Summary
Proper formation of protein phosphatase 2A (PP2A) holoenzymes is essential for fitness of all eukaryotic cells. Carboxyl-methylation of PP2A catalytic subunit plays a critical role in regulating holoenzyme assembly; methylation is catalyzed by PP2A-specific methyltransferase LCMT-1, an enzyme required for cell survival. We determined crystal structures of human LCMT-1 in isolation and in complex with PP2A stabilized by a cofactor-mimic. The structures show that LCMT-1 active site pocket recognizes the carboxyl-terminus of PP2A, and interestingly, PP2A active site makes extensive contacts to LCMT-1. We demonstrated that activation of PP2A active site stimulates methylation, suggesting a mechanism for efficient conversion of activated PP2A into substrate-specific holoenzymes, thus minimizing unregulated phosphatase activity or formation of inactive holoenzymes. A dominant-negative LCMT-1 mutant attenuates cell cycle without causing cell death, likely by inhibiting uncontrolled phosphatase activity. Our studies suggested mechanisms of LCMT-1 in tight control of PP2A function, important for cell cycle and survival.
Introduction
PP2A is the most abundant, tightly regulated Ser/Thr phosphatase with complex composition, and plays important roles in diverse aspects of cellular function in all eukaryotic cells (Virshup 2000; Janssens and Goris 2001). Several conserved proteins or enzymes regulate the phosphatase activity of PP2A by controlling the stability, activation and inactivation of the catalytic subunit (Fellner et al. 2003; Xing et al. 2008; Kong et al. 2009). Substrate-specific activity of PP2A is controlled by diverse holoenzymes, comprising the scaffold A and catalytic C subunits, and a third variable regulatory B subunit from four major families: B, B′, B″, and B‴ (Virshup 2000; Janssens and Goris 2001). Formation of PP2A holoenzymes is a highly-regulated process involving activation of the C subunit by PP2A phosphatase activator (PTPA) (Hombauer et al. 2007) and carboxyl methylation by leucine carboxyl methyltransferase 1 (LCMT-1) (Lee and Stock 1993; De Baere et al. 1999). Proper formation of PP2A holoenzymes is critical for cellular function; the malfunction of PP2A is closely linked to diverse human diseases including cancer and Alzheimer’s disease (Vafai and Stock 2002; Sontag et al. 2004; Arroyo and Hahn 2005; Janssens et al. 2005). Although considerable progress had been made for understanding the atomic architectures of PP2A core enzyme and holoenzymes (Shi 2009), many aspects of PP2A regulation remain unknown. Particularly, the structural basis underlying the mechanisms of activation and methylation of the C subunit remains elusive.
The C subunit of PP2A possesses a flexible peptide motif at the carboxyl terminus (residues 294–309), here we termed “PP2A tail”, which plays an important role in regulating the oligomeric composition of PP2A complexes (Mumby 2001). A wealth of data support that carboxyl methylation of the last residue Leu309 of a highly conserved motif in PP2A tail, “T304PDYFL309”, enhanced the affinity of the PP2A core enzyme for the regulatory subunits (Ogris et al. 1997; Bryant et al. 1999; Tolstykh et al. 2000; Wei et al. 2001; Xing et al. 2006). Changes in PP2A tail and alteration of methylation also affected interaction of the C subunit with the α4 protein, which is essential for cell survival (Chung et al. 1999; Kong et al. 2004). Several lines of evidence suggest that PP2A-specific methyltransferase LCMT-1 is essential for cellular functions. Knockdown of LCMT-1 reduces the cellular level of PP2A methylation and induces apoptosis in mammalian cells (Lee and Pallas 2007; Longin et al. 2007). Loss of LCMT-1 methyltransferase activity was also linked to neurodegenerative diseases (Vafai and Stock 2002; Sontag et al. 2004).
Although the crystal structure of PPM1, the yeast homolog of LCMT-1, has been determined (Leulliot et al. 2004), the molecular mechanism of LCMT-1 remains poorly understood. Many intriguing observations suggest that LCMT-1-mediated PP2A methylation might involve complex mechanisms. For example, a synthetic peptide with the conserved sequence of PP2A tail is not a substrate of LCMT-1 (Xie and Clarke 1994). Okadaic acid and microcystin LR, two highly potent phosphatase inhibitor and tumor-inducing toxins, efficiently blocked PP2A methylation (Floer and Stock 1994; Li and Damuni 1994). Furthermore, mutations in PP2A active site abolished PP2A methylation in cells (Yu et al. 2001). Atomic structures of LCMT-1 and its complex with PP2A will be critical for understanding the structural basis of these observations and the mechanism of LCMT-1 function and PP2A methylation.
Here we report high-resolution crystal structures of LCMT-1 bound to S-adenosyl homocysteine (SAH), and in complex with the PP2A catalytic subunit. Interestingly, the structure revealed that LCMT-1 binds directly to the PP2A active site, which provides a mechanism for stimulating activated PP2A to be methylated and efficiently converted to substrate-specific holoenzymes. This suggests that LCMT-1 minimizes the unregulated phosphatase activity of PP2A core enzyme or free C, or formation of inactive holoenzymes. A dominant negative LCMT-1 mutant blocked the uncontrolled phosphatase activity of the core enzyme and attenuated cell cycle without causing cell death. Our studies revealed important mechanisms of LCMT-1 in precise control of PP2A methylation and function, important for regulating cell cycle and survival.
Results
Structure of LCMT-1 bound to SAH
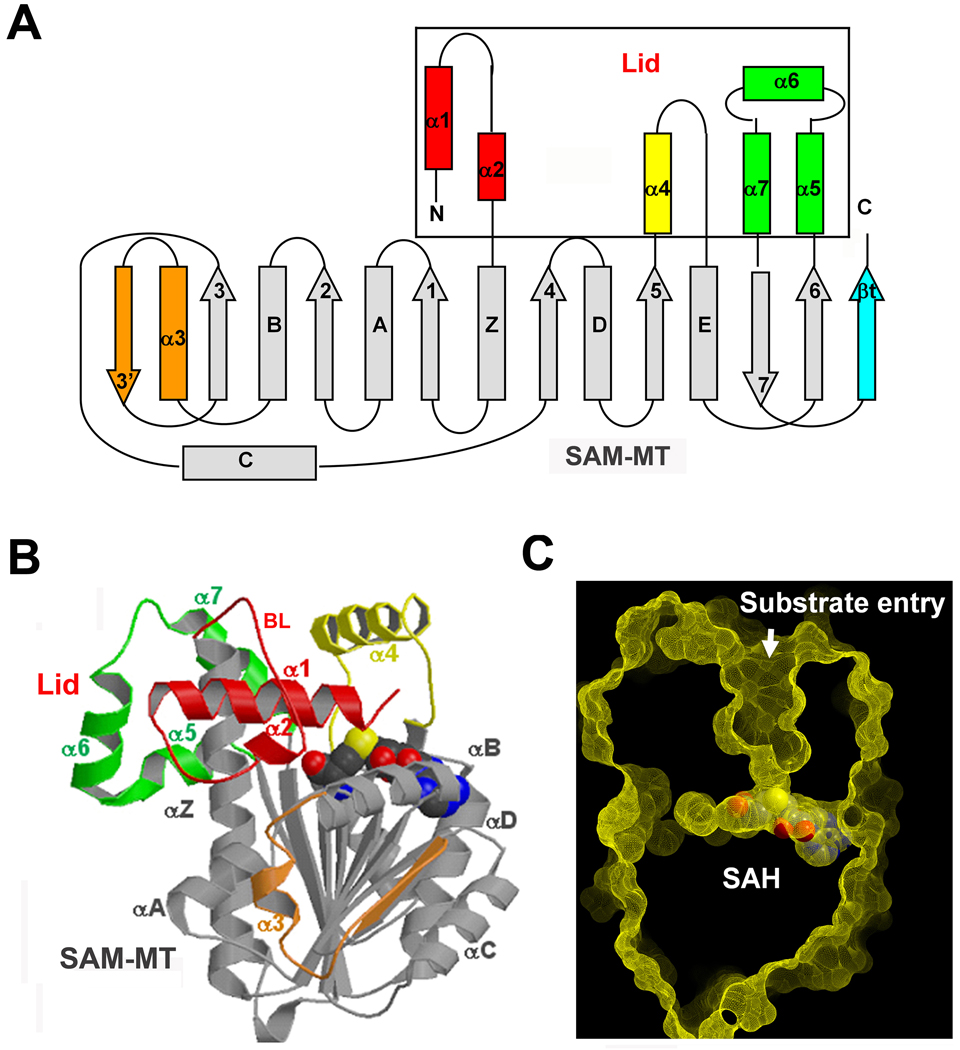
We first determined the crystal structure of LCMT-1 in complex with SAH (S-adenosyl homocysteine), the enzymatic product of cofactor SAM (S-adenosyl methionine). The complex was crystallized at 4°C. The structure was solved by molecular replacement using the model of PPM1, and refined to a resolution of 1.9Å (Leulliot et al. 2004) (Table 1). The structure contains a canonical SAM-dependent methyltransferase (SAM-MT) domain and a unique lid domain consisting entirely of α-helices (Figure 1A, 1B). The MT domain comprises a central β-sheet (β1–7) lying between two groups of α helices: αZ, αA and αB on one side, and αC, αD and αE on the other side (Figure 1B). The lid domain is formed by three of the four unique motifs in LCMT-1 (Figure 1A, 1B) and forms a deep active site pocket that presumably binds to the carboxyl-terminus of PP2A tail (Figure 1C). Alignment of structures of LCMT-1 and PPM1 revealed a key α-helix, α1, within the first non-conserved unique motif (Figure S1A, S1B), which is located next to both the substrate and the cofactor binding pockets (Figure 1B, S1A). Structure-based sequence alignment identified several conserved residues in α1 (Figure S1C), including K37, which participates in cofactor binding, and T29, a catalytic residue near the sulfur atom of the cofactor. As shown later, T29 in α1 is an important catalytic residue.
