Abstract
Somatic mutations contribute to the development of age-associated disease. In earlier work, we found that, at high frequency, aging Saccharomyces cerevisiae diploid cells produce daughters without mitochondrial DNA, leading to loss of respiration competence and increased loss of heterozygosity (LOH) in the nuclear genome. Here we used the recently developed Mother Enrichment Program to ask whether aging cells that maintain the ability to produce respiration-competent daughters also experience increased genomic instability. We discovered that this population exhibits a distinct genomic instability phenotype that primarily affects the repeated ribosomal RNA gene array (rDNA array). As diploid cells passed their median replicative life span, recombination rates between rDNA arrays on homologous chromosomes progressively increased, resulting in mutational events that generated LOH at >300 contiguous open reading frames on the right arm of chromosome XII. We show that, while these recombination events were dependent on the replication fork block protein Fob1, the aging process that underlies this phenotype is Fob1-independent. Furthermore, we provide evidence that this aging process is not driven by mechanisms that modulate rDNA recombination in young cells, including loss of cohesion within the rDNA array or loss of Sir2 function. Instead, we suggest that the age-associated increase in rDNA recombination is a response to increasing DNA replication stress generated in aging cells.
Author Summary
There is a striking correlation between age and the onset of many diseases, such as cancer, suggesting that the aging process itself can contribute to their development. Cancer is a genetic disease caused by the accumulation of a series of deleterious somatic mutations leading to unchecked proliferation. In humans, it is well established that normal mutation rates are not sufficient to account for the sharp increase in cancer rates in aging populations, suggesting a change in mutation rate is a necessary component of cancer development. Here, we find that the aging process in the budding yeast Saccharomyces cerevisiae leads to an increased rate of homologous recombination within a repetitive DNA sequence element, the ribosomal rDNA array. While these mutational events are initiated primarily at this single locus, they are propagated to the end of the chromosome and thus affect hundreds of genes. These results suggest that the aging process itself could contribute to increasing mutation rates and perhaps to the onset of age-associated disease.
Introduction
One of the greatest risk factors associated with carcinogenesis is age. Cancer risk increases exponentially toward the end of life in humans and other mammalian species [1]. Somatic genetic changes contribute significantly to the development of most tumors. However, the rate at which spontaneous mutations arise in normal adult cells has been hypothesized to be too low to generate all the genetic changes necessary to produce tumors at the observed rates [2]–[3]. Consequently, Loeb et al. developed the mutator hypothesis, which postulates an increased mutation rate in precancerous cells [3]. A variety of mechanisms could lead to such an increase, but a favored model is that sporadic mutations in, or epigenetic silencing of, genes responsible for maintaining genome integrity lead to increased rates of mutation. Once acquired, this mutator phenotype may serve as the driving force toward carcinogenesis as individuals age.
When genomes of tumors are examined, loss of heterozygosity (LOH) is observed as a common mechanism in which the sole functional allele of a tumor suppressor gene is inactivated by somatic mutation [4]–[5]. LOH can be generated by many different mutational events, including point mutations, small deletions or inversions, mitotic recombination or chromosome loss. Recent advances in high-resolution single-nucleotide polymorphism arrays (SNP arrays) have revealed that a surprising number of tumors contain long tracts of homozygosity that are not accompanied by a change in gene copy number. This type of LOH arises from somatic mitotic recombination events and is referred to as partial (or acquired) uniparental disomy (UPD) [6]. Importantly, UPD can alter the genotype for hundreds of genes following a single event, thereby amplifying its potential to contribute to cancer development [7].
Within any genome, there are regions that exhibit higher rates of mitotic recombination than the genomic average as the result of their proximity to hotspots or common fragile sites [8]. These regions typically represent slow-replicating sequences, and agents that generate DNA replication stress often reveal their fragility. These regions are also often associated with non-histone protein complexes that inhibit DNA replication fork progression. This generates DNA replication stress that may lead to an increased frequency of DNA damage due to replication fork collapse [9]–[10]. While conservative repair of this DNA damage would have no genetic consequence, the higher incidence of damage results in a greater chance that an alternative repair pathway will be utilized that does confer a genotypic change.
Previously, we found evidence for a mutator phenotype associated with advancing replicative age in a common lab strain of the budding yeast, Saccharomyces cerevisiae. Pedigree analysis revealed that old cells begin to produce offspring that have dramatically higher incidences of genomic instability, which is manifest as an apparent ∼100-fold increase in LOH on at least two different chromosomes [11]. Virtually all the LOH occurred via mitotic recombination and gave rise to UPD genotypes. Subsequent analysis revealed that these LOH events were a consequence of loss of mitochondrial DNA in daughter cells, which led to a transient “crisis” state characterized by cell cycle arrest and high mortality [12]. Cells that survived this crisis showed a high frequency of LOH events in their nuclear genome.
While yeast cells lacking mitochondrial DNA cannot perform oxidative phosphorylation (respiration), they remain viable by relying on aerobic glycolysis (fermentation). In contrast, most eukaryotic cells retain, and even require, respiration. Thus we were interested in determining whether S. cerevisiae cells that retain respiratory competence throughout their replicative life span (RLS) also exhibit a mutator phenotype. The frequency at which functional mitochondria are successfully segregated during cell division varies widely between strains of S. cerevisiae; alleles in over 100 genes can affect mitochondrial DNA transmission frequency [13]–[15]. Therefore, in this study we sought to use a strain in which respiration competence was faithfully transmitted with increasing replicative age.
We recently developed the Mother Enrichment Program (MEP), a genetic program to facilitate replicative aging studies in yeast [16]. The MEP provides an efficient and inducible selection against newborn daughter cells. When the MEP is active, mother cells continue to divide and age normally, while the division of newborn daughter cells is arrested. The MEP provides the opportunity to follow a cohort of mother cells in liquid culture throughout their entire RLS without any requirement for removing progeny cells. Once cells have reached a desired age, the MEP can be switched off and aged mothers will resume production of viable daughters, allowing for colony-based phenotypic analysis. Compared to pedigree analysis, which is done by single-cell micromanipulation, the MEP can dramatically improve the sensitivity of the LOH assay by increasing the sample number of aged cells by over three orders of magnitude. Here we report our finding that replicative age is accompanied by a progressive decline in rDNA array stability, leading to higher incidence of LOH affecting the right arm of chromosome XII.
Results
MEP strain mother cells produce respiration-competent daughters throughout their replicative life span
In order to assess whether MEP strains could be useful for analyzing LOH rates in aging yeast cells, we first examined whether old mother cells produced daughters that retained respiration competence. Specifically, we examined the inheritance of respiration competence in daughter cells of a MEP strain (UCC5185) using pedigree analysis. We found that ∼65% of the mother cells produced daughters with respiration competence through their entire RLS (Median of 36 generations, Figure S1A). This is in striking contrast to the strain we originally examined for age-associated LOH (UCC809), where only ∼5% of the mother cells produced respiration-competent daughters throughout their life span (Figure S1A and [11]–[12]). Furthermore, for those UCC5185 mother cells that did produce respiration-incompetent daughters, the median age at which this occurred was significantly later in the mother's life span (23 generations versus 10 generations in UCC809) (Figure S1B). Together, these results demonstrate that the majority of UCC5185 cells are capable of producing respiration-competent daughters throughout their life span, thus providing the basis for our analysis of LOH in such a population.
An age-associated increase in LOH on chromosome XII but not chromosome IV
LOH events in diploid cells can be visualized using colony color phenotypes and, when combined with half-sector analysis, the rate of LOH can be determined [17]–[20]. We used two heterozygous markers that alter colony color when lost: Loss of MET15 function, resulting in a black colony sector, or loss of ADE2 function, resulting in a red colony sector. LOH events that occur within the first cell division after plating will generate sectors that form one half of a colony and can be used as a direct measure of mutation rate.
