Abstract
Limitations to biomarker discovery are not only technical or bioinformatic but conceptual as well. In our attempt to offer a solution, we are highlighting three issues that we think are limiting progress in biomarkers discovery. First, the confusion stemming from the imposition of a pathology-type immunohistochemical marker (IHCM) concept on omics data without fully understanding the characteristics and limitations of IHCMs as applied in clinical pathology. Second, the lack of serious consideration for the scope of disease heterogeneity. Third, the refusal of the biomedical community to borrow from other biological disciplines their well established methods for dealing with heterogeneity. Therefore, real progress in biomarker discovery will be attained when we recognize that an omics biomarker cannot be assigned and validated without a priori data modeling and subtyping of the disease itself to reveal the extent of its heterogeneity, and its omics' clonal aberrations (drivers) underlying its subtypes and pathways' diversity. To further support our viewpoints, we are contributing a novel a systems biology method such as parsimony phylogenetic approach for disease modeling prior to biomarker circumscription. As an analytical approach that has been successfully used for a half of a century in other biological disciplines, parsimony phylogenetics simultaneously achieves several objectives: it provides disease modeling in a hierarchical phylogenetic classification, identifies biomarkers as the shared derived expressions or mutations—synapomorphies, constructs the omics profiles of specimens based on the most parsimonious arrangement of their heterogeneous data, and permits network profiling of affected signaling pathways as the biosignature of disease classes.
Introduction
With the total number of cancer patients in the United States projected to increase by 55% at 2020, the need for an effective early detection methods and prevention programs becomes more crucial to ameliorate the situation of rising statistics (Roukos, 2009; Warren et al., 2008). Therefore, accurate predictive biomarkers and/or profiling techniques for early detection can play an important role in affecting patients' survival and provide the proper treatment.
The identification, qualification, and application of diagnostic and prognostic biomarkers remain the holy grail of the current omics paradigm. Despite the setbacks, the quest for biomarkers goes on and the expectations are still holding (Morrison and Veenstra, 2008; Rifai et al., 2006). Biomedical researchers keep a watchful eye for any gene, protein, or metabolite expressions that could serve as biomarkers indicative of early disease phenotypes and subphenotypes, or predictive of disease progression and outcome. More highly desirable are biomarkers that can be tagged to drug targets and therapy. The search for biomarkers has not been very fruitful compared to the amount of investment thus far (May, 2010; Morrison and Veenstra, 2008; Nature, 2008, 2010; Sawyers, 2008). Although from our perspective the search is highly justifiable, we think that current omics-based approaches to biomarker discovery face conceptual and bioinformatic challenges that are impeding effective mining of most data types. The need for a conceptual fine-tuning and an integrative analytical approach to data mining has become imperative to move such efforts to success (Abu-Asab et al., 2008a).
We have outlined below conceptual problem and practical challenges, and demonstrated a new systems biology paradigm of omics data mining for biomarkers and profiling of biosignatures. Systems biology here is an integrative framework to data analysis. There are several main issues that, in our opinion, seem to be the largest hurdles in the process of biomarkers' discovery. Central to all other issues is dealing with data heterogeneity that reflects disease heterogeneity. The molecular concept of disease, as it turned out, is more complex and problematic than was thought earlier, and in turn, is reflected in the difficultly of demarcating disease initiation and boundaries in an omics context. Many on the biomarker discovery wagon seem oblivious to the intractable issue of heterogeneity as well as to the history and nature of biomarkers. As we explain below, biomarkers that are in current usage (especially in diagnostic pathology) are far from being perfect, and they are not always applied in a straight forward manner.
Successful search for omics biomarkers will move forward by resolving the conceptual and practical challenges that we have outlined here, especially the need for a utilitarian omics-based biomarker definition, and more disease-relevant bioinformatic tools. As a step forward in this direction, we are proposing the use of parsimony phylogenetic paradigm as a new systems biology paradigm to modeling and analyzing omics data, identify omics biomarkers as the shared clonal aberrations, subtype disease classes, and profile altered signaling pathways as the genotypes of disease classes as demonstrated on the prostate cancer gene microarray dataset presented herein.
Current Problems in Biomarker Discovery
With the expansion of omics applications, it was expected that a new generation of more accurate and ubiquitous biomarkers would be identified with relative ease; however, the task proved to be intractable; only very few biomarkers have been brought to clinical settings, and many proved to be irreproducible (Ransohoff, 2009; Sawyers, 2008). The pathology biomarker paradigms of immunohistochemical markers (IHCM) and blood biomarker have not applied well to the omics realm—mainly because of their own specificities and limitations, and thus far no adjustment of biomarker definition to the new quantitative omics data has been introduced. Many researchers mining omics data do not seem to be familiar with IHCM's features, modes of application, and their limitations. The use of IHCM in diagnostic pathology has been qualitative in nature, and the variability in their detection power, overlapping, and inconsistencies due to the heterogeneity of diseased tissues have always been recognized by pathologists (Heim-Hall and Yohe, 2008; Kashani-Sabet et al., 2009; Lerma et al., 2007; Rosai and Ackerman, 2004).
