methylation levels in HeLa cells. Thus, symmetrical methylation is not required for H3K9.
Figure 2.
Symmetrical methylation is not required for H3K9. (A) Table summarizing the quantified peptides of methylated H3K9. (B) Mass spectra for the quantified peptides, including the backbone peptide and peptides from H3K9me1, H3K9me2 and H3K9me3. AMU, atomic mass unit; pro, propionylation.
Symmetrical methylation is not required for H3K27
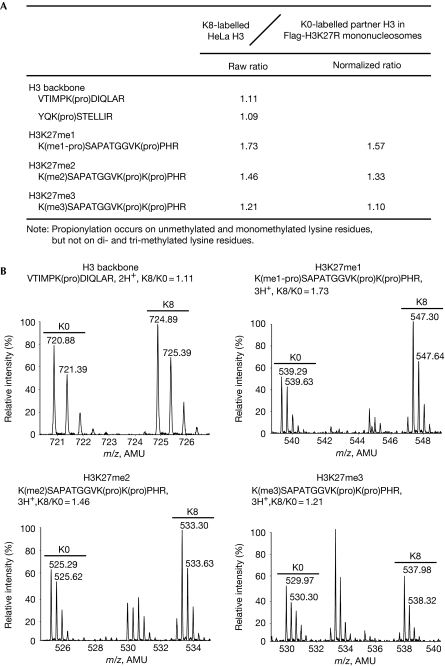
We then expanded the study to another repressive and potentially inheritable histone modification: H3K27 methylation. Examination of two backbone peptides, YQKSTELLIR and VTIMPKDIQLAR, determined the mixing ratio (K8/K0) of histone H3 to be 1.10 (Fig 3). The normalized K8/K0 ratios for H3K27me1, H3K27me2 and H3K27me3 were 1.57, 1.33 and 1.10, respectively (Fig 3), indicating that the endogenous H3 histones copurified with unmethylatable Flag-H3K27R histones had slightly lower levels of H3K27 methylation in comparison to the global H3K27 methylation levels in HeLa cells, particularly for H3K27me1. Nevertheless, the detected K8/K0 ratio of 1.57 for H3K27me1 suggests that nucleosomes with premethylation at K27 might be modestly preferred but are not a globally required symmetrical modification event.
Figure 3.
Symmetrical methylation is not required for H3K27. (A) Table summarizing the quantified peptides of methylated H3K27. (B) Mass spectra for the quantified peptides, including the backbone peptide and peptides from H3K27me1, H3K27me2 and H3K27me3. AMU, atomic mass unit; pro, propionylation.
Symmetrical methylation is not required for H3K79
We then studied H3K79 methylation, the only lysine methylation mediated by a non-Set domain histone lysine methyltransferase, Dot1 (Feng et al, 2002; Lacoste et al, 2002; Ng et al, 2002; van Leeuwen et al, 2002). Analysis of two backbone peptides, VTIMPK and RVTIMPK, led to a K8/K0 ratio of 0.97 for histone H3 mixing (Fig 4). The normalized K8/K0 ratios for H3K79me1, H3K79me2 and H3K79me3 were 0.68, 0.41 and 0.60, respectively (Fig 4). These values occur in the opposite direction of what would be expected if symmetrical methylation is required at H3K79. Instead, endogenous H3 histones copurified with unmethylatable Flag-H3K79R histones had higher levels of H3K79 methylation compared with the global H3K79 methylation levels in HeLa cells, suggesting negative regulation of H3K79 methylations within the nucleosome.
Figure 4.
Symmetrical methylation is not required for H3K79. (A) Table summarizing the quantified peptides of methylated H3K79. (B) Mass spectra for the quantified peptides including the backbone peptide and peptides from H3K79me1, H3K79me2 and H3K79me3. AMU, atomic mass unit; oxi, oxidation.
