Abstract
Rad21L—a new meiotic cohesin described in a recent paper in EMBO reports by the Watanabe group—probably has a role in the establishment of homologous chromosome recognition.
EMBO Rep (2011) advance online publication. doi:10.1038/embor.2011.2
Sexual reproduction is one of the great success stories of evolution, allowing the reshuffling of genetic material from two parents to create new gene combinations for the next generation. To this end, diploid organisms produce gametes containing a randomly selected haploid complement of their genome. The necessary chromosome gymnastics occur in two sequential meiotic cell divisions (Petronczki et al, 2003). The first ‘reductional' division separates the two parental homologues, thereby reducing the number of chromosomes by one-half. A second ‘equational' division splits each homologue into its two sister chromatids. Chromosome behaviour in both divisions is influenced by the chromosomal cohesin complex, which links sister chromatids and mediates their regulated separation. The cohesin complex has been the subject of intense investigation for 14 years and, thus, the identification of a hitherto unknown subunit—by Watanabe and colleagues in EMBO reports, and by Lee and Hirano in a recent issue of the Journal of Cell Biology (Ishiguro et al, 2011; Lee & Hirano, 2011)—comes as a surprise.
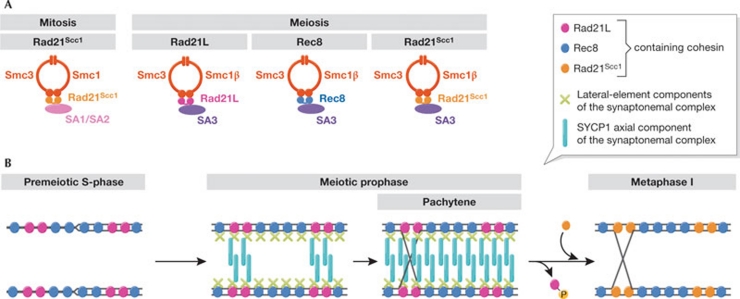
Cohesin is a ring-shaped protein complex that was initially characterized on mitotic chromosomes. It consists of four core subunits: two from the structural maintenance of chromosomes (Smc) family, Smc1 and Smc3; Rad21, which is also known as Scc1; and Scc3, which has two mammalian paralogues—SA1 and SA2. The Smc subunits form most of the ring circumference, whereas the other two stabilize it and probably regulate its function (Fig 1A). Cohesin is loaded onto chromosomes in the G1 phase of the cell cycle and establishes sister chromatid cohesion in S-phase by topologically entrapping the two newly synthesized replication products. It embraces sister chromatids until mitosis; at anaphase onset, proteolytic cleavage of the Rad21Scc1 subunit opens the ring to release sister chromatids (Uhlmann, 2009). During meiosis, this is extended in preparation for the first reductional division and, in most organisms, Rad21Scc1 is mostly replaced by a meiosis-specific paralogue, Rec8. Vertebrates express additional meiosis-specific variants of Smc1 (Smc1β) and Scc3 (SA3). After pre-meiotic DNA replication, meiotic cohesins promote homologue recombination; the basis for genetic exchange between maternal and paternal genomes. The cohesin complex binds to the ‘lateral elements' of the synaptonemal complex, which joins the two homologous pairs of sister chromatids to form the characteristic pachytene chromosomes of meiotic prophase (Fig 1B). Once recombination is complete, the synaptonemal complex disassembles, whereas the two homologous chromosomes remain connected by sister chromatid cohesion, distal to the sites of homologue exchange.
Figure 1.
Rad21L, a new cohesin subunit with unprecedented features. (A) Subunit composition of vertebrate cohesin complexes. The mitotic and three main meiotic complexes are shown, including that containing Rad21L. (B) Model for the localization of cohesin complexes during meiosis. Rec8 and Rad21L cohesin complexes establish sister chromatid cohesion during premeiotic S-phase and act as a basis for lateral-element formation, but only Rad21L recruits SYCP1. Nucleation of synaptonemal-complex assembly is facilitated by the matching patterns of Rad21L on homologous chromosomes. Once recombination is complete, phosphorylation of Rad21L-containing complexes dissociates them from chromosomes, facilitating subsequent synaptonemal-complex disassembly. Rad21Scc1-containing cohesin is then recruited to chromosomes.
At least, that was the picture until the discovery of an additional paralogue of Rad21Scc1 and Rec8 in vertebrates (Ishiguro et al, 2011; Lee & Hirano, 2011). Due to its similarity to Rad21Scc1, it has been called Rad21L (for ‘Rad21-like'). Rad21L is expressed exclusively in cells undergoing meiosis and forms a complex with the other meiosis-specific cohesin subunits (Fig 1A). We cannot be sure of the function of Rad21L, as the development of genetic tools in recombinant mouse strains will take time. Nevertheless, features of the localization pattern of Rad21L have led the authors to speculate about its role. Similarly to Rec8, Rad21L appears on chromosomes at premeiotic S-phase, as would be expected for cohesin subunits that establish sister chromatid cohesion. Surprisingly, once recombination is complete, Rad21L disappears from chromosomes, while homologues remain juxtaposed by the synaptonemal complex and Rec8 persists along chromosome axes. Thus, the Rad21L complex is present on chromosomes during the establishment of sister chromatid cohesion, homologue pairing and synapsis. However, when cells approach division, Rad21L has left the chromosomes, thus a role in sister chromatid cohesion is unlikely. Might Rad21L have a role in homologue pairing? Both studies also highlight the exchange of Rad21L for Rad21Scc1 during meiotic prophase, long after the establishment of replication-coupled cohesion (Prieto et al, 2004). Can Rad21Scc1 substitute for Rad21L in sister chromatid cohesion? Might Rad21Scc1 promote chromosome condensation rather than sister chromatid cohesion?