Provisionality of omics-based biomarkers is a pandemic that is well documented is ample reports (Kaiser, 2009; May 2010). It is a common problem that is linked to the above issues, and can additionally be attributed to a few more factors: the statistically underpowered size of diseased specimens in the majority of reports; use of a limited size (or complete lack) of a nondiseased control group of specimens as a study baseline; and most importantly the use of bioinformatic tools void of biological relevance for data mining and disease modeling. Provisionality extends to some blood biomarkers that are currently in use in clinical practice. For example, the value of CA 125 (cancer antigen 125, also known as mucin 16 or MUC16) and PSA (prostate specific antigen) as markers of early detection remains as of now controversial, for their benefit may be offset by overdiagnosis and unnecessary treatment, as well as poor sensitivity and specificity for early detection (Barry, 2009; Partridge et al., 2009).
Difficulty in locating single biomarkers is giving way to profiling as an alternative to single biomarkers for prognostic and predictive purposes. Small tumors may release low-abundance proteins in the blood that are outside of the dynamic range of detection of most conventional assays, but they also alter the expression pattern of normal proteins (Concato et al., 2009; Oved et al., et al., 2009); profiling of the latter change may therefore be more useful as early disease biomarker than measurement of a single circulating factor that cannot reliably distinguish between individuals with and without cancer (May, 2010; Schaub et al., 2009). Similarly, profiling of gene expression on the basis of two to several thousand genes provides diagnostic, prognostic, or predictive information about tumors (Dowsett and Dunbier, 2008; Loi et al., 2007; Wang et al., 2005).
The current mainstream statistical and mathematical analytical methods need to be contributing toward a solution to these issues. The goal of a bioinformatic analytical tool is to reveal patterns within the data [such as shared-expression aberrations (the clonal changes shared by the specimens of a disease type), modeling heterogeneity, profiling of disease classes and subclasses, etc.) to produce a predictive and seamless model of specimens' classification that can be further utilized in a clinical setting (Abu-Asab et al., 2008b). However, overdependence on mechanistic parametric methods often masks the potential usefulness of the data. As such, they should yield to biologically relevant analytical methods that take into account the evolutionary nature of the disease (especially cancer), capable of processing omics' data heterogeneity, incorporate all expression modes within the data into the analysis (such as biomodal expression), and produce a seamless multidimensional predictive classification of the specimens of interest (Abu-Asab et al., 2008a, 2008b). For example, arbitrary choice of boundaries, such as fold change, and heavy reliance on biostatistics reduce predictivity by suppressing diverse patterns of expressions and their multiple profiles, thus producing artificial interpretations of specimens' relatedness.
Dichotomy between IHCM and Omics Biomarkers
Not all types of biomarkers are equal, and little has been reported on the inherent differences between IHCMs and omics biomarkers (Table 1), as well as their association with the disease process and its boundaries (Knox, 2010). This information vacuum has left room for a mix up of their natures and uses, and created unrealistic expectations. There are many examples of erroneous statements portending the applicability of IHCMs to high throughput omics data (Takikita et al., 2007). What is missing in the biomarker debate is a true understanding of IHCMs' usage and a comprehension of the heterogeneous nature of the disease.
Table 1.
A Comparison Between Immunohistochemical Markers (IHCM) and Omics Biomarkers Showing the Differences and Similarities Between the Two Classes of Biomarkers
| Immunohistochemical markers | Omics biomarkers |
|---|---|
| Qualitative in nature: presence or absence is reported with minimum quantitation | Mostly quantitative in nature: derived from data in absolute numbers but may also be used qualitatively |
| Tissue specific: indicative of the origin of a diseased tissue | Disease specific: used to define the common aberrant omics expressions of a disease |
| Used for diagnosis and often in combinations to narrow down differential diagnosis but of very limited use for early detection of disease | Thus far have not been used for clinical diagnosis or for early detection of disease but search is ongoing for such biomarkers |
| Diagnosis is not affected by their heterogeneous quantitative distribution among specimens | Their heterogeneous quantitative distribution among specimens is significant and affects their statistical designation |
| Based on a few cells of a specimen in a light microscopic field | Based on the cellular extract of many cells of a specimen |
| Are sometimes inconsistent and overlapping | Have not been evaluated for this criterion |
| Their partial staining of tumors indicates polyclonality | They may reveal polyclonality depending on the analytical method (statistical vs. phylogenetic) |
One aspect of this issue is the apparent disconnect between omics data and IHCM data. Researchers are surprised when the IHCMs used in pathology do not turn up as significant in the omics results, and vice versa. For example, positive immunoreactivity of gastric cancer specimens to p53, E-cadherin, and β–catenin was not detected in microarray analysis of the same specimens at the RNA level (Hippo et al., 2002). Omics specimen preparation and common analytical methods impose a stringent quantitative criterion that is incompatible with IHCM's usage. Whereas the IHCM result represents a qualitative assessment of one or a pair of markers at a time in a few cells, the omics result reflects the total content of the cells' extract used in the study. Although an IHCM will most likely fail to transcend into an omics biomarker, the reverse could occasionally take place, that is, an omics biomarker could be a useful IHCM.
