Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region
Laminin polymers are a key component of all basement membranes. The first crystal structure of a laminin amino-terminal domain identifies a conserved surface loop in laminin alpha chains that is involved in polymer formation.
Keywords: basement membrane, LN domain, netrin, X-ray crystallography
Abstract
The polymerization of laminin into a cell-associated network—a key step in basement membrane assembly—is mediated by the laminin amino-terminal (LN) domains at the tips of the three short arms of the laminin αβγ-heterotrimer. The crystal structure of a laminin α5LN–LE1–2 fragment shows that the LN domain is a β-jelly roll with several elaborate insertions that is attached like a flower head to the stalk-like laminin-type epidermal growth factor-like tandem. A surface loop that is strictly conserved in the LN domains of all α-short arms is required for stable ternary association with the β- and γ-short arms in the laminin network.
Introduction
Basement membranes are specialized extracellular matrices that underlie epithelial cell sheets and surround muscle, fat and peripheral nerve cells. Basement membranes are crucially involved in many developmental processes and their integrity is essential for tissue function in adult animals. The main basement membrane proteins are among the most highly conserved extracellular proteins; they include laminins, type IV collagen, nidogen and perlecan. Basement membrane assembly begins with the polymerization of laminin into a cell-associated network (Yurchenco et al, 2004). Consequently, total genetic ablation of laminin leads to early arrest of embryo development. Mutations of tissue-specific laminin genes cause severe diseases in humans, such as congenital muscular dystrophy and epidermolysis bullosa (Miner & Yurchenco, 2004).
Laminins are large, cross-shaped molecules consisting of three polypeptide chains (α, β and γ). The three short arms of the cross are composed of one chain each, whereas the long arm is a coil containing all three chains that terminates in a tandem of five linkage group domains provided by the α-chain (Fig 1A). The human genome encodes five α-, three β- and three γ-chains which are assembled into at least 15 laminin heterotrimers (Aumailley et al, 2005). With the exception of the α3A, α4 and γ2 chains, all short arms have a single laminin amino-terminal (LN) domain at their distal end, followed by a tandem of laminin-type epidermal growth factor-like (LE) domains. The same domain architecture is also found in netrins, an important class of axon guidance molecules (Fig 1A; Dickson, 2002). Laminin-111 (α1β1γ1) polymerization in vitro requires calcium and a full complement of LN domains, suggesting that the nodes in the polygonal laminin-111 network contain one α1, one β1 and one γ1 LN domain (three-arm interaction model; Yurchenco et al, 1985; Paulsson, 1988; Yurchenco & Cheng, 1993; McKee et al, 2007). In support of this model, mutations in the LN domains of the α1 and α2 chains compromise laminin function in vivo (Xu et al, 1994; Patton et al, 2008; Edwards et al, 2010). Binding experiments with short-arm fragments showed heterotypic interactions that were consistent with the three-arm interaction model (α–β, α–γ, β–γ), as well as unexpected homotypic α–α interactions (Odenthal et al, 2004). A molecular understanding of laminin polymerization has been hindered by the lack of structural information about the LN domain. In this study we report the crystal structure and functional analysis of a laminin α5LN–LEa1–2 fragment.
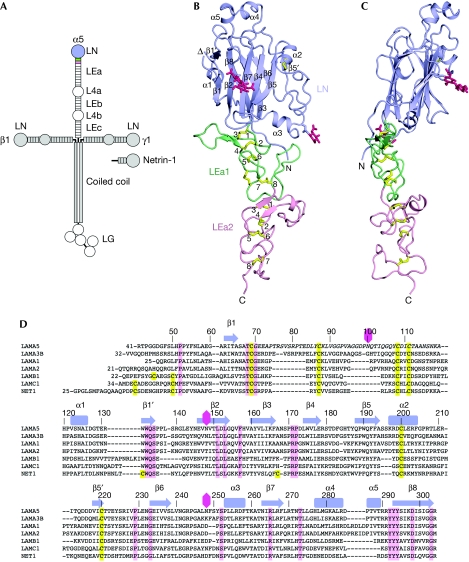
Figure 1.