Table 1.
Data collection and refinement statistics.
| LCMT-1 in complex with SAH (4 copies/unit) |
LCMT-1 in complex with SAH (8 copies/unit) |
LCMT-1 in complex with PP2A |
|
|---|---|---|---|
| Data collection | |||
| Space group | P1 | P1 | P212121 |
| Resolution (Å) | 50-2.2 (2.28-2.20) | 50-1.9 (1.97-1.90) | 50-2.7 (2.75-2.7) |
| Unique observations | 62,057 | 191,558 | 17,595 |
| Data redundancy (outer shell) | 1.9 (1.8) | 3.8 (3.4) | 7.5 (5.2) |
| I/sigma (outer shell) | 12.6 (3.3) | 15.1 (3.7) | 7.5 (4.4) |
| Data coverage (outer shell) | 95.5% (80.0%) | 95.2% (75.4%) | 98.9% (92.9%) |
| Rsym (outer shell) | 0.059 (0.291) | 0.055 (0.319) | 0.136 (0.349) |
| Refinement | |||
| Resolution (Å) | 30–2.2 | 30–1.9 | 48.3-2.7 |
| No. reflections | 56,505 | 181,902 | 16,510 |
| Rwork / Rfree | 21.4%/26.9% | 18.4%/23.2% | 19.3%/25.0% |
| No. atoms | |||
| Protein | 9,860 | 20,046 | 5,001 |
| Ligand/ion | 152 | 304 | 37 |
| Water | 400 | 1851 | 121 |
| B-factors (plus TLS contribution) | 27.6 | 27.6 | 14.7 (43.7) |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.009 | 0.009 | 0.007 |
| Bond angles (°) | 1.11 | 1.15 | 1.03 |
| Ramachandran plot | |||
| Preferred regions (%) | 96.8 | 98.1 | 94.7 |
| Allowed regions (%) | 3.0 | 1.8 | 4.7 |
| Outliers (%) | 0.2 | 0.1 | 0.5 |
X-ray diffraction data were collected on one crystal. Values in parentheses are for highest-resolution shell.
Figure 1.
Overall structure of LCMT-1. (A) Schematic showing the topology of the LCMT-1 structure. The canonical SAM-MT domain is shown in grey, the unique motifs inserted to different locations of the SAM-MT domain are in color, three of which (red, yellow and green) form the lid domain as indicated. Helices are shown as cylinders and strands as arrows. The N and C termini are labeled. (B) Overall structure of LCMT-1 bound to SAH in ribbon. The color scheme for MT and unique motifs is the same as in (A). BL indicates binding loop. As shown later, BL plays an important role in PP2A binding. (C) A slice of LCMT-1 surface showing the binding pockets for PP2A tail and the cofactor. SAH is bound and shown in spacefill. See also Figure S1.
To determine the structural basis for recognition and methylation of PP2A tail by LCMT-1, we synthesized a peptide with the conserved sequence of PP2A tail (RRTPDYFL). The synthetic peptide, however, failed to co-crystallize with LCMT-1, nor did it occupy the active pocket of LCMT-1 when soaked with LCMT-1 crystals, consistent with the previous observation that PP2A tail alone is not a substrate for LCMT-1 (Xie and Clarke 1994). To elucidate the mechanism of LCMT-1 and PP2A methylation, we went on to determine the structure of the PP2A-LCMT-1 complex.
Structure of LCMT-1 in complex with PP2A catalytic subunit
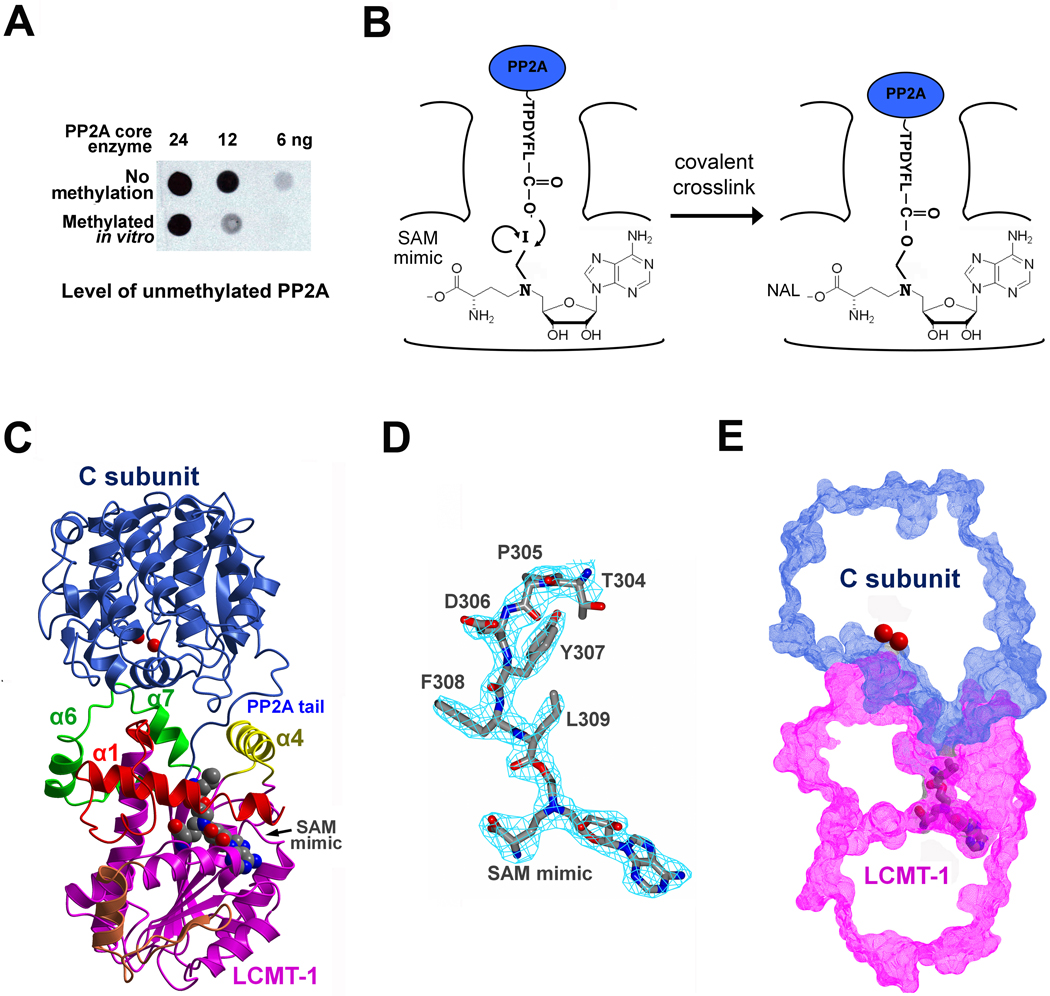
The PP2A-LCMT-1 complexes are not stable and did not yield any protein crystals after extensive efforts. Reducing the flexibility of the A subunit using the mini-A constructs that contain minimal structural elements for folding and binding to the C subunit (Xing et al. 2008), and stabilizing the enzymatic complex using the A-LCMT-1 fusion proteins failed to yield any diffracting crystals. The malleable nature of PP2A active site and heterogeneity of the complex likely hindered crystallization. This notion is reflected by the fact that only a fraction of PP2A could be methylated in vitro (Figure 2A), which is consistent with our observation later that methylation of PP2A tail requires activation of PP2A active site (Figure 5B). To facilitate crystallization of the PP2A-LCMT-1 complex, we synthesized a SAM mimic (Weller and Rajski 2006) that became covalently bound to PP2A tail during catalysis (Figure 2B). Amazingly, this reaction led to a stable PP2A-LCMT-1 complex that could be co-purified by anion exchange chromatography, and separated from the fraction of PP2A that could not be methylated (data not shown). In order to obtain diffracting crystals, an internal truncation in PP2A tail (δ294–298) was introduced to the complex. The structure was solved by molecular replacement and refined to a resolution of 2.7 Å (Table 1 and Figure 2C). This is the first structure of a complex between a methyltransferase and its substrate trapped by a SAM mimic. Note that all functional studies hereafter were performed using the full-length C subunit.
Figure 2.