Tens of IHCMs are used in disease diagnoses, prognoses, and follow-ups. A well trained pathologist has no doubt when a tissue is positive for an antibody marker under the light microscope, and there is minimum quantitation needed in order to reach a conclusion. An IHCM staining of a tumor is usually partial because a marker rarely stains all the cells of a tumor. Thus, in IHCMs evaluations, it is the clear presence or absence and not the quantity that is significant for diagnosis—a qualitative and not quantitative criterion of evaluation. For the pathologist, the positive staining of a few cells in a field of tumor cells is good enough to call the IHCM staining positive, although in this case it is indicative of heterogeneity within the tumor as in many soft tissue sarcomas, blastomas, and mixed nonseminomatous germ cell neoplasia (Maher, 2008; Rosai and Ackerman, 2004). Polyclonality, or heterogeneity, is emerging as the single most important factor in obstructing effective data mining and hindering biomarker discovery, and is responsible for drug resistance as well (Sumer and Gao, 2008).
There are a few additional reasons for the breakdown of the IHCM concept when applied to high-throughput data of microarrays and proteomics; these highlight the specific ways by which IHCMs are applied. In most situations of diagnostic pathology, IHCMs are usually chosen to correlate the tumor immunophenotype with its tissue of origin, so they are markers of normally or partially differentiated tissues (tissue specific) and not of disease type (not disease specific). This is the reverse of the omics' aim because the analysis is done to find common aberrant expressions in diseased tissues. In addition, IHCMs are routinely employed in combinations that are selected by the pathologist. Pathologists occasionally use a combination of IHCMs that are not biologically linked, but are employed for practical reasons in order to narrow down a differential diagnosis; a process that cannot be applied to an omics dataset.
An IHCM among a number of specimens of the same disease may have a widely heterogeneous quantitative distribution when measured by gene-expression or proteomics method, which renders it challenging for consideration as an omics biomarker. Additionally, various omics expressions of genes and proteins have shown a deviation from normal distribution in a group of diseased specimens; a recently recognized phenomenon that has not been taken into consideration when assessing datasets for biomarkers or disease subtyping (Abu-Asab et al., 2008a; Lyons-Weiler et al., 2004). These two forms of heterogeneity are quantitatively measured only by omics methods, and may be utilized for subtyping of disease specimens.
We have outlined the differences underlying the dichotomy between IHCM and omics biomarkers; these differences bolster the argument for a conceptual fine tuning of the omics biomarkers paradigm in order to shake off the misconceptions of the IHCM paradigm. The assumption of one-to-one correspondence between omics' biomarkers and IHCMs, although possible in some cases, should not be expected as the rule. Recognizing that the two approaches stem from different paradigms and may have different utility is important in order to avoid false expectations.
Disease and Specimen Heterogeneities: Roadblocks in the Omics' Biomarker Path
Only recently the term heterogeneity has become widely acknowledged as a phenomenon that is characteristic of pathogenesis and can be detected and quantified within diseased tissues (Deo and Roth, 2009; Fisher et al., 2008). Limitations of conventional anticancer chemotherapy as well as the inadequate efficacy of targeted treatments have been attributed to tumor heterogeneity (Roukos, 2009). Heterogeneity occurs at two levels: one as multiple clones/phenotypes within an individual (such as within a tumor or between several tumors from the same patient—specimen heterogeneity), and the other between individuals (disease heterogeneity).
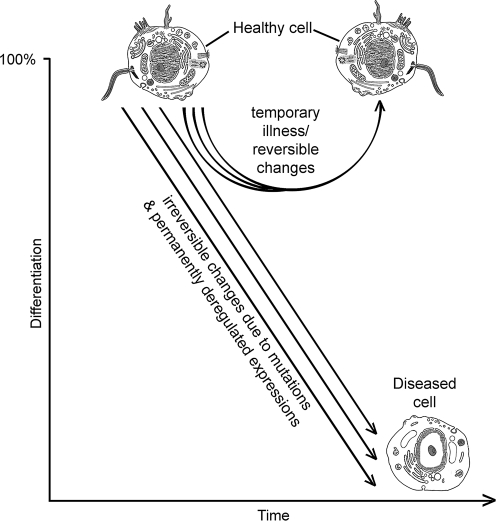
In general, pathogenesis is a downhill disruption of cellular differentiation. The disruption involves the alteration of tissue-specific gene expression (Winter et al., 2004). The alterations are reversible in temporary illnesses, and irreversible due to mutations and permanently dysregulated expressions in cancer and degenerative diseases (Fig. 1). Difficulty with the discovery of valuable omics biomarkers may have to do with the heterogeneous nature of disease initiation and progression where multiple intertwined processes may produce the same disease phenotype (Fig. 1), and the inclusion within the definition of disease the blurry boundaries between normal and abnormal health conditions. The collection of symptoms that we call a disease may be produced by hundreds or thousands of heterogeneous omics variations (Bielas et al., 2006; Maher, 2008). The asynchronous, homoplastic, and heterotachous expression patterns of genes and proteins among diseased specimens further complicate the state of disease heterogeneity (Abu-Asab, 2009).
FIG. 1.
Pathogenesis as a heterogeneous process in cancer, degenerative disease, and temporary illness disturbs the differentiation of the affected cells. The process is reversible in temporary illness, but irreversible in cancer and degenerative diseases. The latter two are associated with mutations and permanently deregulated expressions. Multiple arrows signify that several pathways may produce the same disease phenotype, a condition that complicates biomarkers discovery and calls for a priori modeling of the disease to reveal its classes.