Laminin α5LN–LEa1–2 structure. (A) Schematic drawing of the cross-shaped laminin 511-heterotrimer and of netrin 1. The crystallized α5LN–LEa1–2 region is coloured. (B,C) Two orthogonal views of the α5LN–LEa1–2 structure. The LN, LEa1 and LEa2 domains are shown in lilac, green and light pink, respectively. Disulphide bridges and N-linked glycans are shown in yellow and dark pink, respectively. The secondary structure elements of α5LN are labelled. The chain break due to the disorder of residues 70–119 is indicated (triangle). The eight canonical cysteines in LEa1 and LEa2 are numbered. (D) Sequence alignment of the LN domains of the mouse laminin α5, α3B, α1, α2, β1 and γ1 chains, and of mouse netrin 1. The sequence numbering and secondary structure elements of α5LN are indicated above the alignment. Residues in italics are disordered in the α5LN–LEa1–2 structure. Cysteines and conserved residues are shown in yellow and pink, respectively. Glycosylation sites in α5LN are indicated by dark pink hexagons. LE, laminin-type epidermal growth factor-like; LN, laminin N-terminal.
Results and Discussion
Crystallization of a laminin short-arm fragment
As LN domains cannot be produced in isolation (Ettner et al, 1998; Odenthal et al, 2004), we produced a series of laminin short-arm fragments containing the LN domain, followed by 2–4 LEa domains (supplementary Table S1 online). The best crystals we obtained were of an α5LN–LEa1–2 fragment and diffracted to approximately 3.4 Å resolution. Individual mutations of the four predicted N-linked glycosylation sites (N100A, N148T, N248E, N383E) showed that glycosylation of Asn 148 and Asn 248 was required for protein solubility. A crystal of the α5LN–LEa1–2 N100A/N383E double mutant diffracted to 2.9 Å resolution and was used to determine the structure.
Laminin α5LN–LEa1–2 structure
The laminin α5LN–LEa1–2 structure resembles a flower, with the LN and LE domains forming the head and stalk, respectively (Fig 1B,C). The head measures approximately 40 × 40 × 25 Å and the stalk is approximately 60 Å long. When the head is viewed from its narrow side, it appears tilted by approximately 25° relative to the straight stem. The LE domains of α5LN–LEa1–2 have the canonical LE fold, that was previously established by the crystal structure of laminin γ1LEb2–4 (Stetefeld et al, 1996). The α5LEa1–2 domains almost entirely consist of irregular coils and are stabilized by a continuous ladder of disulphide bridges (Fig 1B). The eight canonical cysteines in each domain are linked 1–3, 2–4, 5–6 and 7–8, creating four loops, which we refer to as the 2–3, 3–4, 5–6 and 7–8 loops. In α5LEa1, there is a fifth disulphide bridge linking the 5–6 and 7–8 loops. The long 3–4 loops of each LE domain contain short antiparallel β-strands and protrude similarly to leaves from the stalk.
The LN domain is a richly elaborated antiparallel β-sandwich, with strands β1–β8 forming a jelly-roll motif (Fig 1B,C). A DALI search for related structures (Holm & Rosenström, 2010) showed that the core of the LN domain is most similar to bacterial galactose binding domains (GBDs; supplementary Fig S1A online). This relationship was recognized in a recent modelling study (Kalkhof et al, 2010). However, the LN fold has several additions to the jelly-roll motif that have no counterpart in GBDs. The two β-sheets of the jelly-roll motif in α5LN are extended by two short strands, β1′ and β5′. The latter strand is part of a larger insertion between β5 and β6 that also contains a conserved disulphide bridge between Cys 200 and Cys 220 (Fig 1D). Additional large insertions between β6, β7 and β8 contain several α-helices that contribute substantially to the bulk of the LN domain. Residues 70–119—another insertion into the jelly-roll motif—are disordered in the α5LN–LEa1–2 structure. The disordered region contains four conserved cysteines, but otherwise its sequence is highly variable in the LN domain superfamily (Fig 1D).