Overall structure of the PP2A-LCMT-1 complex. (A) The level of methylation of PP2A core enzyme by LCMT-1 in vitro was determined by dot blot using an antibody that specifically recognizes the unmethylated PP2A. Three different amounts (24, 12, 6 ng) were spotted and PP2A without methylation was used as standard. The methylation level is around 50% or lower. (B) Illustration of LCMT-1 catalyzed covalent crosslink of the synthesized SAM mimic to PP2A tail. (C) Overall structure of the PP2A-LCMT-1 complex in ribbon diagram. The MT domain is colored magenta, unique motifs colored as in Figure 1A, C in blue, SAM mimic and L309 in spacefill. (D) The 2Fo-Fc electron density map at 1.8σ (cyan) for peptide “TPDYFL” in PP2A tail and the covalently bound SAM mimic (cylinder, colored by atom type). (E) A slice of surface showing that LCMT-1 binds to PP2A active site. Two catalytic metal ions are shown in red spheres.
Figure 5.
Precise control of PP2A methylation by the phosphatase activity of PP2A. (A) Simultaneous loss of LCMT-1 methylation activity and the phosphatase activity of PP2A core enzyme by incubation with PPi for 50 minutes. Neither activity could be recovered by addition of Mn++. (B) PTPA stimulates the phosphatase activity and LCMT-1-mediated methylation of the C subunit. (C) Ceramide stimulates methylation of the C subunit. (D) Polycation peptides stimulate methylation of the PP2A core enzyme. (E) Simultaneous loss of LCMT-1 methylation activity and the phosphatase activity of the PP2A core enzyme caused by okadaic acid, and mild oxidation upon removal of DTT or in the presence of 20µM H2O2. For all panels, the results are representative of three independent experiments. Data values are the average of two independent assays. Error bars show standard deviation. See also Figure S3.
The structure of the PP2A-LCMT-1 complex reveals that the lid domain of LCMT-1 makes extensive dual contacts to both PP2A tail and PP2A active site (Figure 2C, 2E). The carboxyl-terminal six residues of PP2A tail, “T304PDYFL309”, occupy the deep active site pocket of LCMT-1 in the lid domain (Figure 2C). The beginning of PP2A tail is placed near LCMT-1 active site, which allows the carboxyl-terminus of the shortened PP2A tail to readily bind LCMT-1 (Figure 2C). As predicted, the carboxylate group of Leu309 is covalently bound to the SAM mimic (Figure 2D). The extensive contacts of LCMT-1to the PP2A active site (Figure 2E) indicates an important mechanism for strict control of PP2A methylation by regulations of PP2A active site, which would suppress uncontrolled activity of PP2A core enzyme and facilitate formation of active holoenzymes.
Mechanism of recognition and methylation of PP2A tail
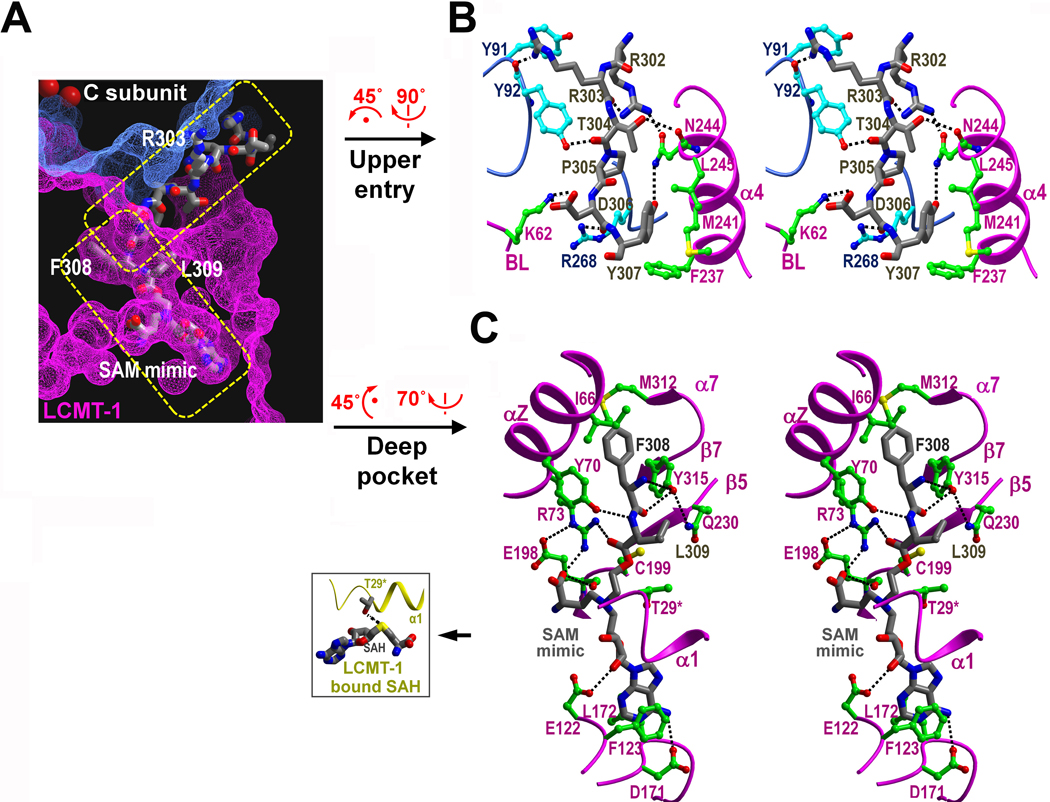
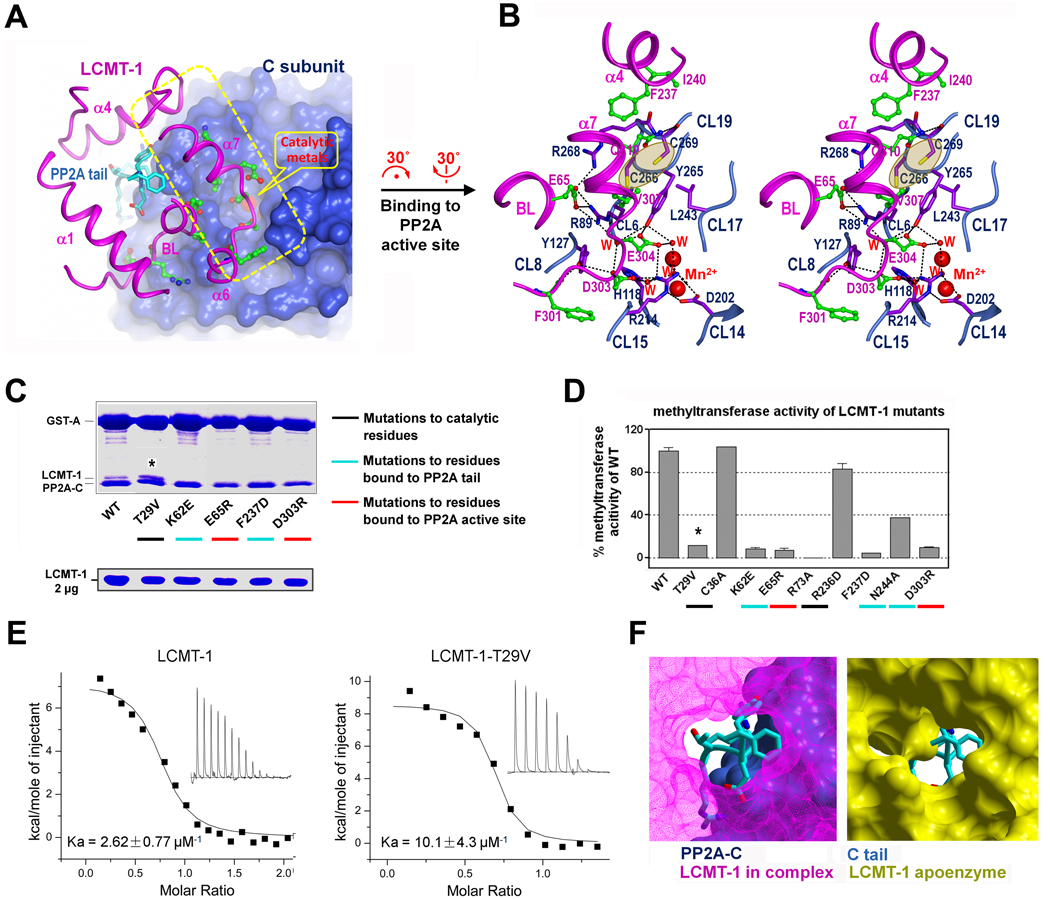
The active site pocket of LCMT-1 divides into two distinct regions: the upper entry and the deep pocket; both make extensive contacts with PP2A tail, burying 1370 Å2 previously exposed surface area (Figure 3A). The upper entry comprises helix α4 and the binding loop (BL) in the lid domain (Figure 3B). K62 in the BL makes key H-bond interactions to D306 in PP2A tail. F237/M241/ L245 in α4 form a well-defined hydrophobic binding site for Y307 in PP2A tail, which explains why mutations or phosphorylation of Y307 can block PP2A methylation (Yu et al. 2001; Longin et al. 2007). The deep pocket comprises α7 in the lid domain, and αZ and several β-sheets in the SAM-MT domain (Figure 3C). Several residues from αZ and α7 make key hydrophobic contacts to F308 in PP2A tail (Figure 3C).
Figure 3.