Several events contribute to homologue pairing during meiotic prophase, but the molecular basis of homologue recognition is poorly understood (Bhalla & Dernburg, 2008). Strikingly, when Watanabe and colleagues and Lee and Hirano visualized Rad21L along meiotic chromosomes, they found a pattern that alternates with Rec8. The Rad21L–Rec8 pattern appears unique to each chromosome, but is reproducible along the two homologues, leading Watanabe and colleagues to suggest it could provide a ‘barcode' for homologue recognition (Fig 1B). While this is an attractive idea, how might Rad21L-containing cohesin complexes on the two homologues recognize each other? No convincing evidence has been obtained so far of interactions between more than one cohesin complex. Instead, Lee and Hirano provide a piece of molecular evidence that could be crucial. They find that Rad21L—but not Rec8 or Rad21Scc1— interacts biochemically with the synaptonemal-complex protein SYCP1, which constitutes its axial element (Liu et al, 1996). After trial and error during early stages of homologue recognition, SYCP1 recruitment by Rad21L could nucleate axial-element formation, and this reaction would progress most efficiently between homologues with complementary Rad21L patterns.
The above hypothesis for Rad21L-mediated homologue recognition merits further investigation, and poses several questions. How are the alternating domains of Rad21L- and Rec8-containing cohesin complexes defined along chromosomes? An analysis of these domains at higher resolution—by chromatin immunoprecipitation experiments—might yield important insight. Is the chromosome binding of Rad21L- and Rec8-containing cohesin catalysed by the same Scc2/Scc4 cohesin-loader? If so, how does the cohesin-loader, or its chromosomal context, distinguish between the two cohesin complexes? Furthermore, clear Rad21L orthologues are only found in vertebrates. If a Rad21L–Rec8 pattern indeed contributes to homologue recognition, how would such a mechanism work in organisms that do not have Rad21L? Watanabe and colleagues suggest that the Rec8 distribution pattern itself could be the barcode that promotes homologue pairing in these cases. In small genomes, like that of budding yeast, Rec8 is preferentially found at convergent transcriptional terminators, the distribution pattern of which is unique to each chromosome (Lengronne et al, 2004). On yeast chromosomes, which condense to a lesser degree in meiosis than vertebrate chromosomes, these patterns could be recognized and provide specificity for homologue-pairing. Such a model would predict that Rec8 interacts with an axial-element component in these organisms.
If Rad21L promotes synaptonemal-complex assembly, what are the consequences of its disassembly after the recombination reactions are complete? The early departure of the Rad21L complex from chromosomes could facilitate synaptonemal-complex disassembly. Watanabe and colleagues report that the dissociation of Rad21L from chromosomes depends on Polo kinase, while Lee and Hirano find that Rad21L- but not Rec8-containing cohesin is preferentially phosphorylated on its SA3 subunit. Similar cohesin phosphorylation on SA1/SA2 subunits has been implicated in the removal of a substantial fraction of cohesin from mitotic chromosomes (Sumara et al, 2002). Notably, Polo kinase has been implicated in cohesin dissociation and synaptonemal-complex disassembly during meiotic prophase in yeast (Clyne et al, 2003; Yu & Koshland, 2005). Although this supports a role for cohesin phosphorylation and dissociation in synaptonemal-complex disassembly, Rad21L is lost from chromosomes during pachytene, when the synaptonemal complex persists. This suggests that additional levels of regulation remain to be found. Clearly, the new kid on the block and its function on meiotic chromosomes will keep us on our toes for some time to come.
References
- Bhalla N, Dernburg AF (2008) Annu Rev Cell Dev Biol 24: 397–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne RK et al. (2003) Nat Cell Biol 5: 480–485 [DOI] [PubMed] [Google Scholar]
- Ishiguro K et al. (2011) EMBO Rep [Epub 28 Jan] doi:10.1038/embor.2011.2 [Google Scholar]
- Lee J, Hirano T (2011) J Cell Biol 192: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A et al. (2004) Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JG et al. (1996) Exp Cell Res 226: 11–19 [DOI] [PubMed] [Google Scholar]
- Petronczki M et al. (2003) Cell 112: 423–440 [DOI] [PubMed] [Google Scholar]
- Prieto IC et al. (2004) Chromosome Res 12: 197–213 [DOI] [PubMed] [Google Scholar]
- Sumara I et al. (2002) Mol Cell 9: 515–525 [DOI] [PubMed] [Google Scholar]
- Uhlmann F (2009) EMBO Rep 10: 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H-G, Koshland D (2005) Cell 123: 397–407 [DOI] [PubMed] [Google Scholar]