Replicative life span is measured in terms of the number of mitotic divisions an individual cell completes before senescence. To measure LOH rates as mother cells divide and age, logarithmically growing MEP cells were inoculated into rich media containing estradiol and aged in liquid culture. Estradiol activates the MEP, allowing mother cells to divide and age normally while daughter cells are rendered incapable of cell division. Throughout the 95-hour aging period, samples were harvested and washed to remove estradiol, then plated to solid medium for half-sector analysis. Once the MEP is inactivated by removal of estradiol, surviving mother cells can produce viable daughter cells and form colonies. Because aged mother cells represent the only cells in the aging liquid culture capable of forming colonies (Figure 1A and [16]), no fractionation of cell populations was required to isolate aged mothers for the analysis. The vast majority of colonies at each time point grew robustly and were respiration-competent. Respiration-competent colonies were easily distinguished from those incapable of respiration, which show a severe growth defect on glucose media and thus could be excluded from our analysis.
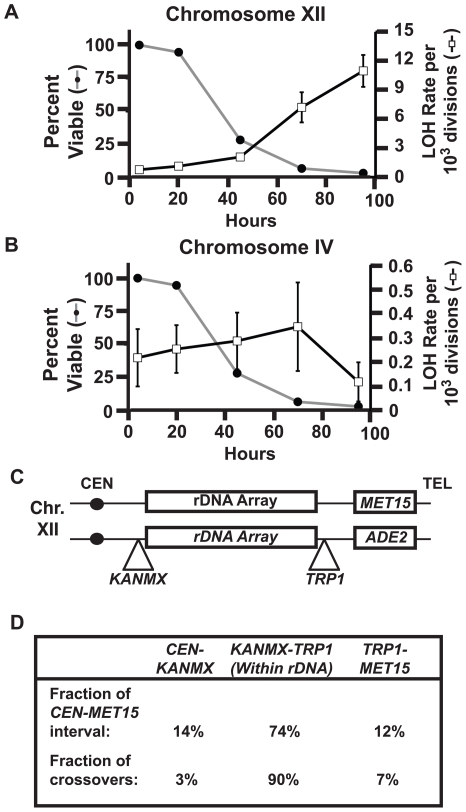
Figure 1. Age-associated LOH events are a result of recombination within the rDNA array.
A) Right y-axis: LOH rates at MET15 (open boxes) reported as total LOH events per cell division. Error bars indicate Standard Error of the Mean (SEM). Left y-axis: Percent viability of mother cells in the aging culture (grey line). B) Right y-axis: LOH rates at Chromosome IV (black line) reported as total LOH events per cell division. Error bars indicate SEM. Left y-axis: Percent viability of mother cells in the aging culture (closed circles). C) A schematic of marker placement on Chromosome XII used to determine homologous recombination break points. D) Table indicating the relative sizes of intervals between markers and the proportion of LOH events that originate within each interval.
We determined the LOH rates in young cells at loci on chromosome IV and chromosome XII simultaneously. By scoring ∼20,000 colonies per time point, we measured an LOH rate of 6.69×10−4 events/cell division at MET15 on chromosome XII and a rate of 1.25×10−4 events/cell division at an intergenic region on chromosome IV (Figure 1A and 1B). The LOH rate at MET15 was significantly higher than the chromosome IV locus (p = 0.0059, Fisher's exact test using contingency tables), suggesting that MET15 LOH rate is affected by proximity to a “hotspot”. As mother cells were aged in liquid culture, a robust and significant increase in LOH rates associated with increasing replicative age was observed at the MET15 locus (Figure 1A). This increase was significant at 45 hours (p<0.0001, Fisher's exact test using contingency tables), a time point where ∼25% of the original mother population retained viability, and continued to increase as the viability of the population declined below 10%. Thus, the increase in LOH rate at MET15 affects cells that have exceeded the median life span potential of the population. In contrast, while there was a trend towards increasing LOH rates with increasing replicative age for the first 72 hours at the chromosome IV locus, it did not reach the level of significance at any age (p>0.05, Fisher's exact test using contingency tables) (Figure 1B). These results suggested that this age-associated increase in LOH was locus-specific, affecting MET15 on chromosome XII.
LOH events at MET15 originate within the rDNA array
The MET15 locus lies ∼250 kbp distal to the rDNA array, a series of ∼150–200 tandemly repeated copies of the genes encoding the structural RNA components of the ribosome [21]. To determine if the rDNA array represented a “hotspot” that was responsible for elevated LOH rates at MET15, we constructed a diploid strain that carried heterozygous markers immediately adjacent to each end of the rDNA array and at MET15 (Figure 1C). In aged populations, we isolated half-sectored colonies based on LOH at MET15 and scored each sector for the presence of the rDNA-linked markers on chromosome XII. We found that 90% of MET15 LOH events were linked to LOH events at the TRP1 marker distal to the rDNA array, but not the KANMX marker proximal to the array. This indicates that most MET15 LOH events originate within the rDNA array, with homozygosity extending ∼250 kbp from the rDNA array to the MET15 locus, and presumably the remaining ∼625 kbp to the telomere (Figure 1D). This LOH pattern is indicative of mitotic recombination events between homologous chromosomes. The fraction of mitotic recombination events originating within the rDNA array was significantly higher than expected based on the sequence distance represented by the rDNA array (based on a 95% confidence interval, binomial distribution), consistent with previous examinations of LOH at MET15 [11], [17]. Thus, MET15 LOH events are primarily serving as a read out for an age-associated increase in mitotic recombination that affects the rDNA array.
Aging populations maintain a constant ratio of reciprocal to non-reciprocal LOH events on chromosome XII
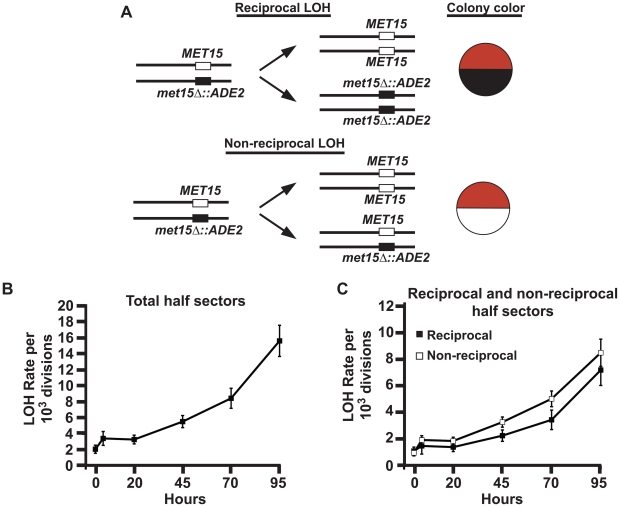
LOH events in S. cerevisiae generating half sectors can be further classified as either reciprocal (LOH occurs in both cells) or nonreciprocal (one cell undergoes LOH while the other remains heterozygous) (see Figure 2A and reviewed in [5]). This classification has facilitated a better mechanistic understanding of LOH events. For instance, in a previous examination of mutations that increase LOH rates in young cells, we found that the ratio of reciprocal to non-reciprocal LOH events reflected the type of defect [17]: Mutations that affect specific DNA repair pathways bias LOH events away from the wild type ratio. By contrast, mutations that increase LOH rates but do not alter the ratio were consistent with a general increase in DNA damage that did not alter normal repair pathways. To examine reciprocal and non-reciprocal events in aging cells, we used the diploid MEP strain UCC5185 carrying ADE2 and MET15 in opposition at the MET15 locus. By examining the color of both half sectors, it is possible to distinguish reciprocal from non-reciprocal events (Figure 2A).
Figure 2. LOH rates at MET15 in aging cells present a constant ratio of reciprocal/non-reciprocal events.