Cancer incipience and progression are driven by random mutations (Abu-Asab, 2009; Heng et al., 2010; Loeb et al., 2008). This randomness, compounded with selective pressure within tumors, produces intra- and interspecimen heterogeneity, which in cancer is the basis for selection to maximize the tumor's success (Abelev and Eraiser, 2008). Heterogeneity is based on a mixture of “clonal” (driver) and “nonexpanded” (passenger) mutations (Campbell et al., 2008; Loeb et al., 2008), and may be also on nongenetic individuality as it has been recently suggested (Brock et al., 2009). Only clonal mutations are the potential biomarkers, because they systematically characterize a larger number of specimens, whereas nonexpanded ones are restricted to fewer specimens and would have limited utility (Abu-Asab et al., 2008a, 2008b; Yaffe, 2008).
Heterogeneity is detected in gene-expression microarrays and proteomic datasets (Abu-Asab et al., 2006, 2008a, 2008b; Dalerba et al., 2007; Heng et al., 2010). It occurs as simple asynchronous pattern of expression, where a group of diseased specimens has a mixture of normal and aberrant expression values for the same genes, or as complex pattern of dichotomous asynchronicity, where the disease gene expressions are both above and below the normal range (i.e., outside the normal range) (Abu-Asab et al., 2008a, 2008b; Lyons-Weiler et al., 2004). As with mutations, gene expression aberrations can be described as clonal and nonexpanded, as well as reversible and irreversible. In cancer, as well as degenerative and chronic diseases, clonal mutations and expressions are most likely irreversible, and will contribute to the pathophysiology throughout the lifespan of the disease (Hoeijmakers, 2001; Kaput and Rodriguez, 2004). This is an important assumption in bioinformatic analysis because it will affect data modeling and subtyping, as well as subsequent biomarker discovery (see Modeling Heterogeneity below).
Lately, heterogeneity in all of its forms has emerged as the most intractable obstacle in biomarker discovery and targeted treatment. Therefore, in an omics biomarker discovery context, the challenge is how to effectively mine heterogeneous data. We argue that a bioinformatic analytical tool should produce an unbiased data-based classification/modeling of the specimens that will simultaneously map the distribution of variant expressions or mutations, and permit the distinction between the clonal and nonexpanded mutations and expressions before any of them can be designated as a potential biomarker. Therefore, modeling of disease and specimens heterogeneities is a prerequisite to omics biomarker discovery.
Modeling Heterogeneity: Biomarkers and Profiles, Two Faces of the Same Coin
Choosing among the different analytical paradigms for the most suitable one to deal with heterogeneity has been the subject of lengthy debate among systematics biologists during the second half of the 20th century (Abu-Asab, 2009). There are a few theoretical aspects to an analytical tool that will successfully tackle heterogeneity and produce meaningful results. Among these is its optimum modeling of complex data in a multidimensional hierarchical classification such as the phylogenetic cladogram (a tree diagram that shows the relatedness/similarity between the specimens); identification of shared derived clonal aberrations—the synapomorphies; subtyping of the study collection—class discovery; ability to reduce multiple occurrences of a change, reversals, and parallelism when modeling the data; and its hierarchical classification reveals direction of change among a group of specimens. In several recent publications, we have detailed our reasoning for the choice of maximum parsimony as the analytical paradigm for systems biology that is most suitable for mining heterogeneity (Abu-Asab, 2009; Abu-Asab et al., 2006; 2008a, 2008b).
Given the inherent uncertainty about data reproducibility between runs and among laboratories, the omics biomarkers discovery may be better approached with a qualitative aspect to data analysis that circumvents the shortcomings of the quantitative approach (Abu-Asab et al., 2006, 2008a, 2008b). Data analysis has been widely focused on specimen-based statistical methods [such as clustering and principal component analysis (PCA)] that are void of attempts to utilize the qualitative aspects of the data and take into consideration the biological and evolutionary nature of the disease. This has hindered a meaningful interpretation of omics data and with it biomarker discovery (Kolaczkowski and Thornton, 2004; Stefankovic and Vigoda, 2007a, 2007b).
Additionally, sorting out clonal from nonexpanded expressions/mutations is a qualitative process that depends on the distribution pattern of the expressions/mutations among the diseased specimens under study. To bring out the qualitative content of a dataset, there are a few required steps. First, the experimental design should always include normal or nondiseased specimens (Abu-Asab et al., 2008a, 2008b). For example, an analysis of cancerous specimens should include noncancerous ones of the same tissue type or a closely related one to be used later in the analysis as the baseline. Second, for an expression value to be considered differentially expressed, its values should be outside of the normals' range (below the minimum or above the maximum) in all of the diseased specimens. We have suggested the normals' range as a baseline reference rather than other mathematical abstracts because it does not force the dismissal from the analysis those expressions that do not have normal distribution among a group of specimens. An expression that occurs outside the normal range is termed derived or abnormal (Abu-Asab et al., 2008a). A shared derived expression state (termed synapomorphy) circumscribes a class into a natural clade of related specimens. Therefore, a synapomorphy is the potential biomarker.