Binding and methylation of PP2A tail by LCMT-1. (A) A slice of surface showing the binding pocket for PP2A tail, divided into the upper entry and the deep pocket. Residues R303, F308 and L309 and SAM mimic are labelled. (B) A stereo view of the upper entry. Residues from LCMT-1, PP2A tail and the PP2A core are shown in green, grey and cyan, respectively. (C) A stereo view of the deep pocket. The color scheme is the same as in (B). The H-bond between T29 in α1 of LCMT-1 and the sulfur atom of SAH is highlighted (inlet).
At the bottom of the active site pocket of LCMT-1, R73 in αZ forms bifurcated H-bonds to both the cofactor and L309 in PP2A tail (Figure 3C), indicating R73 plays a role in positioning the cofactor and PP2A tail for methyl group transfer. In addition, the sidechain hydroxyl group of T29 in α1 forms an H-bond with the sulfur atom of SAH (Figure 3C, inlet), which likely stabilizes the cofactor during or after methyl group transfer. Consistent with these structural observations, mutation of R73 to alanine caused a complete loss of the methyltransferase activity and mutation of T29 to valine dramatically decreased the methyltransferase activity of LCMT-1 (Figure 4D), but appeared to have a higher binding affinity to PP2A, as shown by pull-down assay and measured by isothermal titration calorimetry (ITC) (Figure 4C & 4E). These results supported that R73 and T29 are catalytic residues of LCMT-1. The increased binding of LCMT-1 T29V to PP2A is likely due to an enhanced hydrophobic interaction of this residue to Leu309 (Figure 3C). These data indicated that LCMT-1 T29V is a dominant negative mutant; as shown later, recombinant expression of this mutant hindered the function of endogenous LCMT-1 (Figure 6).
Figure 4.
The contacts to PP2A active site are essential for PP2A methylation. (A) Interface of LCMT-1 to PP2A active site. The lid domain of LCMT-1, the protein core and the tail of the C subunit, are shown in ribbon, surface, and cylinder, respectively. (B) A close-up stereo view of the interface to PP2A active site. Residues from LCMT-1 and the C subunit of PP2A are shown in green and purple, respectively. (C) Pull-down assay using GST-tagged PP2A core enzyme immobilized on GS4B resin, and equal amount of LCMT-1 wild type (WT) or mutants in the mobile phase. Mutations to residues that contact PP2A active site or PP2A tail but do not participate in catalysis abolished binding to the PP2A core enzyme. Same amount of LCMT-1 WT or mutants was shown on SDS-PAGE (lower panel). (D) Methylation of PP2A core enzyme by WT LCMT-1 and LCMT-1 mutants. Mutations to residues that either contact PP2A active site, or bind PP2A tail, as well as mutations to catalytic residues abolished the methyltransferase activity of LCMT-1. (E) ITC measuring the binding affinity of the PP2A core enzyme to LCMT-1 and LCMT-1 T29V. T29V mutation increases the binding affinity by 4-fold. Note that PP2A-LMCT-1 interaction is endothermic. (F) The conformation of LCMT-1 active site pocket changes considerably to allow binding of PP2A tail. Based on structural alignment, PP2A tail overlaps with the LCMT-1 apo structure (the one most similar to the complex is shown). PP2A tail is shown in cylinder and the rest of the model in surface. See also Figure S2.
Figure 6.
LCMT-1 Dominant-Negative Mutant T29V Attenuated Cell Proliferation. (A) Effects of recombinant expression of the human LCMT-1 (hLCMT-1) dominant-negative mutant, T29V, hLCMT-1 wild-type (WT), and the loss-of-function mutant F237D on PP2A methylation in the rat C6 glioma cell line. Expression of hLCMT-1 T29V reduced PP2A methylation without affecting the endogenous level of rat LCMT-1. (B) Images of C6 cells that stably express hLCMT-1 or its mutants cultured for 40 hr after stress. Dividing cells are highlighted by yellow dash cycles. Cells expressing hLCMT-1 T29V had much fewer dividing cells and much lower cell density. (C) The increase of cell numbers of C6 cell cultures expressing hLCMT-1 WT or mutants after stress. (D) The increase of viable cell signals of C6 cell cultures expressing hLCMT-1 WT or mutants after stress. (E) Knockdown of endogenous LCMT-1 by siRNA-induced cell death of C6 glioma cells. Knockdown was confirmed by western blot with an antibody that specifically recognizes rat LCMT-1. (F) LCMT-1 T29V inhibited the phosphatase activity of the PP2A core enzyme with a 4-fold higher efficacy than WT LCMT-1. (G) Structural alignment of the ACB0 g1 holoenzyme and the PP2A-C-LCMT-1 complex. A is shown in green, and C in the holoenzyme and the complex with LCMT-1 are in cyan and blue, respectively. B0 g1 is in yellow and LCMT-1 in magenta. A and C are in worm, B0 g1 in sphere, and LCMT-1 in tube. (H) The effect of PP2A phosphatase activity on cell fitness. The phosphatase activity of free C or the core enzyme is less controlled and might be detrimental to the cell; substrate-specific phosphatase activity of holoenzymes is essential for proper cellular function. Normal cells expressing WT LCMT-1 facilitate efficient transition of activated PP2A to holoenzymes, thus minimizing uncontrolled phosphatase activity. The effect of LCMT-1 knockdown or expression of LCMT-1 T29V is illustrated. For (C), (D), and (F), representative results of three independent experiments are shown. Data values are the average of three (C and D) and two (F) independent assays, respectively. Error bars represent the standard deviation. See also Figure S4.
The contacts to the PP2A active site are essential for methylation of PP2A tail
The rim of the lid domain in LCMT-1 makes extensive contacts to and near PP2A active site grooves (Figure 4A), resulting in an interface larger than that to PP2A tail that buries 1600 Å2 otherwise exposed surface area. The contacts to PP2A active site involve several structural elements critical for the phosphatase activity and regulation of PP2A active site. In particular, helix α7 and the loop between α6 and α7 in the lid domain interacts with several C subunit loops (CL) near PP2A active site, including CL6, CL8, CL14, CL15, CL17 and CL19 (Figure 4B, Table S1). Two conserved acidic residues in α7, D303 and E304, form a rich H-bond network to the catalytic metals bridged by metal-chelated water molecules, and to several residues near PP2A active site, including R89 and R214, residues that are essential for binding PP2A substrates (Figure 4B). Together with R89, R268 in CL19, the longest loop near PP2A active site, makes H-bond interactions to E65 at the carboxyl-terminus of the BL in LCMT-1 (Figure 4B). We recently determined the crystal structure of the PP2A-PTPA complex that demonstrated that R89/R214/R268 of PP2A catalytic subunit also make key contacts to PTPA; the same PP2A active site loops that interact with LCMT-1 bind PTPA as well (unpublished data). Furthermore, two adjacent cysteine residues, Cys266/269, which are prone to oxidation and likely contribute to the loss of the phosphatase activity under oxidative stress (Rao and Clayton 2002), are buried at this interface (Figure 4B, shaded).
To determine whether the contacts to PP2A active site are essential for methylation of PP2A tail, we performed mutagenesis analysis. LCMT-1 mutations at the interface to PP2A active site, E65R and D303R, strongly reduced PP2A binding and methylation, similar to mutations at the interface to PP2A tail, K62E and F237D (Figure 4C, 4D). Mutations located outside the interface have much less effect on the methyltransferase activity of LCMT-1 (Figure 4D). The contacts to PP2A active site likely facilitate binding of PP2A tail to LCMT-1 by increasing the binding affinity and by inducing conformational changes of the LCMT-1 active site to create the binding pocket for PP2A tail. Alignment of all 8 copies of LCMT-1 in the apoenzyme structure and LCMT-1 from the PP2A-LCMT-1 complex showed significant structural variability in helix α4 and the BL (Figure S2A). The apo-structure that is most similar to the complex was compared in detail. The conformation of the apoenzyme active site appears to hinder binding of PP2A tail; the complex undergoes significant conformational rearrangement in the lid domain that allows LCMT-1 active site pocket to accommodate PP2A tail (Figure 4F). These changes are triggered by the repulsive contacts between I240 in helix α4 and a hydrophilic surface on the edge of PP2A active site (Figure S2B), and by interactions of the BL to and near PP2A active site (Figure S2C). The other 7 apo-structures are expected to have larger difference. These structural and biochemical observations demonstrated that the contacts to PP2A active site prime a different and better defined conformation of LCMT-1 essential for methylation of PP2A tail. This explains why the synthetic peptide of PP2A tail alone is not a substrate for LCMT-1(Xie and Clarke 1994).