There is no obvious linker segment between the LN and LEa1 domains (Fig 1B,C). The two domains interact through a large, partly disjointed, interface that buries a total of approximately 2,500 Å2 of solvent-accessible surface area. Half of the buried area is accounted for by the interaction of an extended N-terminal segment with LEa1. Most notably, the side chain of Pro 52 is accommodated in a pocket between the 1–3 and 2–4 disulphide bridges of LEa1 (supplementary Fig S2 online). Pro 52 is strictly conserved in all other LN domains (Fig 1D), suggesting that the LN–LEa1 interface is similarly stabilized in all laminins and netrins. The other contacts between the LN and LEa1 domains of laminin α5 involve the long 3–4 and 7–8 loops of LEa1 and are not as well conserved across the LN superfamily. The α5LN–LEa1–2 crystal structure contains three phosphate ions in the LN–LEa1 interface which interact with the side chains of His 57, His 158, Arg 256, Lys 260, Arg 304 and His 308 (supplementary Fig S2 online). Arg 256 and Lys 260 are involved in proteoglycan-mediated cell adhesion to the α5 short arm (Nielsen & Yamada, 2001). The phosphate ions might thus act similarly to the sulphate groups of heparan sulphate chains.
Bacterial GBDs—which the core of the LN fold is most closely related to—contain a structural calcium binding site at the carboxy terminus of helix α1 (Boraston et al, 2004). There is no metal ion bound to this region in the laminin α5LN–LEa1–2 structure, even though suitable acidic ligands are present in the α5 chain (Asp 127 and Asp 298; supplementary Fig S1B,C online). To exclude the possibility that calcium was lost from α5LN–LEa1–2 during crystallization at pH 4–5, we tested for calcium binding at neutral pH by measuring the intrinsic fluorescence of α5LN–LEa1–2 (the putative calcium binding site is close to two fluorophores, Trp 132 and Trp 133). The fluorescence spectra in the presence and absence of calcium were identical (supplementary Fig S1D online), indicating that the N-terminal region of the α5 chain does not bind to calcium. This conclusion agrees with the results of circular dichroism spectroscopy experiments (Odenthal et al, 2004). The sites responsible for the calcium dependence of laminin polymerization (Yurchenco et al, 1985; Paulsson, 1988) remain unknown.
A motif involved in laminin-network formation
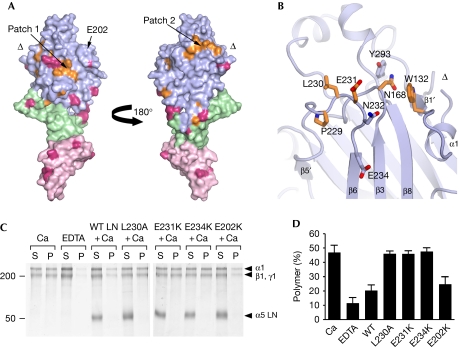
Having determined the first structure of an LN fragment, we sought to identify the αLN domain regions responsible for laminin-network formation. We thought that residues mediating network interactions might be conserved in all α-chains, as laminin heterotrimers with different α-chains can form mixed polymers (Cheng et al, 1997). Mapping the conserved residues of mouse laminin α-chains onto the α5LN domain structure showed that the β6–β3–β8 face of the β-sandwich is more highly conserved than the opposite face (not shown). Remarkably, two conserved surface patches remain even when invertebrate laminin α-chains are included in the analysis (Fig 2A, supplementary Fig S3 online). Patch 1 is situated on the β1–β2–β7–β4–β5 face and comprises Glu 178, Pro 189, Arg 265 and Arg 267. However, the glycan attached to Asn 148 blocks access to these residues in α5LN. All LN domains contain a predicted glycosylation site in this region, either at the start of β2 as in α5LN or at the end of β1 (Fig 1D, supplementary Fig S3 online). This suggests that patch 1 cannot engage in intermolecular interactions. Patch 2 is on the β6–β3–β8 face and comprises Trp 132 and Asn 168, as well as Pro 229, Leu 230 and Glu 231 in the β5′–β6 loop (Fig 2B). Importantly, there are no glycosylation sites obstructing patch 2 in any laminin α-chain (Fig 2A). Further, with the exception of Trp 132, patch 2 residues are not conserved in laminin β- and γ-chains (Fig 1D). These observations do not support a structural role for patch 2 and instead imply that it has a specific role in laminin α-chain function.
Figure 2.