Symmetrical methylation is not required for H4K20
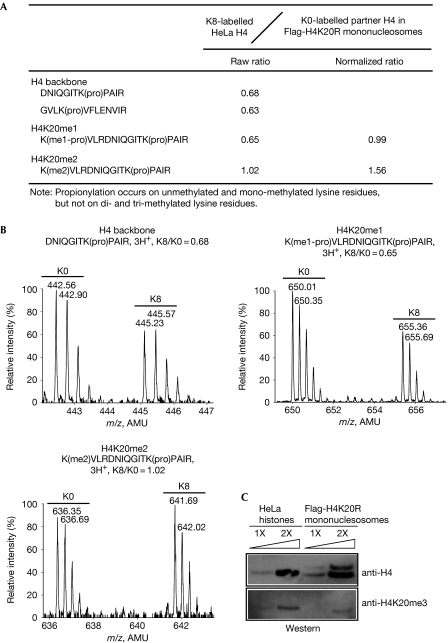
Next, we studied H4K20 methylation. Endogenous H4 histones copurified with unmethylatable Flag-H4K20R histones had similar levels of H4K20me1 to the global levels in HeLa cells (Fig 4A,B). However, bulk HeLa histones had approximately 50% more H4K20me2 (Fig 5A,B) and approximately two times as much H4K20me3 (Fig 5C), compared with the endogenous H4 histones paired with unmethylatable Flag-H4K20R mutants. The relative abundance of H4K20me3 was determined by western blot analysis, as we failed to quantify these peptides by using mass spectrometry, due to the low abundance of this modification.
Figure 5.
Symmetrical methylation is not required for H4K20. (A) Table summarizing the quantified peptides of methylated H3K79. (B) Mass spectra for the quantified peptides including the backbone peptide and peptides from H4K20me1 and H4K20me2. (C) Western blot analysis quantifying relative abundance of H4K20me3. AMU, atomic mass unit; pro, propionylation.
These results indicate that some fractions of H4K20me2 and H4K20me3 might exist symmetrically. However, for a generally required symmetrical methylation event, we would expect the ratio to be higher than 1.56:1 or 2:1. As the reference HeLa cells were labelled with approximately 90% efficiency (supplementary Fig S4 online), the K8/K0 ratio of the methylated peptides is expected to be close to 10 for a full symmetrical pattern and 5 for a semi-symmetrical pattern (Fig 1).
H3K4 and H3K36 methylation do not require symmetry
Finally, we studied the H3K4 and H3K36 sites. We quantified some of the methylation states at these two sites (supplementary Figs S5, S6 online) but did not detect the H3K4me2, H3K4me3 and H3K36me3 peptides. This was probably because of the hydrophilic nature of the methylated H3K4 peptides—which were not retained on the reverse-phase resin during liquid chromatography—and the lack of abundance of H3K36me3. Therefore, we used western blot analysis with antibodies specific for these methylation states and obtained comparable signals between total HeLa histones and histones paired with the FLAG-H3 K–R mutants for these modifications (supplementary Figs S5C, S6C online). The results of quantitative mass spectrometry for the detected peptides, and the results of western blot analysis for the others indicated that these modifications occur at similar levels in the endogenous H3 histones paired with their unmethylatable counterparts and bulk histones (supplementary Figs S5 and S6 online).
Conclusions
We systematically investigated whether any of the 18 main histone methylation states are required to exist in a symmetrical manner within the nucleosome. Our results do not support the idea that symmetrical patterning of methylation is required globally. Instead, our results support the conclusion that, for corresponding residues on the two copies of the same histone within a nucleosome, the methylation states are largely distributive. Together with our recent report (Xu et al, 2010) and early sedimentation analyses (Jackson, 1988, 1990; Yamasu & Senshu, 1990), which demonstrated the conservative segregation of H3–H4 tetramers, our results indicate that epigenetic inheritance does not necessarily involve strict methylation-copying events within the same nucleosome. If there is any methylation copying, neighboring methylated nucleosomes probably act as the templates, as has been implicated in the case of H3K27me3 (Hansen et al, 2008; Margueron et al, 2009).