Precise control of PP2A methylation by the phosphatase activity
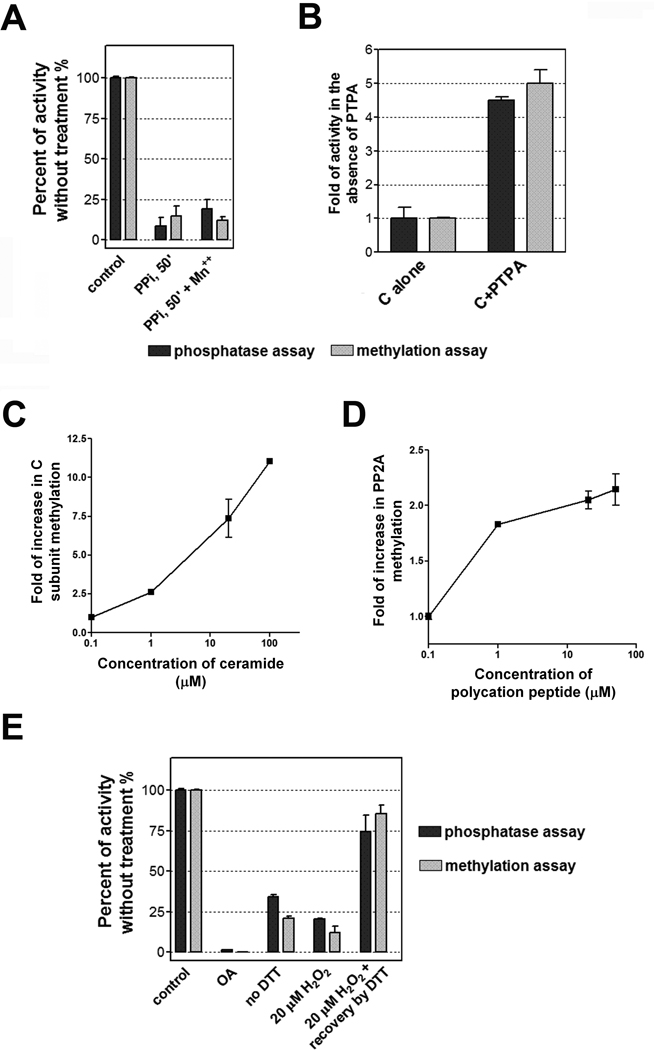
The extensive contacts of LCMT-1 to a handful of critical structural elements of PP2A active site suggest that LCMT-1 binding might be sensitive to the conformational changes of PP2A active site. The C subunit in the PP2A-LCMT-1 complex is almost the same as in the PP2A core enzyme (Figure S3A), indicating that LCMT-1 specifically recognizes an active conformation of PP2A. The indispensable role of these contacts for methylating PP2A tail suggests a checkpoint for PP2A methylation that would rely on activation of PP2A active site. PP2A active site harbors ten flexible protein loops that participate in metal chelation (Figure S3B) and interaction with PTPA (Table S1). Due to the malleable nature of PP2A active site, we reasoned that metal binding and the ability of PTPA to sculpt an active conformation of PP2A active site (unpublished data) would affect PP2A methylation. We recently uncovered that pyrophosphate (PPi) can efficiently evict PP2A catalytic metals and distort the conformation of PP2A active site (unpublished data). Indeed, incubation of PP2A core enzyme with PPi for 50 minutes abolished PP2A methylation; neither the phosphatase activity nor LCMT-1 methylation activity can be restored by the addition of Mn++ (Figure 5A). On contrary, at a molar concentration comparable to that of C, PTPA increased the methylation rate of the C subunit by 4–6 fold, similar in magnitude to the stimulation of the phosphatase activity toward a phosphopeptide, K-R-pT-I-R-R (Figure 5B). These studies demonstrated a tight link of PP2A methylation to the conformation of PP2A active site, which explains the previous observation that mutations in PP2A active site abolished PP2A methylation in cells (Yu et al. 2001). Since both catalytic metals and PTPA play roles in stabilizing the active conformation of PP2A, mutations to metal-chelating residues or residues that interact with PTPA and LCMT-1 would abolish PP2A methylation both by interfering maturation of PP2A active site and by interfering LCMT-1 binding, which agrees with previous speculations (Yu et al. 2001).
Besides PTPA, PP2A phosphatase activity is highly regulated by a variety of cellular factors and environmental cues, such as polycations, apoptosis signals, tumor inducing toxins and oxidative stress (Hermann et al. 1988; Galadari et al. 1998; Rao and Clayton 2002). We examined the rate of PP2A methylation under these different contexts. Ceramide, the small molecule released from mitochondrion during apoptosis that was reported to stimulate the phosphatase activity of PP2A (Galadari et al. 1998), increased the methylation rate of the C subunit in a concentration-dependent manner (Figure 5C). Similarly, consistent with the well known fact that polycations stimulate PP2A phosphatase activity (Hermann et al. 1988), a peptide with multiple positively charged residues, KRTIRR, accelerated the methylation rate of PP2A core enzyme by LCMT-1 (Figure 5D). These observations reflect the malleable nature of PP2A active site, and support that methylation of PP2A tail relies on activation of PP2A phosphatase activity. On contrary, PP2A methylation was blocked by inhibition and inactivation of PP2A phosphatase activity. As previously described (Floer and Stock 1994; Li and Damuni 1994), tumor-inducing toxins such as okadaic acid that bind directly to PP2A active site (Xing et al. 2006) inhibited both the phosphatase activity and LCMT-1-mediated methylation of PP2A core enzyme (Figure 5E). The two activities were also simultaneously reduced when PP2A core enzyme was incubated with a DTT-free buffer or a buffer containing 20 µM H2O2 (Figure 5E), presumably due to a disulfide crosslink between Cys266 and Cys269 that are located near PP2A active site and at the interface to LCMT-1. Addition of DTT to reduce the disulfide crosslink recovered both activities (Figure 5E).
These data demonstrated that LCMT-1-mediated PP2A methylation can function as a signalling “hub” that integrates a variety of signals regulating the level of PP2A phosphatase activity. This would prevent ambiguous PP2A function in two ways: selective enhancement of activated PP2A to form holoenzyme would minimize formation of inactive holoenzymes; efficient conversion of activated PP2A into substrate-specific holoenzymes would prevent uncontrolled phosphatase activity. In light of the role of LCMT-1 and PP2A methylation in cell cycle (Lee and Pallas 2007; Longin et al. 2007), this provides a mechanism for LCMT-1 to integrate various signals for controlling cell cycle progression.
LCMT-1 dominant negative mutant attenuates glioma cell proliferation after stress
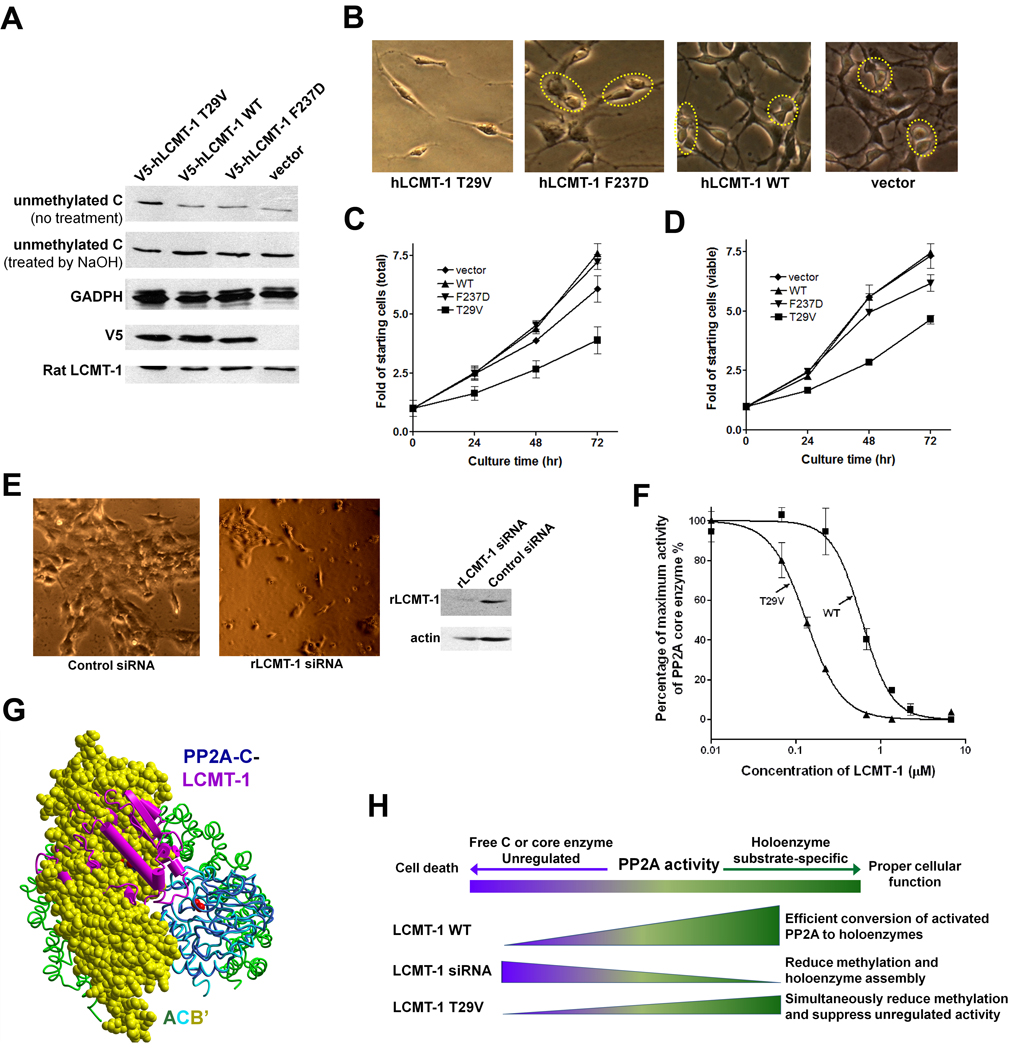
Defect of LCMT-1 T29V in PP2A methylation without affecting PP2A binding (Figure 4C–E) indicated that T29V is a dominant negative mutant. To test whether it mimics the phenotype of LCMT-1 knockdown, we transfected human LCMT-1 T29V into C6, a mouse glioma cell line, using a retrovirus vector. Wild type LCMT-1 and a loss of function mutant, F237D, were used as controls. Recombinant LCMT-1 and its mutants were expressed as V5-tagged proteins and their expression level is determined by western blot using anti-V5 antibody (Figure 6A). The level of PP2A methylation was determined by western blot using an antibody that specifically recognizes the unmethylated PP2A prior to and after the methyl group was removed by NaOH treatment (Figure 6A). We showed that after the cells underwent nutritional stress by culturing consecutively for 6 days without changing medium, C6 cells that stably express human LCMT-1 T29V reduced PP2A methylation without affecting the level of endogenous rat LCMT-1; while cells that express wild type human LCMT-1 or loss of function LCMT-1 mutant, F237D, exhibited little effect on PP2A methylation (Figure 6A). These data supported that T29V has dominant negative effect on PP2A methylation.