Functionally important residues in laminin α5LN–LEa1–2. (A) Two views of the laminin α5LN–LEa1–2 surface related by a rotation of 180° about the vertical axis. The direction of the view on the left is the same as in Fig 1B. The LN, LEa1 and LEa2 domains are shown in lilac, green and light pink, respectively. Residues that are identical in vertebrate and invertebrate laminin α-chains are shown in orange. Residues that are predicted to be modified by N-linked glycosylation in at least one α-chain are in dark pink. The locations of the two conserved surface patches, of Glu 202 and of the chain break (triangle) are indicated. (B) Close-up view of conserved surface patch 2. Selected residues are shown in atomic detail (orange, conserved in all α-chains). (C) Inhibition of laminin-111 polymerization by α5LN–LEa1–2 wild-type and mutant proteins. Laminin-111 was incubated in 1 mM calcium or in 10 mM EDTA. α5LN–LEa1–2 proteins were added as indicated. Polymerized laminin-111 was pelleted by centrifugation and the supernatant (S) and pellet (P) fractions were analysed by SDS–PAGE. The positions of laminin chains and molecular weight markers (in kD) are indicated. (D) Densitometric quantification of three independent polymerization experiments carried out as in C. Results are expressed as percentages of polymer (P/(S+P)) and are shown as mean±s.d. Ca, calcium; EDTA, ethylenediaminetetraacetic acid; LE, laminin-type epidermal growth factor-like; LN, laminin N-terminal; PAGE, polyacrylamide gel electrophoresis; WT, wild type.
To test whether patch 2 is important for α5LN–LEa1–2 function, we introduced three mutations into the β5′–β6 loop: L230A, E231K and E234K. The first two residues are strictly conserved in all α-chains, whereas Glu 234 is conserved in all α-chains except in Drosophila wing blister (supplementary Fig S3 online). As a control, we mutated a surface-exposed glutamic acid on the opposite face of the LN domain (E202K). All mutants were secreted as efficiently as wild-type protein by HEK 293 cells, indicating correct folding. To determine the effects of the mutations, we used an established functional assay in which laminin-111 polymerization is inhibited by soluble short-arm fragments (Yurchenco & Cheng, 1993; Cheng et al, 1997; Garbe et al, 2002). As expected, wild-type α5LN–LEa1–2 protein robustly inhibited laminin-111 polymerization, so that most of the laminin-111 protein remained in the soluble fraction (Fig 2C,D). The E202K control mutant was a similarly potent inhibitor. By contrast, all three α5LN–LEa1–2 proteins with mutations in the β5′–β6 loop did not inhibit laminin-111 polymerization, demonstrating that the β5′–β6 loop of laminin α5LN interacts with a crucial network-forming site on laminin-111. As β5′–β6 loop residues are strictly conserved in all laminin α-chains of invertebrates and vertebrates (Fig 2A), it is likely that this αLN region is involved in the network interactions of other laminins.
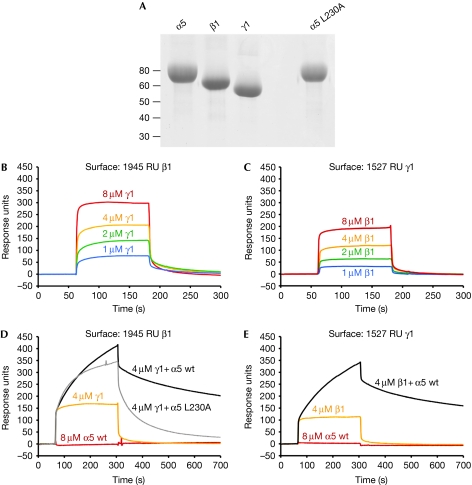
Which interaction in the laminin-111 network is blocked by α5LN–LEa1–2? Binding experiments with recombinant LN–LEa1–4 fragments of the α1, α5, β1 and γ1 chains detected all possible heterotypic interactions, as well as homotypic α1–α1 and α5–α5 interactions (Odenthal et al, 2004). However, this binding pattern is inconsistent with results from an earlier study using proteolytic laminin-111 fragments, which found only a single interaction of measurable affinity (binary β1–γ1 or ternary α1–β1–γ1; Yurchenco & Cheng, 1993). We used surface plasmon resonance (SPR) binding experiments to study the interactions between the LN–LEa1–4 fragments of the α5, β1 and γ1 chains (Fig 3). We detected a weak interaction (estimated Kd of ⩾5 μM) between β1 and γ1 (Fig 3B,C), but, in contrast to the results of Odenthal et al (2004), α5 did not bind to β1, γ1 or to itself in our experiments (Fig 3D,E and data not shown). Most interestingly, when we flowed a 1:1 mixture of α5 and γ1 over a β1 surface, we observed a larger response than for γ1 alone, indicative of a ternary α5–β1–γ1 complex (Fig 3D). The same ternary complex could also be assembled on a γ1 surface (Fig 3E). The ternary α5–β1–γ1 interaction showed slower kinetics than the binary β1–γ1 interaction, possibly indicating that α5 binds to a β1–γ1 heterodimer. Although the underlying equilibria are probably more complex, the association and dissociation phases of the ternary complex (Fig 3D) could be fitted (χ2=1.9) by a 1:1 Langmuir model with a Kd of 0.8 μM. The α5 L230A mutant—which was inactive in the polymerization assay (Fig 2C,D)—also formed a ternary complex with β1 and γ1, but the mutant complex dissociated substantially faster than the wild-type complex (Fig 3D). Thus, mutations in the β5′–β6 loop of α5LN seem to destabilize the ternary α5–β1–γ1 complex. As the dissociation of the mutant complex could not be described by a single rate constant, it was not possible to quantify the destabilizing effect.