A diagram of UCC5185 colony color markers located on Chromosome XII, with expected results of reciprocal and non-reciprocal LOH events. Non-reciprocal events can also lead to black/white half-sectored colonies. (Note: The normal chromosomal copies of ADE2 have been deleted.) B) LOH rates for UCC5185 at MET15 reported as total LOH events per cell division. Error bars indicate SEM. The sample size at each time point ranges from 21 to 28. LOH rates are significantly increased at 45, 70 and 95 hours, (unpaired t-test; P values are 0.0003, <0.0001, and <0.0001 respectively). C) LOH events from panel B segregated into reciprocal and non-reciprocal LOH rates. Error bars indicate SEM.
Consistent with the first MEP strain examined above, we observed a significant increase in total LOH events at MET15 in populations of aging UCC5185 cells (Figure 2B). By 45 hours, when populations have passed their median viability, median LOH rates were increased 2-fold compared to the rate observed in the young population (Figure 2B) (p<0.0001, Fisher's exact test based on contingency tables). Between 70 and 95 hours, when population viability had fallen below ∼10%, median LOH rates were increased by four to eight-fold (p<0.0001, Fisher's exact test based on contingency tables). We found both reciprocal and non-reciprocal events increased with similar kinetics in aging populations (Figure 2C), indicating that cells maintain a stable ratio of these events throughout the aging process. This result suggests that aging cells may experience an increased frequency of DNA damage within the rDNA array, rather than an age-associated defect in a particular repair pathway.
Age-associated rDNA recombination events are FOB1-dependent
In haploid cells, double stranded breaks (DSBs) within the rDNA are often initiated by the DNA replication fork-blocking (RFB) activity of Fob1 [22]–[23]. Replication forks traveling opposite to the direction of transcription of the 35S rRNA are blocked by the specific interaction of Fob1 with sequences within the non-transcribed region 1 (NTS1) [24]. This source of DNA replication stress can cause fork collapse to generate DSBs, which can be repaired by homologous recombination to yield a variety of products [25]–[26].
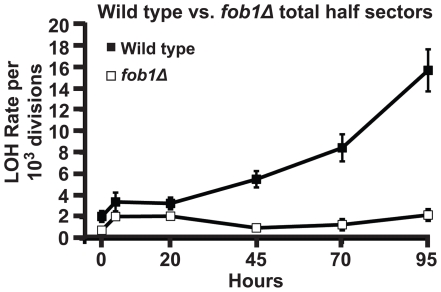
We tested whether Fob1 was required for age-associated LOH at the MET15 locus. In young cells, fob1Δ led to a reduction in the MET15 LOH rate of ∼2-fold (Figure 3), but this rate was still significantly higher than the LOH rate on chromosome IV, suggesting that other mechanisms must contribute to the hotspot activity affecting MET15. When we measured MET15 LOH in aging fob1Δ cells we found the age-associated increase in rate of LOH was completely suppressed (Figure 3). This result indicated that Fob1 activity is required for this aging phenotype and suggests that Fob1-mediated DSBs are a critical intermediate for these LOH events.
Figure 3. Age-associated LOH events depend on FOB1.
Total LOH rates at MET15 from individual aging cultures of UCC526 (fob1Δ). Error bars indicate SEM.
A FOB1-independent aging process acts cooperatively with Fob1 to generate LOH events in aged cells
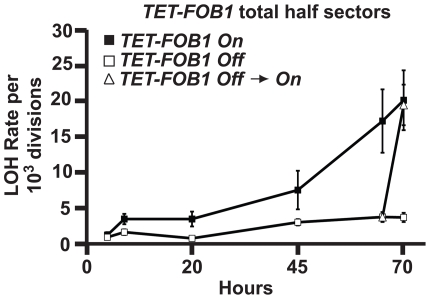
We offer two simple models to explain the requirement for FOB1 in the age-associated increase in LOH at MET15. Fob1 activity may be required to generate an aging factor or process that leads to increased LOH. Alternatively, an aging process may occur independent of Fob1, while Fob1 remains necessary for this process to manifest as LOH events in old cells. To distinguish between these modes of action, we generated a diploid MEP strain in which FOB1 is expressed from a tetracycline-repressible promoter [27]. When the TET-FOB1 strain is aged over a 70-hour time course in the absence of repressor, cells exhibit a normal increase in LOH (Figure 4; Fob1 ON). In contrast, when the TET-FOB1 strain is aged over a 70-hour time course in the presence of doxycycline (a tetracycline analog), it behaves as a fob1 null allele and age-associated LOH is suppressed (Figure 4; Fob1 Off). Critically, after the TET-FOB1 strain is aged over a 65-hour time course in the presence of doxycycline, removal of the repressor results in a rapid ∼5-fold increase in LOH levels within 5 hours (Figure 4; Fob1 Off → On). This result suggests that the process responsible for increased LOH rates in aging cells occurs independently of Fob1, but that Fob1 activity is required in old cells for this aging process to generate LOH events.
Figure 4. An inducible allele of FOB1 reveals the accumulation of aging factors in the absence of Fob1.
Total MET15 LOH rates in aging UCC8912 cells exposed to no doxycycline (FOB1 On; filled squares) or 20 µg/mL doxycycline (FOB1 Off; open squares). At 65 hours, a portion of the Fob1 Off culture was harvested and transferred to media with no doxycycline (FOB1 Off → On; triangles).
We hypothesize that the aging process may involve attenuation of an activity that normally keeps the rate of homologous recombination relatively low within the rDNA array. By extension, elimination (e.g. by gene mutation) of such an activity in young cells should phenocopy the age-associated increase in LOH. Our characterization of the age-associated LOH phenotype has delineated two criteria such candidate activities must possess: Deletion of a candidate activity must show a FOB1-dependent increase in LOH at the MET15 locus, and the ratio of reciprocal-to-nonreciprocal LOH events should be the same as in wild type cells.
Disruption of cohesion increases LOH rates independent of Fob1
One mechanism by which LOH could increase in old cells is through an age-associated defect in sister chromatid cohesion, which normally restricts template selection during DSB repair to favor sister chromatids [28]–[31]. Such a defect would allow more recombination to occur between homologs during repair of Fob1-mediated DSBs, thus giving rise to increased LOH.
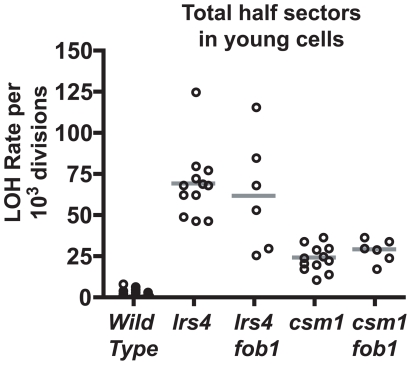
The effect of disrupting sister chromatid cohesion was examined by deleting the genes encoding the cohibin complex proteins Csm1 and Lrs4. The cohibins physically interact with both Fob1 and cohesins, anchoring cohesin rings within the rDNA array and inhibiting both unequal sister chromatid exchange (USCE) in haploid cells and homologous recombination in diploids [17], [31]. Consistent with these reports, we found that csm1Δ or lrs4Δ alleles resulted in high rates of MET15 LOH in young cells (Figure 5). But importantly, when csm1Δ or lrs4Δ were combined with fob1Δ, rDNA recombination rates in young cells remained high (Figure 5). Thus, loss of cohesion leads to Fob1-independent rDNA recombination, which strongly suggests that this is unlikely to be the mechanism underlying the increased rDNA recombination observed in aging cells.
Figure 5. Double mutant analysis of fob1 and cohibin mutants.
Scatter plots of total LOH rates at the MET15 locus in young cells with genotypes as indicated.