Knockdown of LCMT-1 was previously shown to affect both cell cycle progression and survival of HeLa and colon cancer cells (Lee and Pallas 2007; Longin et al. 2007). We showed that stable expression of human LCMT-1 T29V in C6 glioma cells did not cause significant cell death (Figure 6B); trypan blue staining barely detected any dead cells (data not shown). After nutritional stress as described above, C6 cells expressing LCMT-1 T29V had a lower ratio of proliferating cells than control cells (Figure 6B). Consistently, C6 cells expressing LCMT-1 T29V showed a slower rate of increase in the cell number (Figure 6C). In contrast, expression of human LCMT-1 wild type or loss of function mutant, F237D, in C6 glioma cells exhibited little effect on cell division or cell doubling rate under the same condition (Figure 6B, 6C). Similar results were obtained from viable cell assay (Figure 6D).
The ability of LCMT-1 T29V to attenuate the proliferation rate of glioma cells without causing cell death is different from the phenotype of LCMT-1 knockdown reported previously (Lee and Pallas 2007; Longin et al. 2007). To prove that LCMT-1 knockdown indeed gave different phenotypes than expression of LCMT-1 T29V, the endogenous rat LCMT-1 was knocked down by siRNA. Cell death was observed when the cellular LCMT-1 was diminished (Figure 6E). As illustrated in Figure 6H, we reasoned that, LCMT-1 knockdown would unleash the uncontrolled phosphatase activity of free C or core enzyme, and lead to cell death due to reduced methylation and holoenzyme assembly; expression of LCMT-1 T29V likely blocks the uncontrolled phosphatase activity although methylation and holoenzyme assembly were reduced. Indeed, purified LCMT-1 T29V inhibited the phosphatase activity of the core enzyme with an efficacy 4-fold higher than that of wild type LCMT-1 (Figure 6F), which agrees well with its 4-fold higher binding affinity (Figure 4E). This inhibition selectively targets the core enzyme or free C but not holoenzymes (Figure S4A), because LCMT-1 overlaps with regulatory subunits based on structural alignment of the PP2A-LCMT-1 complex and the B’ holoenzyme (Figure 6G). Since holoenzyme assembly is essential for proper cellular function and cell cycle progression, decrease in methylation, holoenzyme assembly, and uncontrolled phosphatase activity by LCMT-1 T29V attenuated cell cycle progression without causing cell death (Figure 6H).
Discussion
PP2A is the most abundant protein phosphatase in all eukaryotic cells and methylation is critical for the proper biogenesis and function of numerous PP2A holoenzymes. Despite its essential cellular function, the structural and biochemical basis of LCMT-1 function remained elusive. Using a synthetic SAM analog to trap the enzymatic intermediate, we crystallized the PP2A-LCMT-1 complex, and elucidated the structural basis for the interaction between LCMT-1 and PP2A catalytic subunit, and the enzymatic mechanism of LCMT-1. The structure of the complex provided unambiguous proof that LCMT-1 binds directly to the PP2A active site. We further demonstrated that PP2A methylation is highly sensitive to regulation of the phosphatase activity. This provides a structural and biochemical basis for LCMT-1 to integrate numerous signals that regulate the level of PP2A phosphatase activity for precise control of PP2A methylation.
In light of complex regulations of malleable PP2A active site in cells, the significance of this checkpoint function of LCMT-1 in PP2A methylation is 2-fold: 1) ensure assembly of functional PP2A holoenzymes and prevent formation of dominant negative inactive holoenzymes; 2) facilitate efficient transition of the activated PP2A to substrate-specific holoenzymes, thus prevent the uncontrolled phosphatase activity of PP2A core enzyme or free C subunit. The different consequences of LCMT-1 knockdown versus expression of a dominant negative LCMT-1 mutant in glioma cells reflect the role of LCMT-1 in minimizing the unregulated activity of PP2A core enzyme or free C. The ability of LCMT-1 dominant negative mutant to hamper cell cycle progression without causing cell death likely relies on its ability to block the uncontrolled phosphatase activity of PP2A (Figure 6F, S4A & S4B). Reduced PP2A holoenzyme formation, caused by reduced methylation (Figure 6A), likely contributes to the attenuated cell cycle. It remains to be determined whether inactive holoenzymes are formed and contributes to the functional defects under LCMT-1 knockdown. Furthermore, it can not be excluded that cell death caused by LCMT-1 knockdown is the combined effects of reduced holoenzyme formation and increase in the uncontrolled phosphatase activity.
Previous studies showed that PP2A methylesterase (PME-1) blocks methylation of inactive PP2A in yeast (Hombauer et al. 2007). It is conceivable that LCMT-1, PME-1 and PTPA work in a concerted manner to control PP2A activation, methylation, and binding of regulatory subunits to the core enzyme in strictly-ordered consecutive steps to ensure proper biogenesis of functional PP2A holoenzymes and minimize ambiguous PP2A function. Uncontrolled phosphatase activity and inactive holoenzymes are likely detrimental to cell survival and proper cellular functions (Figure 6H). Together, these studies demonstrated that, despite the huge complexity, different aspects of PP2A regulation and function are precisely controlled and coordinated, an emerging concept important for understanding PP2A function and its role in cellular signalling (Virshup and Shenolikar 2009).
The level of PP2A methylation was previously shown to vary during cell cycle (Turowski et al. 1995), indicating that cell cycle regulators might participate in the regulation of PP2A methylation. While this can be modulated by the cellular level of LCMT-1 and PME-1, this manuscript revealed another mechanism for regulating PP2A methylation by alteration of PP2A active site. Because of the highly regulated nature of PP2A active site, PP2A methylation might represent a signalling “hub” that integrates various signals for controlling cell cycle and likely other aspects of cellular functions. Modulating PP2A activity and LCMT-1 function provides a potential strategy to control cell cycle and survival of cancer cells. The structure of the PP2A-LCMT-1 complex provides an important basis for further investigation of the cellular function of LCMT-1 and PP2A methylation in PP2A holoenzyme biogenesis, and in cell cycle and survival.
Experimental procedures
Synthesis of SAM analogue
Synthesis of the SAM analogue is carried out in three steps, as reported previously (Weller and Rajski 2006). In brief: First, the 5’-hydroxyl group of adenosine was converted to 5’-hydroxylethylamino group following the Mitsunobu condensation reaction using o-NBS-protected α-amino esters. Second, the aldehyde group of the protected γ-aldehyde α-amino acid was converged to the amino group of 5’-hydroxylethylamino adenosine obtained from the previous step; the resulting crosslink became stabilized following reduction. The aldehyde was produced from aspartate following the Rapoport sequence of thioester formation and reduction. Finally, iodination and deprotection led to the final product.
Protein preparation
All constructs and point mutations were generated using standard PCR-based cloning strategy. LCMT-1δ19 (20–338) and missense mutants were cloned in pET15b (Invitrogen) and overexpressed at 37°C in E. coli BL21(DE3). N-terminal truncation (δ19) improved LCMT-1 solubility with no loss of enzyme activity (data not shown). For convenience, LCMT-1δ19 is referred to as LCMT-1. The soluble fraction of E.coli cell lysate was purified over Ni-NTA (Qiagen), further fractionated by cation exchange chromatography (Source 15S, Amersham) after removal of His6-tag. Cloning, expression and purification of human PP2A A(α), C(α) subunits or mutants, and assembly of PP2A A–C core dimer was described previously (Xing et al. 2006). Note that full-length C(α) was used for functional studies.
The stable PP2A-LCMT-1 complexes were assembled by mixing PP2A core enzyme or the C subunit with LCMT-1 and SAM analog at 1:2:10 molar ratio, incubated at 30°C for 1h, and purified by anion exchange chromatography (Source 15Q, Amersham) and gel filtration chromatography (Superdex200, Amersham). Due to difficulty of PP2A methylation in vitro, excess amount of LCMT-1 was used to improve the level of covalent crosslink of PP2A to SAM analog.