Figure 3.
Surface plasmon resonance analysis of laminin α5, β1 and γ1 short-arm interactions. (A) Reducing SDS–PAGE of the α5, β1 and γ1 LN–LEa1–4 fragments used in the analysis. The positions of molecular weight markers (in kD) are indicated. (B,C) Sensorgrams showing binding of β1 to γ1. (D,E) Sensorgrams showing lack of binding of α5 to β1 and γ1 (red) and formation of ternary α5–β1–γ1 complexes (black and grey). The concentrations of soluble analytes are indicated. LE, laminin-type epidermal growth factor-like; LN, laminin N-terminal; PAGE, polyacrylamide gel electrophoresis; WT, wild type.
In conclusion, the results of our SPR binding studies with laminin-511 short-arm fragments support the original three-arm interaction model of laminin-111 polymerization (Yurchenco & Cheng, 1993; McKee et al, 2007). We propose that transient contacts between the β- and γ-short arms are consolidated by the slower incorporation of an α-short arm, and that the conserved β5′–β6 loop of αLN domains has an important function in the cooperative assembly of the ternary αβγ nodes in the laminin network.
Methods
Protein production. Complementary DNAs coding for LN–LEa1–2 and LN–LEa1–4 fragments of the mouse laminin α1, α2, α5, β1 and γ1 chains (supplementary Table S1 online) were obtained by PCR amplification from plasmids kindly provided by Takako Sasaki (University of Erlangen, Germany). The PCR products were cloned into a modified pCEP-Pu vector coding for proteins with a C-terminal His6-tag (Carafoli et al, 2009). Site-directed mutagenesis was performed by strand overlap-extension PCR. All expression constructs were verified by DNA sequencing. Proteins were purified from the conditioned medium of episomally transfected HEK293 c18 cells (Carafoli et al, 2009) by HisTrap affinity chromatography (GE Healthcare), followed by size exclusion chromatography on a S200 HR10/30 column (GE Healthcare) at 4°C using a running buffer of 20 mM Na-HEPES, pH 7.5, 150 mM NaCl, 2 mM CaCl2.
Crystal structure determination. Crystals of fully glycosylated laminin α5LN–LEa1–2 were obtained by 200 nl sitting drop vapour diffusion using a protein concentration of 8 mg/ml and 4% PEG6000, 0.1 M MgCl2, 0.1 M sodium acetate, pH 5.2 as precipitant. The crystals contain one α5LN–LEa1–2 molecule in the asymmetrical unit and 75% solvent. Heavy atom derivatives were prepared by soaking crystals for 18 h in 2 mM UO2(NO3)2 or 1 mM K2PtCl4. Crystals were flash-frozen in liquid nitrogen in mother liquor supplemented with 20% glycerol. Diffraction data were collected at the Diamond Light Source (native data, beamline IO2, λ=0.980 Å) and at the Swiss Light Source (derivative data, beamline PXIII, λ=1.000 Å). The diffraction limit was anisotropic (approximately 3.2 Å in the a*b* plane and approximately 4 Å along c*). The images were processed with iMOSFLM (www.mrc-lmb.cam.ac.uk/harry/imosflm) and with programs of the CCP4 suite (CCP4, 1994). A MIRAS-phased electron density map at 3.4 Å resolution was obtained with autoSHARP (Vonrhein et al, 2007). A partial model was built with O (Jones et al, 1991) and refined with CNS (Brünger et al, 1998). The model was completed using a native data set to 2.9 Å resolution (approximately 3.2 Å along c*) collected from an isomorphous crystal of the laminin α5LN–LEa1–2 N100A/N383E double mutant obtained at 6 mg/ml protein concentration and 0.4 M NH4H2PO4 (pH 4.0). Crystallographic statistics are summarized in Table 1. Figures were generated using PYMOL (www.pymol.org).