A decline in Sir2 protein levels in aging cells does not correlate with increased LOH rates
The protein deacetylase Sir2 has been implicated in the yeast aging process and life span determination [32]. Furthermore, loss of Sir2 function leads to high rates of USCE in haploid cells, resulting in dramatic copy-number instability within the rDNA array [33]–[35], and increased rates of homologous recombination between rDNA arrays in diploid cells [11]. Therefore, we examined the potential role of Sir2 in age-associated LOH in detail.
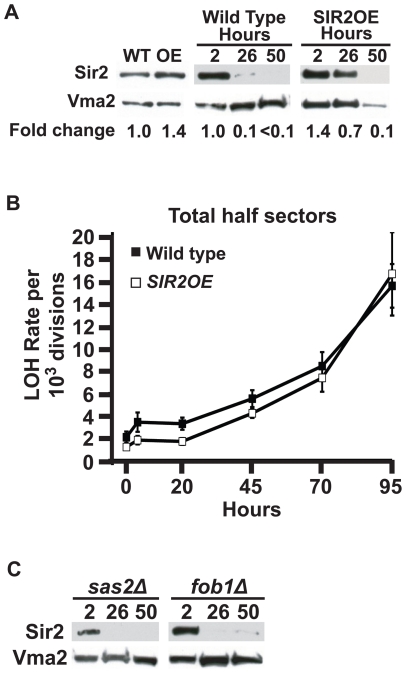
It was recently shown that Sir2 protein levels decline in aging haploid cells [36]. To determine if Sir2 levels are reduced in aging diploid cells, we purified aged mother cells from liquid MEP cultures and prepared total protein extracts. When the MEP is activated, the time a mother cell spends in culture corresponds to her replicative lifetime [16]. Hence we can age-match different strains by aging these MEP cultures for equivalent periods of time (the strains analyzed here divide at the same approximate rate; data not shown). To confirm age matching between strains, purified 26-hour populations were stained with calcofluor white to count bud scars (see Figure 6 legend). Western blotting with an anti-Sir2 antibody confirmed that Sir2 protein levels were dramatically reduced in aging diploid cells when normalized to total protein (Figure 6A). Sir2 levels in aging populations had fallen 10-fold by 26 hours, and were further reduced by 50 hours. This same decline was also observed with a polyclonal antiserum raised to a different region of the Sir2 protein (data not show). However, this behavior was not a universal feature of proteins in aging cells: Western blotting against the vacuolar protein Vma2 (Figure 6A) and the kinase Pkc1 (data not shown) both showed no decline in aging cells. Therefore, Sir2 protein levels specifically decline in aging cells, however this decline precedes any significant change in LOH rates by 20 hours, or approximately 12 generations.
Figure 6. Declining Sir2 levels do not correlate with the onset of age-associated LOH events.
A) Western blotting against Sir2 and Vma2 in total protein extracts prepared from cells aged for hours as indicated at top. Sir2 levels were normalized to Vma2 and quantified by densitometry, with wild type Sir2 levels in log cultures set to 1x Fold. Mean bud scar counts at 26 hours: WT = 15.5, SIR2OE = 18.0. B) Total LOH rates at MET15 of wild type UCC5185 (filled squares) compared to the SIR2OE strain UCC8910 (open squares). Error bars indicate SEM. C) Western blotting against Sir2 and Vma2 in total protein extracts prepared from aged cells. Genotype and hours of aging indicated at top. Mean bud scar counts at 26 hours: sas2Δ = 16.8, fob1Δ = 15.7.
Modulation of Sir2 protein levels in aging cells does not affect age-associated LOH
The observation that Sir2 protein levels decline precipitously 20 hours before rDNA recombination rates increase significantly led us to further examine the relationship between Sir2 depletion and age-associated LOH. We tested the effect of SIR2 gene copy number on rDNA recombination using diploid strains homozygous for a tandem repeat of SIR2 under control of its native promoter (SIR2OE). Previous studies indicated that this modest level of over-expression is sufficient to increase silencing of RNA Polymerase II-dependent transcription within the rDNA array [34], [37] and extend RLS [32], while higher expression of Sir2 is toxic [38]. Sir2 over-expression in young (log-phase) diploid cells was verified by Western blot (Figure 6A). The decline in Sir2 protein levels was partially suppressed in the SIR2OE strain: Sir2 levels in SIR2OE populations were elevated at 26 hours compared to wild type, but declined precipitously to wild type levels by 50 hours (Figure 6A).
Next, we examined LOH rates at MET15 in the SIR2OE strain to determine if increased Sir2 levels suppressed the age-associated change in LOH rates. Although MET15 LOH rates in young SIR2OE cells were significantly lower than the wild type strain (p<0.0001, Fisher's exact test based on contingency tables), we observed no difference in the timing or magnitude of the age-associated increase in MET15 LOH rates in SIR2OE cells compared to wild type (Figure 6B). This result offers further support that Sir2 protein levels are not the determining factor regulating rDNA recombination in aging cells.
To confirm this result, we attempted to stabilize Sir2 protein levels by a second method. In a previous study, Dang, et al. found that deletion of SAS2, the gene encoding the catalytic histone acetyltransferase subunit of the SAS complex that acts antagonistically to Sir2 for modification of histone H4 at lysine 16, stabilized Sir2 levels in aging cells [36]. When we constructed a sas2 deletion strain in the MEP background and examined Sir2 protein levels, we saw no evidence of Sir2 stabilization in this mutant background (Figure 6C). Finally, we asked whether deletion of fob1, which does suppress age-associated rDNA recombination, might also stabilize Sir2 protein levels in aging cells. Again, we observed no stabilization of Sir2 protein levels (Figure 6C). Thus, we found no evidence of a correlation between Sir2 protein levels and LOH rates in aging populations.
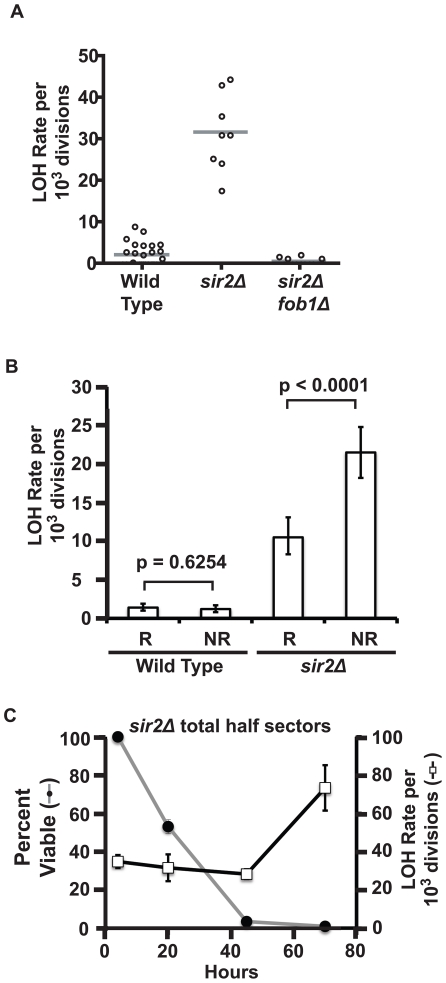
Following the same logic behind our examination of cohesion, we characterized the pattern of LOH events observed in young cells when sir2 was deleted. We found that elevated MET15 LOH rates in young sir2Δ cells were dependent on FOB1 (Figure 7A). In this regard it shows similarity to the age-associated LOH phenotype. However, the ratios of reciprocal to non-reciprocal events in sir2Δ cells was significantly shifted to favor non-reciprocal events (Figure 7B), indicating that loss of Sir2 function does not accurately phenocopy the age-associated LOH phenotype.
Figure 7. Characterization of LOH events in young sir2Δ cells.