However, we cannot exclude cross-regulation between the two identical methylation sites within the same nucleosome. For example, in the case of H3K79, we observed elevated H3K79 methylation levels on endogenous H3 histones copurified with the unmethylatable Flag-tagged H3K79R histones. This might indicate that the methylation of H3K79 on one H3 downregulates the methylation of its partner H3 within the same nucleosome. This effect could be indirectly achieved by recruiting binding proteins to limit accessibility for Dot1L. By contrast, we reproducibly observed reduced levels of H4K20me2, H4K20me3 and H3K27me1 on histones copurified with the Flag-tagged K–R mutants. These observations indicate that symmetrical methylation of these modifications might be favored in certain chromatin regions, and enzymes mediating H3K27me1, H4K20me2 and H4K20me3 might favour nucleosome substrates that have been methylated previously. As this study was conducted on a global scale, further studies with increased spatial resolution will help to address whether this is the case.
Methods
Stable cell lines. Complementary DNA fragments carrying a Flag-tagged mutant H3 variant (H3K4R, H3K9R, H3K27R, H3K36R, H3K79R or H4K20R) or Flag-H4K20R were inserted into pcDNA4/TO (Invitrogen) and transfected into HeLa cells. Stable clones were selected against Zeocin (100 μg/ml; Invitrogen).
Mononucleosome preparation and affinity purification. Cells were pelleted, resuspended in lysis buffer containing 10 mM Tris–HCl (pH 8.0), 250 mM sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.5 mM dithiothreitol (DTT) and 0.5% Triton X-100, and kept on ice for 10 min. Crude nuclei were collected and resuspended in a digestion buffer containing 10 mM Tris–HCl (pH 8.0), 250 mM sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.5 mM DTT and 1 mM CaCl2. The nuclei were then incubated at 37°C for 80 min with MNase (TaKaRa) at 40 U/107 nuclei. Digestion was stopped by adding ethylenediaminetetraacetic acid (EDTA) to a final concentration of 20 mM, followed by chilling at 4°C. After centrifuging at 10,000 g for 5 min, the nuclear pellet was resuspended in 5 mM EDTA and incubated at 4°C overnight. We have previously shown that this protocol recovers similar amounts of histones as the acidic-extraction protocol, suggesting that this protocol offers nearly quantitative recovery of the nucleosomes (Xu et al, 2010).
After a second centrifugation step of 10,000 g for 10 min, the supernatant fraction was subjected to further fractionation with a 24-ml Superose 6 gel filtration column (GE) in a buffer containing 10 mM Tris–HCl (pH 8.0), 100 mM KCl, 0.5 mM EDTA, 1 mM DTT and 10% glycerol. Fractions containing mononucleosomes were pooled for affinity purification. The mononucleosome suspension was then incubated with M2 anti-Flag agarose (Sigma) for 3 h at 4°C. The beads were washed extensively with a buffer containing 10 mM Tris–HCl (pH 8.0), 500 mM KCl, 0.5 mM EDTA, 1 mM DTT, 10% glycerol and 0.5% NP40. Affinity-purified mononucleosomes were then eluted with the addition of 1 mg/ml Flag peptides.
Stable isotope labelling of HeLa cells. Stable isotope labelling of HeLa cells was performed as described previously (Xu et al, 2010). Briefly, HeLa cells were cultured in DME/F-12-deficient medium (Sigma, D9785) containing 10% dialysed fetal bovine serum (Invitrogen) and supplemented with 0.365 mg/ml L-glutamine (Invitrogen), 0.059 mg/ml L-leucine (Sigma), 0.017 mg/ml L-methionine (Sigma) and 0.091 mg/ml [13C6, 15N2] heavy isotope-labelled L-lysine (Cambridge Isotope Laboratories Inc.). These cells were cultured in this medium for 1 month to reach satisfactory labelling efficiency (supplementary Fig S2 online). Histones were then extracted from these cells as the K8-labelled reference sample.