Table 1. Crystallographic statistics of the laminin α5LN–LEa1–2 structure.
| Native (N100A/N383E) | Native (wild type) | UO2(NO3)2 (wild type) | K2PtCl4 (wild type) | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P62 | P62 | P62 | P62 |
| Cell dimensions | ||||
| a, b, c (Å) | 116.4, 116.4, 112.3 | 116.7, 116.7, 112.8 | 116.4, 116.4, 112.7 | 117.0, 117.0, 110.5 |
| α, β, γ (deg) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 25–2.9 (3.06–2.90)* | 20–3.4 | 20–3.7 | 20–3.7 |
| Rmerge | 0.135 (0.679) | 0.137 | 0.102 | 0.151 |
| <I/σ(I)> | 10.7 (3.1) | 11.3 | 16.4 | 10.3 |
| Completeness (%) | 100 (100) | 100 | 100 | 100 |
| Redundancy | 11.0 (11.2) | 10.9 | 11.5 | 11.2 |
| Phasing | ||||
| Rderiv | 0.231 | 0.269 | ||
| Phasing power (acentric/centric) | 1.37/1.60 | 1.37/1.33 | ||
| Refinement | ||||
| Resolution (Å) | 25–2.9 | |||
| No. of reflections | 19173 | |||
| Rwork/Rfree | 0.254/0.292 | |||
| No. of atoms | ||||
| Protein | 2682 | |||
| Ion | 30 | |||
| Average B-factor (Å2) | 76.0 | |||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.008 | |||
| Bond angles (deg) | 1.8 | |||
| Ramachandran plot† | 68.6/28.6/2.4/0.3 | |||
| LE, laminin-type epidermal growth factor-like domain; LN, laminin N-terminal. *Values in parentheses are for the highest resolution shell. †Percentage of residues in most favoured/additionally allowed/generously allowed/disallowed regions (program PROCHECK). | ||||
Laminin polymerization assay. The assay was carried out as described (Cheng et al, 1997). Briefly, mouse laminin-111 (Sigma) at a concentration of 0.25 mg/ml (0.35 μM) was incubated with 1 mM CaCl2 or 10 mM EDTA in 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1% Triton X-100 for 3 h at 37°C in a total volume of 100 μl. Some reactions also contained 2.1 μM laminin α5LN–LEa1–2 proteins. Polymerized laminin-111 was pelleted by centrifugation (approximately 10,000 g, 15 min). The pellet and supernatant fractions were analysed by SDS–polyacrylamide gel electrophoresis and Coomassie Blue staining. Band intensities (α1 and β1/γ1 combined) were quantified with ImageJ (rsbweb.nih.gov/ij).
SPR experiments. The SPR experiments were performed at 25°C using a Biacore 3000 instrument (GE Healthcare). Proteins were coupled to CM5 chips in 10 mM sodium acetate, pH 4.5, according to the manufacturer's instructions. Reference cells without protein were treated identically. The running buffer was 20 mM Na-HEPES, pH 7.5, 150 mM NaCl, 2 mM CaCl2 at a flow rate of 30 μl/min. Binary interactions were measured without regeneration of the sensor surfaces between runs. Ternary interactions on the β1LN–LEa1–4 surface were measured with a brief pulse of 10 mM glycine–HCl, pH 2.5, between runs. No suitable regeneration buffer could be found for the γ1LN–LEa1–4 surface. The sensorgrams were analysed with Biacore software.
Accession codes. The coordinates of the laminin α5 LN–LEa1–2 fragment have been deposited in the Protein Data Bank under accession code 2y38.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Takako Sasaki (University of Erlangen, Germany) for laminin complementary DNAs, the beamline staff at the Diamond and Swiss Light Sources for help with data collection, Geoff Baldwin for help with fluorescence spectroscopy and Jan Gebauer for help with ImageJ software. This work was supported by a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science to E.H. (ref 083942).