A) Scatter plots of total LOH rates at the MET15 locus in young cells with genotypes as indicated. B) Rates of total LOH events at MET15 in young cells for wild type (UCC5185) and sir2Δ (UCC8836) strains. Rates of reciprocal/non-reciprocal events were significantly different in the sir2Δ strain but not in wild type (Fisher's exact test by contingency tables, p values indicated above columns). C) Rates of total LOH events at MET15 in replicatively aging sir2Δ cultures. Right y-axis: LOH rates at MET15 (open boxes) reported as total LOH events per cell division. Error bars indicate SEM. Left y-axis: Percent viability of mother cells in the aging culture (grey line).
Lastly, we examined LOH rates in aging sir2Δ cells to determine if there is an age-associated increase in the absence of Sir2. Consistent with our earlier results [11], there was a very high level of LOH in young cells. Nevertheless, after 70 hours of replicative aging, LOH rates at MET15 increased significantly (p = 0.0003, Fisher's exact test based on contingency tables; Figure 7C). It is worth noting that sir2Δ cells have a short RLS [32]; thus at 70 hours only the longest-lived 1% of the total population are represented. When taken together, these data - the lack of correlation between Sir2 protein levels and rDNA recombination rates in aging cells, the difference in reciprocal/non-reciprocal LOH events between aging wild type and sir2Δ cells, and the Sir2-independent increase in LOH in aging cells - indicate that declining Sir2 levels are insufficient to explain the age-associated rDNA recombination phenotype.
ERCs accumulate to high levels in fob1Δ cells
Intrachromosomal recombination between the rDNA repeats can produce extrachromosomal rDNA circles (ERCs) that have been postulated to induce senescence in aging mothers by an undefined mechanism [39]. Each repeat contains an origin of DNA replication, allowing ERCs to replicate independently, but lacks a centromeric sequence to ensure equal partitioning of ERCs between mother and daughter cells during mitosis [39]–[40]. Consequently, ERCs preferentially accumulate within aging mother cells. It has been suggested recently that ERCs can influence rDNA array stability in haploid cells [41]. Given these findings, we explored whether ERC levels play a role in age-associated LOH in diploid cells.
In order to examine this issue, we found it necessary to further develop methods employed to examine ERC levels with age. Previous analyses of ERC levels have been technically limited to the examination of relatively young cells (∼10 generations) [42], whereas we observe age-associated increases in LOH after the median life span of our diploid strains (36 generations for UCC5185) [16]. To better understand the kinetics of ERC accumulation, we purified aged mother cells from MEP cultures and quantified ERC levels by Southern blotting. To improve ERC quantitation, we removed the linear rDNA array by digestion with RecBCD exonuclease, a highly processive exonuclease that does not degrade intact or nicked circular DNA [43] (Figure S2). Monomeric, dimeric, and multimeric ERC species were identified based on their migration rates [44] (Figure 8A) and quantified by densitometry (Figure 8B).
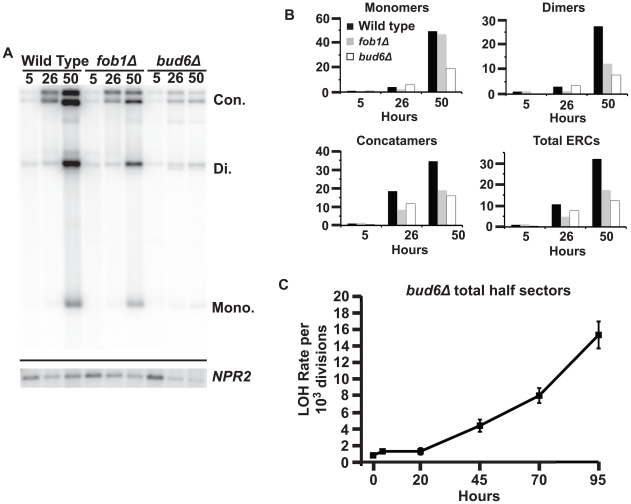
Figure 8. ERC accumulation in aging diploid cells.
A) Southern blot of total genomic DNA isolated from aging populations and digested with BamHI and RecBCD. Genotypes and replicative age in hours indicated at top. ERC species indicated at right: ERC Concatamers- Con.; Dimers- Di.; Monomers- Mono. Mean bud scar counts at 26 hours: WT = 15.3 fob1Δ = 15.6, bud6Δ = 12.9. Lower panel: Southern blot of total genomic DNA isolated from aging cell populations, digested with BamHI and probed for NPR2. B) ERC levels normalized to NPR2 (panel A). For individual ERC species, wild type 5-hour lanes are set as 1x fold. Total ERCs were calculated as the sum of the integrated density of each ERC species, with the wild type 5-hour lane set as 1x fold. C) Total LOH rates at MET15 in aging cultures of the bud6Δ strain UCC8904. Error bars indicate SEM.
First, we compared ERC levels in wild type and fob1Δ cells. A modest increase in all three species of ERCs was evident in wild type cells after aging replicatively for 26 hours, a relatively young age when the population maintains ∼90% viability (Figure 1A and [16]). As the population approaches its median RLS by 50 hours, ERC levels continued to increase dramatically. Similar to previous reports, in relatively young cells, ERC levels in a fob1Δ diploid were low compared to wild type cells (Figure 8B and [42]), but their accumulation was not completely prevented. After 26 hours of replicative aging, total ERC levels in the fob1Δ strain had increased ∼5-fold above young wild type levels, but were still reduced ∼50% compared to the age-matched wild type population. Interestingly, as the fob1Δ population continued to age, ERC accumulation continued: Total ERC levels in fob1Δ cells were still ∼50% lower than wild type cells after aging replicatively for 50 hours, but by this age ERC levels in the fob1Δ strain had increased ∼17-fold above young wild type levels. These results indicate that ERC accumulation is an aging process that is not strictly dependent on Fob1 activity. The delayed accumulation of ERCs could reflect a reduction in the rate of formation of ERCs from the rDNA array in the fob1Δ strain, while amplification of ERCs by replication is unaffected [39].
Age-associated LOH is not suppressed by reduced ERC accumulation
While the fob1Δ strain permitted us to determine the utility of our ERC measurements with age, it was not useful for examining the effect of ERC accumulation on LOH because, as shown above, Fob1 is required for the age-associated LOH events. For this, we examined a genetic background that could reduce ERC accumulation independently of Fob1. Deletion of BUD6 has been reported to disrupt the asymmetric segregation of ERCs, resulting in extended life span and a prediction of reduced ERC accumulation in mother cells [45]. Consistent with this prediction, we found that deletion of bud6Δ reduced the level of ERCs throughout the life span of a population. The bud6Δ cells showed a 50% reduction in ERCs after 50 hours of replicative aging: A level that was achieved in wild type populations aged for only 26 hours (Figure 8B). Remarkably, we saw no change in the age-associated increase in LOH rate at MET15 in the bud6Δ strain (Figure 8C). This indicated that in the presence of Fob1 activity, a 50% reduction in ERC accumulation was not sufficient to suppress age-associated LOH.
Discussion
Age-associated genomic instability in respiration-competent cells
Previously we discovered an age-associated, genome-wide increase in LOH in yeast that results from the loss of mtDNA and respiratory function in the progeny of aging cells [11]–[12]. Here we used our recently developed MEP [16] to examine LOH in cells that retain respiratory function throughout their life span. Although the MEP strains are in the same S288C strain background as strains used in previous studies, they display a greater capacity to maintain mitochondrial function both in logarithmically growing cultures and during aging [11], [46]. This greater stability of mitochondrial function in the MEP strain is similar to that found for most natural isolates of S. cerevisiae [14]. The faithful maintenance of functional mitochondria depends on over 100 genetic loci [15] and we have not determined the precise genetic basis for the phenotypic differences we observe.