There could be tens, hundreds, or even thousands, of synapomorphies within a group or the subgroups of diseased specimens. Some synapomorphies may be universally shared by all specimens of a disease, but others have a restricted distribution among these specimens; thus, an analytical algorithm that can optimally model such a heterogeneous distribution is needed here. Parsimony phylogenetics has been recognized as the most suitable method to analyze heterogeneity among specimens (Abu-Asab et al., 2006, 2008a, 2008b; Kolaczkowski and Thornton, 2004; Stefankovic and Vigoda, 2007a, 2007b); it is known to produce the most plausible hypothesis of relationships for the study set in a hierarchical classification—usually presented in a graphical tree-like format termed cladogram (see Fig. 2 for an example of a cladogram). At the same time, parsimony identifies the shared derived states of expressions for each clade of specimens when present—the simultaneously deregulated expressions/mutations that form a module driving disease pathogenesis. Because expressions/mutations in cancers and chronic diseases are irreversible, a parsimony phylogenetic analysis based on Camin-Sokal algorithm is a more appropriate method because it is built on such assumption (Camin and Sokal, 1965).
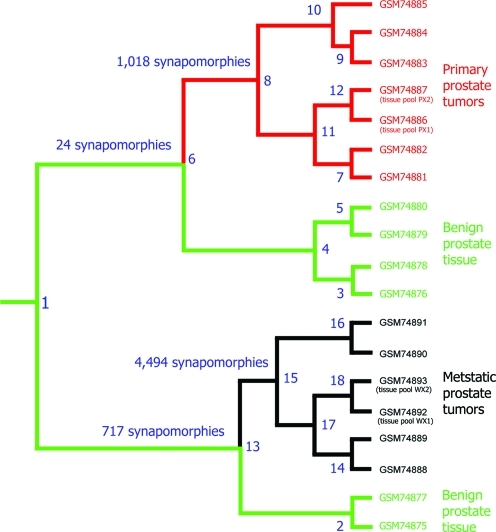
FIG. 2.
A most parsimonious cladogram produced by PHYLIP's MIX using Camin-Sokal parsimony algorithm. Dataset GDS1439 (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gds) is comprised of six benign specimens, as well as seven primary and six metastatic prostate tumors. The cladogram shows a major bifurcation that delineates two clades; the first composed of all primary tumors and four benign specimens (node 6, supported by 24 synapomorphies), and the second composed of all metastatic tumors and two benign specimens (node 13, supported by 717 synapomorphies). A clade of primary tumors is delimited by 1,018 synapomorphies (node 8), whereas a clade of the metastatic tumors is delimited by 4,494 synapomorphies (node 15). Synapomorphies at nodes 6, 8, 13, and 15 are considered clonal (driver) expression aberrations. Pooled primary tumor specimens PX1 and PX2 grouped into a clade (node 12), whereas pooled metastatic specimens, WX1 and WX2 formed a clade (node 18).
To illustrate our premise by an example, we selected dataset GDS1439 from NCBI (http://www.ncbi.nlm.nih.gov/sites/entrez?db = gds). GDS1439 contains the gene-expression microarray data of a set of specimens composed of six benign specimens, as well as seven primary and six metastatic prostate carcinoma specimens (Varambally et al., 2005). The first step of the analysis was sorting the expression values into derived (abnormal) and ancestral (normal) by comparing the values of the cancerous specimens against the range of the benign specimens for every gene in the dataset. This transformed the original data matrix into a new qualitative matrix of 0s (ancestral/normal) and 1s (derived/abnormal). The new matrix was processed with MIX (the parsimony program of the PHYLIP package) using Camin-Sokal parsimony method (Felsenstein, 1989), which produced one most parsimonious cladogram (Fig. 2).
The cladogram shows a major dichotomy of two clades; the first clade encompasses all seven primary tumors and four of the six benign specimens (Fig. 2, node 6), and the second clade composed of all metastatic tumors and the remaining two benign specimens (Fig. 2, node 13). Each of these two major clades is supported by a list of synapomorphies that are shared by their respective specimens; the number of synapomorphies for each clade appears close to its node in Fig. 2 (for a list of synapomorphies see Supplementary Table 1). Furthermore, the two sets of cancerous specimens, the primary and metastatic, each form a clade separate from their respective benign sister group (nodes 8 and 15, respectively) that are also supported by their own synapomorphies. The two sets of synapomorphies of the primary and metastatic prostate cancer are the potential biomarkers for each group.
The two benign specimens forming the sister group to the metastatic specimens share 717 synapomorphies with the metastatic tumors (node 13), a far larger number than what the other benign specimens (node 4) share with the primary cancer clade—a total of 24 synapomorphies. Additional 4,944 synapomorphies separate the metastatic from their benign sister group (node 15), whereas those of the primary cancer clade is defined by only 1,018 synapomorphies.