Stable isotope labelling-based quantitative mass spectrometry. Stable isotope labelling-based quantitative mass spectrometry was performed using a QSTAR XL mass spectrometer (MDS SCIEX) with a nano-electrospray ionization (ESI) ion source. Histone samples from affinity-purified mononucleosomes and the reference K8-labelled HeLa cells were mixed in an approximate 1:1 ratio and separated by 13% SDS–polyacrylamide gel electrophoresis. Except for H3K79 peptides, which were easily detected, the histone samples were chemically propionylated (Garcia et al, 2007) before mass spectrometry, to improve the detection of methylated peptides. Propionylated histones were digested with trypsin (Promega) overnight at 37°C, at a substrate/enzyme ratio of 20:1. Peptides generated by trypsin digestion were separated by an analytical capillary column (50 μm × 10 cm) packed with C18 reverse-phase material (Yamamura Chemical Research Institute 5-μm spherical particles). The eluted peptides were sprayed into a QSTAR XL mass spectrometer (MDS SCIEX), which was operated in Information Dependent Acquisition mode. The ion-spray voltage was 2.1 kV. The mass spectrometry scan was from 300–2,000 m/z . From each mass spectrometry scan, the top three most abundant peaks were selected for mass spectrometry/mass spectrometry scans. Each scan was taken for 0.5 s. The dynamic exclusion time was set to 20 s. The search results from data analysis using Mascot were analysed by MSQuant (Mortensen et al, 2010) to calculate the ratios for the heavy/light peptide pairs.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank N. Yang at the National Institute of Biological Sciences, Beijing, for the colour illustration. This work was supported by the Chinese Ministry of Science and Technology through the following grants: 973 grant 2011CB965300 (to B.Z.) and 863 grants 2007AA02Z1A6 (to B.Z.) and 2007AA02Z1A3 (to S.C.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Allis CD, Jenuwein T, Reinberg D (2006) Overview and concepts. In Epigenetics, Allis CD, Jenuwein T, Reinberg D (eds) pp 23–56. New York: Cold Spring Harbor Laboratory [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD (2002) Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet 30: 73–76 [DOI] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43: 559–599 [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y (2002) Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 12: 1052–1058 [DOI] [PubMed] [Google Scholar]
- Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF (2007) Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc 2: 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Lüscher B, Amati B (2007) Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449: 933–937 [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K (2008) A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300 [DOI] [PubMed] [Google Scholar]
- Jackson V (1988) Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry 27: 2109–2120 [DOI] [PubMed] [Google Scholar]
- Jackson V (1990) In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry 29: 719–731 [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bähler J, Green RD, Kouzarides T (2007) Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449: 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Côte J (2002) Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem 277: 30421–30424 [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D (2010) Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11: 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R et al. (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y (2007) Mechanisms of epigenetic inheritance. Curr Opin Cell Biol 19: 266–272 [DOI] [PubMed] [Google Scholar]
- Mortensen P et al. (2010) MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J Proteome Res 9: 393–403 [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Ray-Gallet D, Quivy JP, Tagami H, Almouzni G (2004) Two distinct nucleosome assembly pathways: dependent or independent of DNA synthesis promoted by histone H3.1 and H3.3 complexes. Cold Spring Harb Symp Quant Biol 69: 273–280 [DOI] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K (2002) Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev 16: 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293: 1150–1155 [DOI] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G (2009) Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206 [DOI] [PubMed] [Google Scholar]
- Rea S et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Shilatifard A (2006) Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269 [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3–K9. Genes Dev 19: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, Zhu B (2010) Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328: 94–98 [DOI] [PubMed] [Google Scholar]
- Xu M, Zhu B (2010) Nucleosome assembly and epigenetic inheritance. Protein Cell 1: 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasu K, Senshu T (1990) Conservative segregation of tetrameric units of H3 and H4 histones during nucleosome replication. J Biochem 107: 15–20 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343–2360 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.