Using the MEP, we have discovered a distinct age-associated LOH phenotype in cells that retain respiration competence. This new LOH phenotype is distinguished by an apparent specificity for the rDNA array and dependence on the replication fork-block protein Fob1. As cells pass their median life span, they experience a significant increase in homologous recombination within the rDNA array which leads to LOH along a ∼875 kbp span from the rDNA to the telomere of the right arm of chromosome XII. These genomic alterations are mechanistically equivalent to events that generate partial uniparental disomy in mammalian cells, which has recently been found to occur at high frequency in many human cancers [6], [47]–[48].
We observed a significant increase in both reciprocal and non-reciprocal recombination events, which likely report two different routes of DSB repair. Reciprocal LOH events likely result from strand invasion leading to Holliday junction formation and resolution with crossing over [49], while the non-reciprocal events we observed are consistent with break-induced replication (BIR) initiated within the rDNA and propagated to the telomere of chromosome XII [17], [49]. These recombination events also differ from the age-associated genomic instability described in cells that have lost respiration competence [11], where LOH events were almost uniformly non-reciprocal. The phenotypic differences between the earlier studies and those reported here support the conclusion that a different mechanism leads to age-associated LOH events in cells that maintain respiration competence. Because most eukaryotic cells cannot tolerate the loss of mtDNA, it is likely that the findings we report here about genomic instability may be relevant to the aging process in other organisms.
Loss of cohesion or Sir2 protein does not appear to lead to increased age-associated LOH rates
Taking advantage of the requirement for Fob1, we investigated potential mechanisms leading to the age-associated LOH phenotype. Disruption of cohesion, via the deletion of the cohibins LRS4 and CSM1, was eliminated as a potential mechanism because it leads to Fob1-independent LOH events. While SIR2 deletion generated Fob1-dependent LOH events in young cells, the LOH events observed in this strain were significantly biased towards a non-reciprocal pathway compared to the LOH events generated in young or old wild type cells. Additionally, when sir2Δ cells were aged they also displayed an age-associated increase in LOH in the longest-lived fraction of the population. Both the difference in repair bias and presence of an age-associated increase in LOH independent of SIR2 suggests that loss of Sir2 function in aging cells is likely not the driver of age-associated LOH.
Using the MEP for biochemical analysis of populations at their median RLS
Aging is typically accompanied by an exponentially increasing hazard rate of death – i.e. the probability that an individual within a population will die within a given time interval. Previous biochemical analyses of aging cells focused on cells aged for only ∼10 generations due to technical limitations [50]. At this age, a diploid cell population in the common laboratory S288C strain is still relatively young: It retains >95% viability and has yet to experience the dramatic increase in hazard rate of death that affects cells near their median RLS [16], [51]. Using the MEP, we developed an effective purification method to isolate age-matched populations near their median RLS which allowed us to make new observations that change our understanding of age-associated processes. Earlier reports concluded that fob1Δ effectively suppressed the accumulation of ERCs [32], [42]. By examining populations at their median life span we found that ERC levels in fob1Δ cells actually increase significantly as cells approach their median RLS, although their accumulation was reduced/delayed compared to wild type cells. The extension of our ability to observe age-associated genetic and biochemical changes with the MEP allows us to begin to develop an understanding of the order of events that affect an aging population.
Declining Sir2 protein levels in aging cells
It was recently reported that Sir2 protein levels decline in aging haploid cells after only 7-9 generations, a relatively early point in their life span [36]. Our findings are consistent with this earlier work: Sir2 protein levels in aging diploid cells show a precipitous decline by 24 hours. However, this occurs at a time in which LOH rates have not yet increased and ∼95% of cells remain viable. Furthermore, when Sir2 was over expressed, no suppression of age-associated LOH rates was observed. Taking all these results together, we interpret these data to indicate that declining Sir2 protein levels do not correlate with the increase in age-associated LOH and offer further evidence that Sir2 is unlikely to be responsible for the age-associated increase in LOH. One potential caveat to our interpretation is that if there is significant cell-to-cell heterogeneity in levels of Sir2 over-expression, then LOH may occur in a subpopulation of cells with lower Sir2 levels.
Why might reduced Sir2 protein levels in aging cells fail to alter rDNA recombination rates in a fashion similar to young cells? One possible explanation is a phenotypic lag of the increased rDNA recombination that lasts for >10 generations after the reduction of Sir2 protein. For instance, if Sir2 is completely absent in most cells, then one of it substrates (e.g. K16 of histone H4) may not immediately become acetylated [52]. In fact, newly synthesized histones start out in an unacetylated state, and how a heritable switch between acetylated and unacetylated states of histones occurs remains mysterious [53]. Another possibility is that a very low level of Sir2 protein may be sufficient to suppress rDNA recombination. Relevant to this idea, different regions of silent chromatin can compete for recruitment of Sir2 [37], [54]. If the rDNA array is dominant in such a competition, the effect of declining Sir2 levels could be masked.
Are ERCs responsible for age-associated LOH?
If ERC accumulation is the aging process that leads to age-associated LOH, their influence on LOH must be Fob1-dependent. Indeed, it has previously been shown that plasmids containing the RFB sites from an rDNA repeat can integrate into the chromosomal rDNA array in a Fob1-dependent manner [55]. Thus, ERCs may be capable of initiating recombination events in a Fob1-dependent manner that somehow increases the frequency of homologous recombination in diploid cells. Alternatively, accumulating ERCs could simply titrate away factors that affect the frequency or stability of stalled DNA replication forks and thus increase their conversion to DSBs within the rDNA array [26].
In order to determine whether ERCs play a role in age-associated LOH, it is necessary to eliminate ERC formation and/or accumulation in aging cells. Using the bud6Δ strain we reduced ERC levels at a median RLS by approximately 50%, down to a level that was equivalent to a younger wild type population (26 hours of replicative aging; Figure 8B). It is significant that at this younger age, wild type cells show no increase in LOH rate at the MET15 locus. Despite this reduction in ERC levels, the bud6Δ strain showed no suppression of age-associated LOH with age. While we interpret this as evidence that ERC accumulation does not drive age-associated LOH at MET15, it remains formally possible that this reduced level of ERCs is still above a threshold required to increase LOH, or that a subpopulation of cells contain higher ERC levels that drive age-associated LOH events. A clearer understanding of the contribution of ERCs to age-associated LOH will require more refined control of ERC initiation and accumulation.
ERCs, LOH, and life span
Previously, life span extension by fob1Δ was interpreted as a result of reduced ERC accumulation to a degree at which RLS becomes limited by an alternative mechanism [32], [42]. However, we found that ERCs accumulate to high levels even in fob1Δ cells (Figure 8). This result could be interpreted in two ways that support opposing models for how life span is limited in fob1Δ cells. First, the delayed accumulation of ERCs early in life could extend, but still limit, RLS in fob1Δ cells. Alternatively, the life span-limiting function of ERCs may be Fob1-dependent, suggesting that RLS in fob1Δ cells is limited by an alternative mechanism.
Recently it has been argued that rDNA stability, rather than ERC accumulation, limits life span [41]. However, because life span potential is ‘re-set’ in daughter cells [56], we cannot conclude that an LOH event (which would be heritable) directly limits RLS: Instead, a reversible aging process that impacts rDNA stability, and can generate LOH events at some frequency, may limit RLS.
Curiously, we find that the highest rates of age-associated LOH are observed at the latest time points, where the population consists of the longest-lived survivors (<10% of the population remains viable), and when the hazard rate of death has declined from its peak near the median life span [16]. This lack of correlation between LOH rate and hazard rate of death suggests that rDNA recombination is not limiting for life span of most cells. Nevertheless, the longest-lived individuals in the population no longer experience the same hazard rate of death, and thus their life span may be limited by a different mechanism (e.g. rDNA recombination) than the average population.