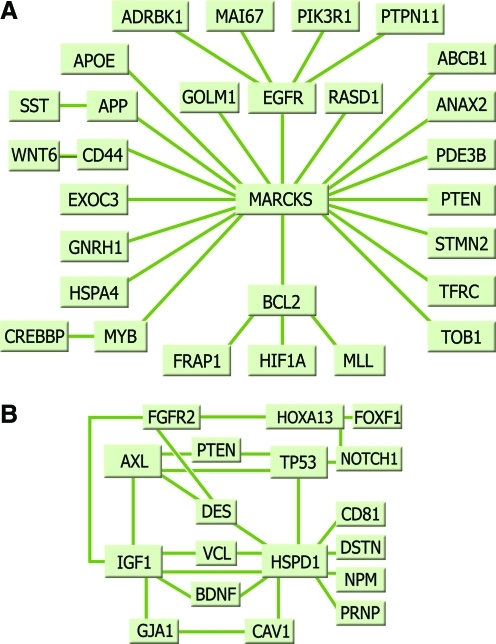
In addition to being the potential biomarkers, the synapomorphies of a clade permit the construction of an interaction or linkage network, which simulates the altered signaling processes of a tissue—the genotypic profile of a disease class (Schadt et al., 2009). Cluster-centered analysis approach using the synapomorphies of nodes 8 and 13 with Genomatix's BiblioSphere (www.genomatix.com), set at abstract level filtering, showed that the primary prostate tumors share a set of affected signaling pathways that was different from the metastatic tumors and their benign sister specimens (Fig. 3A and B, detailed graphs are in Supplementary Figs. 1 and 2). BiblioSphere identified MARCKS (actin filament crosslinking protein) as a central node for the primary tumors (Fig. 3A), and designated several central nodes for the metastatic tumors that include AXL (AXL receptor tyrosine kinase), DES (desmin), HSPD1 (heat-shock protein 1), IGF1 (insulin-like growth factor 1), and TP53 (tumor protein p53) (Fig. 3B). Although the network analysis showed star-connected pathways centered on a cytoskeltal gene in the primary tumors, it produced a complex multidirectional network in the metastatic specimens and their related benign tissue.
FIG. 3.
Schematic summary of network analyses produced by Genomatix's BiblioSphere. (A) A summary of affected nodal pathways in primary prostate tumors at node 8 of Figure 2, and (B) in metastatic prostate tumors at node 13 of Figure 2. More details are provided in Supplementary Figures 1 and 2.
Parsimony phylogenetic analysis permits further insights into the biological implications of the expression data. The cladogram and network analysis clearly indicated that early tumor transformation within benign cells can be detected by knowing the location of the specimen within the topology of the cladogram, shared synapomorphies, or by plotting its linkage network analysis as in Figure 3. Although it has been established that early tumor cell dissemination occurs in prostate tissue (Klein and Stoecklein, 2009), direct cellular transformation into a metastatic cancer phenotype without the formation of a primary tumor has not been shown before—that is, whether the biological stage of metastasis is set at initiation. In 3% of all cancer cases and in 5–10% of all cancer patients with metastases, a metastatic tumor is diagnosed but the primary tumor cannot be found (Briasoulis and Pavlidis, 1997; Mareel and Leroy, 2003). Podsypanina et al. (2008) have shown by injecting precancerous pancreatic cells into mice that direct transformation into metastatic phenotype can take place, and our analysis as illustrated in the cladogram (Fig. 2) seems to lend support to this hypothesis. The sharing of a good number of clonal expressions (synapomorphies) between the metastatic cancerous specimens and their benign sister clade that were different from those of the primary tumors' clade and its respective benign sister clade suggested that the metastatic phenotype follows an independent pathway from primary tumor formation; this point was further illustrated by network analysis (Fig. 3). It may be also congruent with our early prediction of two major developmental pathways within pancreatic, prostate, and ovarian cancers based on the phylogenetic analysis of their serum mass spectrometry proteomics (Abu-Asab et al., 2006).
Conclusions
We have argued that the qualitative nature of pathology's IHCMs cannot be extrapolated to the realm of omics biomarkers, and the latter should be defined within their own paradigm preferably through a systems biology approach. Therefore, we proposed that only shared derived mutations/expressions (also known as clonal aberrations or synapomorphies) in relation to normal conditions are the potential omics biomarkers. Within the evolutionary paradigm, we demonstrated how a parsimony phylogenetic analysis models a disease onto a tree-like diagram—the cladogram, that maps heterogeneous multigene expression profiles and at the same time shows the major shared clonal expressions at various levels of the hierarchical classification. Shared clonal expressions are the synapomorphies and the potential omics biomarkers that can be translated to a clinical setting in order to provide specimen characterization for early detection, diagnosis, prognosis, and posttreatment assessment. Additionally, using a set of synapomorphies for pathway profiling produces a genotype profile for a clade of specimens. These two points support our premise that a phylogenetic modeling of the disease datasets should be a priori step to biomarker exploration.
This parsimony analysis is also a systems biology approach that permits the recognition of shared aberrations within several datasets and among some diseases, and may allow us in the future to construct a comprehensive cancer tree of all cancer types to show the commonalities among them as well as the differences. Furthermore, stratification of the patients' population through parsimony phylogenetics into subpopulations will allow a better design of randomized clinical trials to reveal the effectiveness of treatment within various subpopulations, and determines future personalized therapeutic decisions.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
Supplementary Material
References
- Abelev G.I. Eraiser T.L. On the path to understanding the nature of cancer. Biochemistry (Moscow) 2008;73:487–497. doi: 10.1134/s0006297908050015. [DOI] [PubMed] [Google Scholar]
- Abu-Asab M. Microarrays need phylogenetics. 2009. http://stkesciencemagorg/cgi/eletters/sigtrans;1/51/eg11 http://stkesciencemagorg/cgi/eletters/sigtrans;1/51/eg11 Sci STKE (E-Letter, 30 January 2009).