Crystallization and data collection
Crystals of LCMT-1 bound to SAH were grown at 4°C using the hanging-drop vapor-diffusion method by mixing the LCMT-1-SAH complex (~10mg/ml LCMT-1/5mM SAH) with an equal volume of reservoir solution containing 17–19% PEG2000 monomethyl ether (v/v), 150mM triethylamine N-oxide, and 5mM DTT. The crystals are in space group P1, with unit cells containing either 4 molecules or eight molecules in each asymmetric unit. Crystals were equilibrated in a reservoir buffer with 20% glycerol (v/v), and flash frozen in liquid nitrogen. Native datasets were collected at NSLS beamline X25 and processed using the software DENZO and SCALEPACK (Otwinowski 1997).
Crystals of the PP2Ac(δ294–298) -LCMT-1 complex were grown at 18°C using the sitting-drop vapor-diffusion method by mixing the complex (~5 mg/ml) with an equal volume of reservoir solution containing 100mM Tris pH8.2, 22–26% PEG4000 (v/v), 200 mM sodium acetate and 5 mM DTT. Diffracting crystals were generated by microseeding. The crystals were flash frozen as described above. The native datasets for the PP2Ac-LCMT-1 complex were collected at APS LS-CAT and processed using the software HKL2000 (Otwinowski 1997).
Structure determination
Using the dataset with 4 molecules of LCMT-1 per asymmetric unit, the structure of the LCMT-1-SAH complex was solved by molecular replacement using PHASER (McCoy et al. 2005) and the coordinates of the yeast homolog PPM1 (accession code 1RFG) with non-conserved loop regions removed. The electron density map was improved by non-crystallographic symmetry (NCS) averaging using AVE (Kleywegt and Read 1997). The structure was examined and modified using Coot (Emsley and Cowtan 2004) and O (Jones et al. 1991), and refined using CNS with NCS restraints (Brunger et al. 1998) to 2.2 Å. This structure was used for molecular replacement on the dataset with larger unit cell; 8 molecules were found in the asymmetric unit. The structure was built and refined by REFMAC (Murshudov et al. 1997) to 1.9 Å.
The structure of the PP2A-LCMT-1 complex was solved by molecular replacement using two models: LCMT-1 and the C subunit (residues 6–293) from the structure of the PP2A core enzyme (accession code 2IE3), and found 1 complex per asymmetric unit. The structure was built using Coot (Emsley and Cowtan 2004) and refined using REFMAC restraints with TLS (Winn et al. 2003) and weights adjusted based on R-free. PP2A tail was built after the electron density map was improved. The SAM analog was built using Sybyl (Tripos), and topology file generated using Dundee (Schuttelkopf and van Aalten 2004) for REFMAC refinement. Two TLS groups were used: one is the C subunit (1–293), and the other is LCMT-1 together with the SAM analog and the last six residues of the PP2A tail. The structure was refined to 2.7 Å.
GST-Mediated Pull-Down Assay
The pull-down assay was performed following a previously described procedure (Chao et al. 2006). Briefly, LCMT-1 or LCMT-1 mutants (10µg in 100 µl binding buffer) was mixed 20µl glutathione resin with immobilized GST-tagged PP2A A–C dimer (10µg) in the presence of 1mg/ml BSA. The protein bound to resin was separated from unbound protein and analyzed by SDS-PAGE gels and Coomassie blue staining.
Methylation assay
Methylation assays were performed by mixing LCMT-1 (4–10nM), PP2A (10–100nM) and [3H]-SAM (3µM) in a reaction buffer containing 50mM MOPS, pH7.2, 20mM DTT (omitted for oxidized PP2A), 50µM MnCl2 (omitted for PPi-treated PP2A). After incubation at 37°C for 2–20min, the reactions were stopped and precipitated by addition of 25% ice-cold trichloroacetic acid and 1mg/ml BSA. The radioactivity of precipitate was quantified by scintillation counting. The results were used to estimate the rate of methylation. For dot blot, the indicated amount of PP2A core enzyme before and after in vitro methylation by LCMT-1 was spotted onto PVDF membrane, and blotted by an antibody that specifically recognizes the unmethylated PP2A tail to determine the level of PP2A methylation (Millipore, clone 4b7).
Isothermal titration calorimetry
To obtain a direct binding affinity, 15 µM PP2A core enzyme was titrated with 300 µM LCMT-1 or LCMT-1 T29V using a VP-ITC microcalorimeter (MicroCal). All proteins were prepared in a buffer containing 20 mM Tris (pH 7.5), 50 mM NaCl and 50 µM MnCl2. The data were fitted by Origin 7.0.
Mammalian cell culture and westernblot
The rat glioma cell line C6 was kindly provided by Dr. Andreas Friedl (University of Wisconsin-Madison). The cell was cultured in DMEM containing 1% FBS. V5-tagged human LCMT-1 or mutants were cloned into a retroviral vector harboring a GFP marker. The same titer of retroviruses packaged using LCMT-1 expression vectors or the empty vector was used to infect C6 culture with 50–80% confluence. Infection efficiency was monitored by GFP marker, and expression of recombinant proteins was monitored by westernblot against V5 tag. Knockdown of endogenous LCMT-1 was mediated by transfection of rat LCMT-1 siRNAs: CAGAAUGCCGAUGUCAAAU, CACUCCUGAUAACCGAAUG, and GAAGGAGAUAACCUAUUGA. Retrovirus-infected cells or cells transfected with siRNAs were collected after 48 hours. Cell extracts were examined by westernblots using antibodies that specifically recognize the unmethylated C subunit of PP2A (Millipore, clone 4b7), the V5 tag (Millipore), the rat LCMT-1 (Santa Cruz Biotechnology, 4A4) and the loading control GADPH (Santa Cruz Biotechnology, A3) or actin (Millipore, clone C4), respectively. The total C subunit of PP2A was determined by antibody 4b7 after treated by 0.1N NaOH for 10min at RT, and neutralized by equal molar amount of HCl, similar to previously described (Favre et al. 1994).
Cell proliferation and viability assay
After nutritional stress, the same number of C6 cells infected by retroviruses packaged with LCMT-1 expression vectors or empty vectors were placed into 24-well and 96-well plates, respectively, and cultured in DMEM containing 1% FBS. Cell numbers of the 24-well cultures in triplicates were counted after trypsin digestion at the indicated culture time with and without trypan blue staining. The growth of viable cells was determined using CellTiter-Glo luminescent cell viability assay (Promega) at the indicated culture time. The experiments were performed in triplicates and repeated three times. Values were normalized to the starting culture.
Phosphatase assays
PP2A phosphatase activity was measured using a Ser/Thr Phosphatase Assay Kit 1 (Upstate Biotechnology) according to manufacturer's instructions. 2µl of 1mM phosphopeptide substrate (K-R-pT-I-R-R) was added to 20µl of PP2A sample (40–100nM). The reaction was carried out at 30°C for 15 min and stopped by the addition of 50µl malachite green solution. The absorbance at 620 nm was measured after 10 min incubation at room temperature.