DNA replication stress and age-associated LOH
If pathways that have been previously identified to modulate rDNA recombination do not adequately account for the age-associated increase in LOH rates, can this phenotype be a response to a change in a more general biological process that is most readily manifested at the rDNA? Since DSBs within the rDNA array normally arise through the interaction between DNA replication forks and Fob1 [57], we speculate that DSB rates in the rDNA are modulated by DNA replication stress. This model is supported by several lines of evidence: A hypomorphic allele of the essential DNA replication helicase encoded by DNA2 results in an increased frequency of DSBs within the rDNA, which can be suppressed by deletion of FOB1 [57]–[58]. Similarly, mutations in DNA polymerase α and δ subunits can also lead to increased DSBs within the rDNA array [26], [59]. Deletion of RRM3, a helicase that functions to remove non-histone protein barriers from DNA, also affects rDNA recombination [60]–[61]. Further evidence comes from a screen for deletion mutants that increase LOH in young cells [17], which classified mutations based on locus specificity, magnitude and ratio of reciprocal/non-reciprocal events. The group of deletion mutants that showed a bias toward increasing LOH at MET15 on chromosome XII (and presumably originate in the rDNA) included five genes implicated in the regulation of nucleotide pools, which can be a source of DNA replication stress [62].
While the effects of DNA replication stress on the rDNA may be dependent on Fob1, the mode of action may not necessarily be specific to the rDNA. Ivessa, et al. identified genomic regions prone to DNA replication fork pausing in an rrm3Δ mutant which included centromeric regions, tRNAs and sub-telomeric sequences in addition to the rDNA array [9]. Similarly, RRM3 was originally identified in a genetic screen for mutations that induce gene duplication at the tandemly repeated CUP1 locus [60], which suggests that other regions of the genome that combine replication fork blocks with repetitive sequence elements could also generate age-associated LOH events. While repetitive elements are uncommon in the S. cerevisiae genome, the ubiquitous nature of these features in mammalian genomes suggests great potential for age-associated genomic instability generated by a similar mechanism.
Materials and Methods
Subcloning and strain construction
Genotypes of all yeast strains used in this study are provided in Table S1, oligonucleotide sequences are provided in Table S2, and plasmids are listed in Table S3.
The two-chromosome LOH reporter strain UCC8918 was generated by mating UCC8917 x UCC5181. UCC8917 was derived from UCC5179 by one-step integration of a PCR fragment carrying ADE2 into an intergenic region of chromosome IV (coordinates 1,515,634–1,515,738), which was generated using oligos MarthaN/H2L and MarthaN/H2R with pRS402 [63] as a template.
The diploid strain with multiple heterozygous markers on chromosome XII (UCC8915) was generated by mating UCC8914 x UCC5179. UCC8914 was derived from UCC8913 by one-step integration of a PCR fragment carrying TRP1 into an intergenic region distal to the right end of the rDNA array (coordinates 486062-486189), which was generated using oligos RDNRF and RDNRR with pRS304 [64] as a template. Integration was verified by PCR from genomic DNA using oligos RDNRconF and RDNRconR. UCC8913 was derived from UCC5181 by one-step integration of a PCR fragment carrying KANMX into an intergenic region proximal to the left end of the rDNA array (coordinates 450191-450372), which was generated using oligos RDNLF and RDNLR with pRS400 [63] as a template. Integration was verified by PCR from genomic DNA using oligos RDNLconF and RDNLconR.
MEP deletion strains for bud6Δ, csm1Δ, lrs4Δ, sir2Δ and sas2Δ were generated by one-step gene disruption and verified by PCR using the following oligonucleotides and DNA templates: BUD6: deletion- Bud6delF/bud6delR, template- pRS306, confirmation- Bud6delcheck/Bud6delchkDN. CSM1: deletion- CSM1A/CSM1D, template- UCC7629-1 genomic DNA, confirmation- CSM1A/CSM1B. LRS4: deletion- LRS4A/LRS4D, template- UCC7598-1 genomic DNA, confirmation- LRS4A/LRS4B. SIR2: deletion- SIR2KO1/SIR2KO2, template- pRS400 [63], confirmation- 5′SIR2/3′SIR2. SAS2: deletion- SAS2KOF/SAS2KOR, template- pRS400 [63], confirmation- SAS2conF/SAS2conR.
The sir2Δ fob1Δ double mutant was constructed by transforming UCC8832 with plasmid pRS314-SIR2 [65] to complement the mating defect of sir2Δ, followed by mating to UCC524 and sporulation. Because both deletions are marked with KANMX, PCR was used to identify double mutant spore products to generate UCC8839 and UCC8840. These haploids were mated to generate UCC8844, which was subsequently cured of the pRS314-SIR2 plasmid. Standard mating and sporulation, followed by PCR to identify double mutant spore products, was used to generate csm1Δ fob1Δ and lrs4Δ fob1Δ double mutant strains.
To generate SIR2OE strains, a genomic clone of SIR2 along with ∼500 bp of upstream sequence and ∼250 bp of downstream sequence was subcloned by PCR amplification with SIR2ecoriF and SIR2ecoriR primers, digested with EcoRI, and ligated into pRS306 cut with EcoRI to create the integration plasmid pRS306-SIR2. The plasmid was cut with BglII and transformed into UCC5179 and UCC5181 to generate strains UCC8908 and UCC8909. Correct integration was confirmed by PCR. These haploids were mated together to generate the diploid strain UCC8910. The SIR2hemi diploid was generated by transforming UCC8832 with pRS314-SIR2, mating to UCC5179, and subsequently curing the diploid of the plasmid.
TET-FOB1 construction
The TET-OP2 promoter was inserted upstream of the endogenous FOB1 locus by one-step integration of a PCR fragment generated using primers FOB1tetF and FOB1tetR with pKAN-TETO2 as a template. The TETR'-SSN6 cassette was inserted by one-step integration into the met15Δ0 locus by digesting plasmid pLMI-tetR'S with PacI. The LEU2 marker was subsequently exchanged for ADE2 or MET15 using the primers LEU2swapF and LEU2swapR with the appropriate plasmid templates (pRS402 and pRS401, respectively [63]). The VP16 activation domain from the tTA activator was replaced with activation domain A of GCN4 by generating a GCN4A PCR product using GCN4salAF and GCN4ascAR primers with plasmid pGCN4 (a gift from S. Hahn) as a template. The PCR product was digested with SalI and AscI and subcloned into pUI-tTA-ADH1term-URA3 digested with the same enzymes to create pUI-tTA-GCN4A-ADH1term-URA3. The tTA-GCN4A-ADH1term-URA3 cassette was integrated into a neutral site on Chromosome I (coordinates 17030-17205) by one-step integration of a PCR product using the primers URA3-tTA-intCHRIF and URA3-tTA-intCHRIR. Subsequent strain construction to generate haploid and diploid MEP strains with the appropriate genotypes was performed by standard methods.
Pedigree analysis
Pedigree analysis was performed as previously described [11]. Briefly, daughter cells born to individual mothers were micromanipulated to new positions on a YEPD plate and allowed to form colonies. Colonies were scored for mitochondrial function by replica plating to YEP+glycerol.
Liquid aging and LOH assay
Diploid cells were grown to saturation overnight in YC media lacking adenine and methionine. Cells were used to inoculate YEPD cultures, which were grown to log phase by incubation with shaking, 30°C for 3 hours. Cells were counted and used to inoculate 25 ml YEPD +1 µM estradiol cultures at 2×104 cells/ml and incubated with shaking, 30°C for 95 hours. At indicated times, samples were harvested, washed, and plated to lead nitrate media. Sample volumes were adjusted appropriately to maintain a colony density of ∼500–1000 colonies/150 mm plate. Plates were incubated at 30°C for 3 days and colonies were counted using a Geldoc XR+ imaging system (Biorad). Plates were further incubated at room temperature for 2–3 days for color development before scoring for half sectors. Reciprocal and non-reciprocal colonies were counted separately, and rates of total half sectors were calculated as (2*reciprocal + non-reciprocal/total colonies) [17].