- Abu-Asab M. Chaouchi M. Amri H. Phyloproteomics: what phylogenetic analysis reveals about serum proteomics. J Proteome Res. 2006;5:2236–2240. doi: 10.1021/pr0504485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab M. Chaouchi M. Amri H. Evolutionary medicine: a meaningful connection between omics, disease, and treatment. Proteomics Clin Appl. 2008a;2:122–134. doi: 10.1002/prca.200780047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab M.S. Chaouchi M. Amri H. Phylogenetic modeling of heterogeneous gene-expression microarray data from cancerous specimens. Omics. 2008b;12:183–199. doi: 10.1089/omi.2008.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry M.J. Screening for prostate cancer—the controversy that refuses to die. N Engl J Med. 2009;360:1351–1354. doi: 10.1056/NEJMe0901166. [DOI] [PubMed] [Google Scholar]
- Bielas J.H. Loeb K.R. Rubin B.P. True L.D. Loeb L.A. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briasoulis E. Pavlidis N. Cancer of unknown primary origin. Oncologist. 1997;2:142–152. [PubMed] [Google Scholar]
- Brock A. Chang H. Huang S. Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10:336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- Camin J. Sokal R. A method for deducing branching sequences in phylogeny. Evolution. 1965;19:311–326. [Google Scholar]
- Campbell P.J. Pleasance E.D. Stephens P.J. Dicks E. Rance R., et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci USA. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concato J. Jain D. Uchio E. Risch H. Li W.W. Wells C.K. Molecular markers and death from prostate cancer. Ann Intern Med. 2009;150:595–603. doi: 10.7326/0003-4819-150-9-200905050-00005. [DOI] [PubMed] [Google Scholar]
- Dalerba P. Cho R.W. Clarke M.F. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- Deo R.C. Roth F.P. Pathways of the heart. Circ Cardiovasc Genet. 2009;2:303–305. doi: 10.1161/CIRCGENETICS.109.892588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett M. Dunbier A.K. Emerging biomarkers and new understanding of traditional markers in personalized therapy for breast cancer. Clin Cancer Res. 2008;14:8019–8026. doi: 10.1158/1078-0432.CCR-08-0974. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: phylogeny inference package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fisher B. Redmond C.K. Fisher E.R. Evolution of knowledge related to breast cancer heterogeneity: a 25-year retrospective. J Clin Oncol. 2008;26:2068–2071. doi: 10.1200/JCO.2007.14.1804. [DOI] [PubMed] [Google Scholar]
- Heim-Hall J. Yohe S.L. Application of immunohistochemistry to soft tissue neoplasms. Arch Pathol Lab Med. 2008;132:476–489. doi: 10.5858/2008-132-476-AOITST. [DOI] [PubMed] [Google Scholar]
- Heng H.H. Stevens J.B. Bremer S.W. Ye K.J. Liu G. Ye C.J. The evolutionary mechanism of cancer. J Cell Biochem. 2010;6:1072–1084. doi: 10.1002/jcb.22497. [DOI] [PubMed] [Google Scholar]
- Hippo Y. Taniguchi H. Tsutsumi S. Machida N. Chong J.M. Fukayama M., et al. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–240. [PubMed] [Google Scholar]
- Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Kaput J. Rodriguez R.L. Nutritional genomics: the next frontier in the postgenomic era. Physiol Genomics. 2004;16:166–177. doi: 10.1152/physiolgenomics.00107.2003. [DOI] [PubMed] [Google Scholar]
- Kaiser J. Researcher, two universities sued over validity of prostate cancer test. Science. 2009;5947:1484. doi: 10.1126/science.325_1484. [DOI] [PubMed] [Google Scholar]
- Kashani-Sabet M. Rangel J. Torabian S. Nosrati M. Simko J. Jablons D.M., et al. A multi-marker assay to distinguish malignant melanomas from benign nevi. Proc Natl Acad Sci USA. 2009;15:6268–6272. doi: 10.1073/pnas.0901185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C.A. Stoecklein N.H. Lessons from an aggressive cancer: evolutionary dynamics in esophageal carcinoma. Cancer Res. 2009;69:5285–5288. doi: 10.1158/0008-5472.CAN-08-4586. [DOI] [PubMed] [Google Scholar]
- Knox S.S. From “omics” to complex disease: a systems biology approach to gene–environment interactions in cancer. Cancer Cell Int. 2010;10:11. doi: 10.1186/1475-2867-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowski B. Thornton J.W. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature. 2004;431:980–984. doi: 10.1038/nature02917. [DOI] [PubMed] [Google Scholar]
- Lerma E. Peiro G. Ramón T. Fernandez S. Martinez D. Pons C., et al. Immunohistochemical heterogeneity of breast carcinomas negative for estrogen receptors, progesterone receptors and Her2/neu (basal-like breast carcinomas) Mod Pathol. 2007;11:1200–1207. doi: 10.1038/modpathol.3800961. [DOI] [PubMed] [Google Scholar]