Footnotes
The authors declare that they have no conflict of interest.
References
- Aumailley M et al. (2005) A simplified laminin nomenclature. Matrix Biol 24: 326–332 [DOI] [PubMed] [Google Scholar]
- Boraston AB, Bolam DN, Gilbert HJ, Davies GJ (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Carafoli F, Clout NJ, Hohenester E (2009) Crystal structure of the LG1-3 region of the laminin α2 chain. J Biol Chem 284: 22786–22792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cheng YS, Champliaud MF, Burgeson RE, Marinkovich MP, Yurchenco PD (1997) Self-assembly of laminin isoforms. J Biol Chem 272: 31525–31532 [DOI] [PubMed] [Google Scholar]
- Dickson BJ (2002) Molecular mechanisms of axon guidance. Science 298: 1959–1964 [DOI] [PubMed] [Google Scholar]
- Edwards MM et al. (2010) Mutations in Lama1 disrupt retinal vascular development and inner limiting membrane formation. J Biol Chem 285: 7697–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettner N, Göhring W, Sasaki T, Mann K, Timpl R (1998) The N-terminal globular domain of the laminin α1 chain binds to α1β1 and α2β1 integrins and to the heparan sulfate-containing domains of perlecan. FEBS Lett 430: 217–221 [DOI] [PubMed] [Google Scholar]
- Garbe JH, Göhring W, Mann K, Timpl R, Sasaki T (2002) Complete sequence, recombinant analysis and binding to laminins and sulphated ligands of the N-terminal domains of laminin α3B and α5 chains. Biochem J 362: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kalkhof S, Haehn S, Paulsson M, Smyth N, Meiler J, Sinz A (2010) Computational modeling of laminin N-terminal domains using sparse distance constraints from disulfide bonds and chemical cross-linking. Proteins 78: 3409–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee KK, Harrison D, Capizzi S, Yurchenco PD (2007) Role of laminin terminal globular domains in basement membrane assembly. J Biol Chem 282: 21437–21447 [DOI] [PubMed] [Google Scholar]
- Miner JH, Yurchenco PD (2004) Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol 20: 255–284 [DOI] [PubMed] [Google Scholar]
- Nielsen PK, Yamada Y (2001) Identification of cell-binding sites on the Laminin α5 N-terminal domain by site-directed mutagenesis. J Biol Chem 276: 10906–10912 [DOI] [PubMed] [Google Scholar]
- Odenthal U, Haehn S, Tunggal P, Merkl B, Schomburg D, Frie C, Paulsson M, Smyth N (2004) Molecular analysis of laminin N-terminal domains mediating self-interactions. J Biol Chem 279: 44504–44512 [DOI] [PubMed] [Google Scholar]
- Patton BL, Wang B, Tarumi YS, Seburn KL, Burgess RW (2008) A single point mutation in the LN domain of LAMA2 causes muscular dystrophy and peripheral amyelination. J Cell Sci 121: 1593–1604 [DOI] [PubMed] [Google Scholar]
- Paulsson M (1988) The role of Ca2+ binding in the self-aggregation of laminin-nidogen complexes. J Biol Chem 263: 5425–5430 [PubMed] [Google Scholar]
- Stetefeld J, Mayer U, Timpl R, Huber R (1996) Crystal structure of three consecutive laminin-type epidermal growth factor-like (LE) modules of laminin γ1 chain harboring the nidogen binding site. J Mol Biol 257: 644–657 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G (2007) Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Xu H, Wu XR, Wewer UM, Engvall E (1994) Murine muscular dystrophy caused by a mutation in the laminin α2 (Lama2) gene. Nat Genet 8: 297–302 [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, Cheng YS (1993) Self-assembly and calcium-binding sites in laminin. A three-arm interaction model. J Biol Chem 268: 17286–17299 [PubMed] [Google Scholar]
- Yurchenco PD, Tsilibary EC, Charonis AS, Furthmayr H (1985) Laminin polymerization in vitro. Evidence for a two-step assembly with domain specificity. J Biol Chem 260: 7636–7644 [PubMed] [Google Scholar]
- Yurchenco PD, Amenta PS, Patton BL (2004) Basement membrane assembly, stability and activities observed through a developmental lens. Matrix Biol 22: 521–538 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.