Activation, inhibition, inactivation and oxidation of PP2A
Free C or core enzyme of PP2A was mixed with PTPA at a 1:1 molar ratio, with C2 ceramide (N-Acetyl-D-sphingosine, Sigma) or synthetic peptide (KRTIRR) at indicated concentration and incubated at 37°C for 5 min for PP2A activation. Okadaic acid was added to the core enzyme at a 2:1 molar ratio for PP2A inhibition. PP2A core enzyme (1mg/ml) was incubated at 37°C with 1mM PPi for 50 min for inactivation, and after desalting, diluted 100-fold into buffer containing 25mM Tris, pH8.0, 150mM NaCl, and 20mM DTT for phosphatase activity and methylation assay. PP2A core enzyme was changed to DTT-free buffer by gel filtration chromatography to remove DTT (superdex 200, Amersham); the peak fraction (1.5 mg/ml) was incubated with 20 µM H2O2 at 37°C for 30 min, and after desalting, diluted at least 100-fold into reaction buffer containing 25mM Tris, pH8.0, 150mM NaCl, and 50µM MnCl2 for the phosphatase activity and methylation assay. Phosphatase activity and methylation assay were performed as described above. All assays were performed in duplicates and repeated three times. Average values and standard error were calculated. Where indicated, values were normalized to the positive control.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anand Saxena (NSLS) and David Smith (APS LS-CAT) for assistance, Dr. Yigong Shi (Tsinghua University), Dr. David Pallas (Emory University), and Dr. David Brautigan (University of Virginia) for discussion, Dr. Andreas Friedl (University of Wisconsin-Madison) for rat C6 glioma cell line, and Dr. Bill Sugden (University of Wisconsin-Madison) for discussion and the retrovirus vector. This work was supported by University of Wisconsin-Madison (Y. Xing), NIH fellowship K01 CA124856 (Y. Xing) and ACS Research Scholar Grant RSG-10-153-01-DMC (Y. Xing). The atomic coordinates of LCMT-1-SAH and PP2A-LCMT-1 complexes were deposited in the Protein Data Bank with accession codes 3IEI and 3P71, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arroyo JD, Hahn WC. Involvement of PP2A in viral and cellular transformation. Oncogene. 2005;24(52):7746–7755. doi: 10.1038/sj.onc.1209038. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Bryant JC, Westphal RS, Wadzinski BE. Methylated C-terminal leucine residue of PP2A catalytic subunit is important for binding of regulatory Balpha subunit. Biochem J. 1999;339(Pt 2):241–246. [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Xing Y, Chen Y, Xu Y, Lin Z, Li Z, Jeffrey PD, Stock JB, Shi Y. Structure and mechanism of the phosphotyrosyl phosphatase activator. Mol Cell. 2006;23(4):535–546. doi: 10.1016/j.molcel.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Chung H, Nairn AC, Murata K, Brautigan DL. Mutation of Tyr307 and Leu309 in the protein phosphatase 2A catalytic subunit favors association with the alpha 4 subunit which promotes dephosphorylation of elongation factor-2. Biochemistry. 1999;38(32):10371–10376. doi: 10.1021/bi990902g. [DOI] [PubMed] [Google Scholar]
- De Baere I, Derua R, Janssens V, Van Hoof C, Waelkens E, Merlevede W, Goris J. Purification of porcine brain protein phosphatase 2A leucine carboxyl methyltransferase and cloning of the human homologue. Biochemistry. 1999;38(50):16539–16547. doi: 10.1021/bi991646a. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fellner T, Lackner DH, Hombauer H, Piribauer P, Mudrak I, Zaragoza K, Juno C, Ogris E. A novel and essential mechanism determining specificity and activity of protein phosphatase 2A (PP2A) in vivo. Genes Dev. 2003;17(17):2138–2150. doi: 10.1101/gad.259903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre B, Zolnierowicz S, Turowski P, Hemmings BA. The catalytic subunit of protein phosphatase 2A is carboxyl-methylated in vivo. J Biol Chem. 1994;269(23):16311–16317. [PubMed] [Google Scholar]
- Floer M, Stock J. Carboxyl methylation of protein phosphatase 2A from Xenopus eggs is stimulated by cAMP and inhibited by okadaic acid. Biochem Biophys Res Commun. 1994;198(1):372–379. doi: 10.1006/bbrc.1994.1052. [DOI] [PubMed] [Google Scholar]
- Galadari S, Kishikawa K, Kamibayashi C, Mumby MC, Hannun YA. Purification and characterization of ceramide-activated protein phosphatases. Biochemistry. 1998;37(32):11232–11238. doi: 10.1021/bi980911+. [DOI] [PubMed] [Google Scholar]
- Hermann J, Cayla X, Dumortier K, Goris J, Ozon R, Merlevede W. Modulation of the substrate specificity of the polycation-stimulated protein phosphatase from Xenopus laevis oocytes. Eur J Biochem. 1988;173(1):17–25. doi: 10.1111/j.1432-1033.1988.tb13961.x. [DOI] [PubMed] [Google Scholar]
- Hombauer H, Weismann D, Mudrak I, Stanzel C, Fellner T, Lackner DH, Ogris E. Generation of active protein phosphatase 2A is coupled to holoenzyme assembly. PLoS Biol. 2007;5(6):e155. doi: 10.1371/journal.pbio.0050155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353(Pt 3):417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J, Van Hoof C. PP2A: the expected tumor suppressor. Curr Opin Genet Dev. 2005;15(1):34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Read RJ. Not your average density. Structure. 1997;5(12):1557–1569. doi: 10.1016/s0969-2126(97)00305-5. [DOI] [PubMed] [Google Scholar]
- Kong M, Ditsworth D, Lindsten T, Thompson CB. alpha4 is an essential regulator of PP2A phosphatase activity. Mol Cell. 2009;36(1):51–60. doi: 10.1016/j.molcel.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M, Fox CJ, Mu J, Solt L, Xu A, Cinalli RM, Birnbaum MJ, Lindsten T, Thompson CB. The PP2A-associated protein alpha4 is an essential inhibitor of apoptosis. Science. 2004;306(5696):695–698. doi: 10.1126/science.1100537. [DOI] [PubMed] [Google Scholar]
- Lee J, Stock J. Protein phosphatase 2A catalytic subunit is methylesterified at its carboxyl terminus by a novel methyltransferase. J Biol Chem. 1993;268(26):19192–19195. [PubMed] [Google Scholar]
- Lee JA, Pallas DC. Leucine carboxyl methyltransferase-1 is necessary for normal progression through mitosis in mammalian cells. J Biol Chem. 2007 doi: 10.1074/jbc.M704861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulliot N, Quevillon-Cheruel S, Sorel I, de La Sierra-Gallay IL, Collinet B, Graille M, Blondeau K, Bettache N, Poupon A, Janin J, van Tilbeurgh H. Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity. J Biol Chem. 2004;279(9):8351–8358. doi: 10.1074/jbc.M311484200. [DOI] [PubMed] [Google Scholar]
- Li M, Damuni Z. Okadaic acid and microcystin-LR directly inhibit the methylation of protein phosphatase 2A by its specific methyltransferase. Biochem Biophys Res Commun. 1994;202(2):1023–1030. doi: 10.1006/bbrc.1994.2031. [DOI] [PubMed] [Google Scholar]
- Longin S, Zwaenepoel K, Louis JV, Dilworth S, Goris J, Janssens V. Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic Subunit. J Biol Chem. 2007;282(37):26971–26980. doi: 10.1074/jbc.M704059200. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Mumby M. A new role for protein methylation: switching partners at the phosphatase ball. Sci STKE. 2001;2001(79) doi: 10.1126/stke.2001.79.pe1. PE1. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Ogris E, Gibson DM, Pallas DC. Protein phosphatase 2A subunit assembly: the catalytic subunit carboxy terminus is important for binding cellular B subunit but not polyomavirus middle tumor antigen. Oncogene. 1997;15(8):911–917. doi: 10.1038/sj.onc.1201259. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Rao RK, Clayton LW. Regulation of protein phosphatase 2A by hydrogen peroxide and glutathionylation. Biochem Biophys Res Commun. 2002;293(1):610–616. doi: 10.1016/S0006-291X(02)00268-1. [DOI] [PubMed] [Google Scholar]
- Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139(3):468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Sontag E, Hladik C, Montgomery L, Luangpirom A, Mudrak I, Ogris E, White CL., 3rd Downregulation of protein phosphatase 2A carboxyl methylation and methyltransferase may contribute to Alzheimer disease pathogenesis. J Neuropathol Exp Neurol. 2004;63(10):1080–1091. doi: 10.1093/jnen/63.10.1080. [DOI] [PubMed] [Google Scholar]
- Tolstykh T, Lee J, Vafai S, Stock JB. Carboxyl methylation regulates phosphoprotein phosphatase 2A by controlling the association of regulatory B subunits. Embo J. 2000;19(21):5682–5691. doi: 10.1093/emboj/19.21.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski P, Fernandez A, Favre B, Lamb NJ, Hemmings BA. Differential methylation and altered conformation of cytoplasmic and nuclear forms of protein phosphatase 2A during cell cycle progression. J Cell Biol. 1995;129(2):397–410. doi: 10.1083/jcb.129.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai SB, Stock JB. Protein phosphatase 2A methylation: a link between elevated plasma homocysteine and Alzheimer's Disease. FEBS Lett. 2002;518(1–3):1–4. doi: 10.1016/s0014-5793(02)02702-3. [DOI] [PubMed] [Google Scholar]
- Virshup DM. Protein phosphatase 2A: a panoply of enzymes. Curr Opin Cell Biol. 2000;12(2):180–185. doi: 10.1016/s0955-0674(99)00074-5. [DOI] [PubMed] [Google Scholar]
- Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33(5):537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Wei H, Ashby DG, Moreno CS, Ogris E, Yeong FM, Corbett AH, Pallas DC. Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J Biol Chem. 2001;276(2):1570–1577. doi: 10.1074/jbc.M008694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller RL, Rajski SR. Design, synthesis, and preliminary biological evaluation of a DNA methyltransferase-directed alkylating agent. Chembiochem. 2006;7(2):243–245. doi: 10.1002/cbic.200500362. [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- Xie H, Clarke S. Protein phosphatase 2A is reversibly modified by methyl esterification at its C-terminal leucine residue in bovine brain. J Biol Chem. 1994;269(3):1981–1984. [PubMed] [Google Scholar]
- Xing Y, Li Z, Chen Y, Stock JB, Jeffrey PD, Shi Y. Structural mechanism of demethylation and inactivation of protein phosphatase 2A. Cell. 2008;133(1):154–163. doi: 10.1016/j.cell.2008.02.041. [DOI] [PubMed] [Google Scholar]
- Xing Y, Xu Y, Chen Y, Jeffrey PD, Chao Y, Lin Z, Li Z, Strack S, Stock JB, Shi Y. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell. 2006;127(2):341–353. doi: 10.1016/j.cell.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Yu XX, Du X, Moreno CS, Green RE, Ogris E, Feng Q, Chou L, McQuoid MJ, Pallas DC. Methylation of the protein phosphatase 2A catalytic subunit is essential for association of Balpha regulatory subunit but not SG2NA, striatin, or polyomavirus middle tumor antigen. Mol Biol Cell. 2001;12(1):185–199. doi: 10.1091/mbc.12.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.