- Loeb L.A. Bielas J.H. Beckman R.A. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–3557. doi: 10.1158/0008-5472.CAN-07-5835. discussion 3557. [DOI] [PubMed] [Google Scholar]
- Loi S. Haibe-Kains B. Desmedt C. Lallemand F. Tutt A.M. Gillet C., et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- Lyons-Weiler J. Patel S. Becich M.J. Godfrey T.E. Tests for finding complex patterns of differential expression in cancers: towards individualized medicine. BMC Bioinformatics. 2004;5:110. doi: 10.1186/1471-2105-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Mareel M. Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol Rev. 2003;83:337–376. doi: 10.1152/physrev.00024.2002. [DOI] [PubMed] [Google Scholar]
- May M. Biomarkers still off the mark for detecting breast cancer. Nat Med. 2010;16:3. doi: 10.1038/nm0110-3. [DOI] [PubMed] [Google Scholar]
- Morrison R.S. Veenstra T.D. Biomarker discovery: Has it been worth it so far? Proteomics Clin Appl. 2008;2:1375–1376. doi: 10.1002/prca.200890031. [DOI] [PubMed] [Google Scholar]
- Nature. The big ome. Nature. 2008;452:913–914. doi: 10.1038/452913b. [DOI] [PubMed] [Google Scholar]
- Nature. Valid concerns. Nature. 2010;463:401–402. doi: 10.1038/463401b. [DOI] [PubMed] [Google Scholar]
- Oved K. Eden E. Akerman M. Noy R. Wolchinsky R. Izhaki O., et al. Predicting and controlling the reactivity of immune cell populations against cancer. Mol Syst Biol. 2009;5:265. doi: 10.1038/msb.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge E. Kreimer A.R. Greenlee R.T. Williams C. Xu J.L. Church T.R., et al. Results from four rounds of ovarian cancer screening in a randomized trial. Obstet Gynecol. 2009;113:775–782. doi: 10.1097/AOG.0b013e31819cda77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K. Du Y.C. Jechlinger M. Beverly L.J. Hambardzumyan D. Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff D.F. Promises and limitations of biomarkers. Rec Results Cancer Res. 2009;181:55–59. doi: 10.1007/978-3-540-69297-3_6. [DOI] [PubMed] [Google Scholar]
- Rifai N. Gillette M.A. Carr S.A. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- Rosai J. Ackerman L.V. Mosby, Edinburgh; New York: 2004. Rosai and Ackerman's surgical pathology. [Google Scholar]
- Roukos D.H. Mea Culpa with cancer-targeted therapy: new thinking and new agents design for novel, causal networks-based, personalized biomedicine. Expert Rev Mol Diag. 2009;9:217–221. doi: 10.1586/erm.09.7. [DOI] [PubMed] [Google Scholar]
- Sawyers C.L. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- Schadt E.E. Friend S.H. Shaywitz D.A. A network view of disease and compound screening. Nat Rev Drug Discov. 2009;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- Schaub N.P. Jones K.J. Nyalwidhe J.O. Cazares L.H. Karbassi I.D. Semmes O.J., et al. Serum proteomic biomarker discovery reflective of stage and obesity in breast cancer patients. J Am Coll Surg. 2009;208:970–978. doi: 10.1016/j.jamcollsurg.2008.12.024. discussion 978–980. [DOI] [PubMed] [Google Scholar]
- Stefankovic D. Vigoda E. Phylogeny of mixture models: robustness of maximum likelihood and non-identifiable distributions. J Comput Biol. 2007a;14:156–189. doi: 10.1089/cmb.2006.0126. [DOI] [PubMed] [Google Scholar]
- Stefankovic D. Vigoda E. Pitfalls of heterogeneous processes for phylogenetic reconstruction. Syst Biol. 2007b;56:113–124. doi: 10.1080/10635150701245388. [DOI] [PubMed] [Google Scholar]
- Sumer B. Gao J. Theranostic nanomedicine for cancer. Nanomedicine. 2008;3:137–140. doi: 10.2217/17435889.3.2.137. (London, England) [DOI] [PubMed] [Google Scholar]
- Takikita M. Chung J.Y. Hewitt S.M. Tissue microarrays enabling high-throughput molecular pathology. Curr Opin Biotechnol. 2007;18:318–325. doi: 10.1016/j.copbio.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Varambally S. Yu J. Laxman B. Rhodes D.R. Mehra R. Tomlins S.A., et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B. How to “cell” a genomic or proteomic screen. Sci Signal. 2008;1:eg9. doi: 10.1126/scisignal.147eg9. [DOI] [PubMed] [Google Scholar]
- Wang Y. Klijn J.G. Zhang Y. Sieuwerts A.M. Look M.P. Yang F., et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Warren J.L. Mariotto A.B. Meekins A. Topor M. Brown M.L. Current and future utilization of services from medical oncologists. J Clin Oncol. 2008;26:3242–3247. doi: 10.1200/JCO.2007.14.6357. [DOI] [PubMed] [Google Scholar]
- Winter E.E. Goodstadt L. Ponting C.P. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.