Western blot analysis
Lysates were prepared using NaOH lysis followed by TCA precipitation [66]. TCA pellets were resuspended in SUME buffer (1% SDS, 8 M Urea, 10 mM MOPS, pH 6.8, 10 mM EDTA) and total protein concentration was determined using the BCA protein assay (Thermo Scientific). 10 µg total protein per lane was run on 10% tris-glycine polyacrylamide gels (PAGEr gold, Lonza) and transferred to Immobilon P membrane (Millipore) using a semi-dry transfer apparatus. Western blot analysis was performed by standard methods and developed with Supersignal West Pico (Thermo Scientific). Antibodies: Goat α-Sir2 (yN-19; Santa Cruz Biotechnology), mouse α-Sir2 (sc-25753; Santa Cruz Biotechnology) mouse α-Vma2 (Invitrogen), mouse α-Pkc1 (Invitrogen), and HRP-conjugated secondary antibodies (Jackson Immunoresearch Laboratories). Quantitation of Western blots was performed by densitometry using ImageJ comparing exposures that fell within a linear range of detection.
Purification of aged populations
Log phase cultures of cells were harvested and labeled with NHC-Biotin as previously described [16]. Labeled populations were transferred to YEPD and incubated for 30°C, 2 hours to ensure that most labeled cells were mothers (have completed at least one cell division). Cells were counted and used to inoculate 1.5 liter cultures of YEPD +1 µM estradiol +100 µg/ml ampicillin at a density of 2×104 cells/ml. Cultures were incubated at 30°C, shaking at 100 rpm for the indicated times before purification. Cells were harvested by centrifugation, resuspended at 6×108 cells/ml in RNAlater (Ambion) and fixed at room temperature, 45 minutes. Cells were harvested by centrifugation and resuspended at 2×108 cells/ml in PBS +2 mM EDTA and incubated with 1/20 volume streptavidin beads (Miltenyi Biotec), 4°C, 30 minutes. Cells were harvested by centrifugation and resuspended in 4 ml 40 mM Tris HCl pH 7.4 and layered onto Percoll Plus gradients (GE Healthcare). Gradients were spun at 4°C, 20 minutes at 2000 RPM in a GS-6R tabletop centrifuge (Beckman). A brown, flocculent layer of cell debris was removed from the top of the gradients with a pipette, and the remainder of the gradient was pooled with 40 ml PBS +2 mM EDTA. Cells were harvested by centrifugation, resuspended in 45 mL PBS +2 mM EDTA and purified on an Automacs Pro separator system (Miltenyi Biotek).
ERC Southern blot analysis
Genomic DNA was isolated from purified aged populations by standard methods. 1 µg of genomic DNA was digested overnight with BamHI + RecBCD (a gift from G. Smith, FHCRC) and separated by gel electrophoresis (0.8% agarose, 2 V/cm for 36 hours). DNA was visualized by staining with ethidium bromide and transferred to nitrocellulose membranes by standard methods. Membranes were hybridized with a 32P labeled double-stranded probe specific to the rDNA generated with oligos RDN5S-2 and RDN25S-2 using plasmid pDL05 as a template, visualized on a Typhoon phosphorimager (GE Health Sciences), and quantified using ImageJ. For normalization of DNA samples, 1 µg of genomic DNA was digested with BamHI, separated by gel electrophoresis (0.8% agarose, 10 V/cm, 3 hours), transferred and hybridized with a probe specific to NPR2 generated with oligos 5_NPR2 and 3_NPR2.
TET-fob1 time courses
Because over expression of Fob1 also increases rDNA recombination rates [67], we generated a weaker version of the tet-activator by replacing the VP16 activation domain with activation domain A of Gcn4 [27], [68]. By carefully titrating Fob1 expression levels using a single copy of both TETOP2-Fob1 and the tTA-GCN4A activator in diploid cells, we could express FOB1 at normal levels in the absence of doxycycline, while effectively suppressing expression in the presence of 20 µg/ml doxycycline (Figure 5A).
Diploid cells were grown to saturation overnight in YC media lacking adenine and methionine. Cells were used to inoculate YEPD +/- 20 µg/ml doxycycline cultures, which were incubated with shaking, 30°C for 5 hours. Cells were counted and used to inoculate 40 ml YEPD +1 µM estradiol +/- 20 µg/ml doxycycline cultures at 2×104 cells/ml and incubated with shaking, 30°C for 95 hours. Samples were harvested and plated as described above for the LOH assay. To shift FOB1 expression from repression to activation, 15 ml of the + doxycycline culture was harvested at the 65 hour time point, washed, and resuspended in 15 ml fresh YEPD +1 µM estradiol.
Supporting Information
Loss of respiratory function is less frequent in MEP strains. A) Examples of inheritance of respiratory function during pedigree analysis of UCC5185. 68% of mothers generate pedigrees with all daughters maintaining respiratory function (upper panel). 32% of mothers lose the ability to produce daughters with respiratory function (small colonies; lower panel). Respiration competence of daughter cells could be inferred from their growth rate and was confirmed by replica plating to media containing a non-fermentable carbon source (data not shown). The upper panel was photographed 2 days after placement of last daughter, while the lower panel was photographed 10 days after placement of last daughter. B) A comparison of the age of onset for the subpopulation of mother cells that lose the ability to produce daughters with respiratory function.
(TIF)
Preparation and normalization of ERC southern blotting. ERC Southern blot of total genomic DNA isolated from wild type cells and digested with BamHI and with or without RecBCD. Arrow indicates the linear rDNA array.
(EPS)
Yeast strains used in this study.
(DOC)
Oligonucleotides used in this study.
(DOC)
Plasmids used in this study.
(DOC)
Acknowledgments
We thank Jessica Hsu, Zara Nelson, and Steve Hahn for providing plasmids used in this study and Gerry Smith and Andrew Taylor for providing purified RecBCD enzyme.
Footnotes
The authors have declared that no competing interests exist.
This work was supported in part by postdoctoral fellowship #PF-04-041-01-GMC from the American Cancer Society (http://www.cancer.org) to DLL, a Fellow's Career Development Program Award from The Leukemia and Lymphoma Society (http://www.leukemia-lymphoma.org/hm_lls) to KAH, National Institutes of Health (http://www.nih.gov/) grant AG023779 and a Glenn Award for Research in Biological Mechanisms of Aging (http://glennfoundation.org/) to DEG, and National Cancer Institute (http://www.cancer.gov/) grant T32 CA09657. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.DePinho RA. The age of cancer. Nature. 2000;408:248–254. doi: 10.1038/35041694. [DOI] [PubMed] [Google Scholar]
- 2.Renan MJ. How many mutations are required for tumorigenesis? Implications from human cancer data. Mol Carcinog. 1993;7:139–146. doi: 10.1002/mc.2940070303. [DOI] [PubMed] [Google Scholar]
- 3.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 4.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr LL, Gottschling DE. Does age influence loss of heterozygosity? Exp Gerontol. 2008;43:123–129. doi: 10.1016/j.exger.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Bacolod MD, Schemmann GS, Giardina SF, Paty P, Notterman DA, et al. Emerging paradigms in cancer genetics: some important findings from high-density single nucleotide polymorphism array studies. Cancer Res. 2009;69:723–727. doi: 10.1158/0008-5472.CAN-08-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuna M, Knuutila S, Mills GB. Uniparental disomy in cancer. Trends Mol Med. 2009;15:120–128. doi: 10.1016/j.molmed.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 9.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, et al. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 11.McMurray MA, Gottschling DE. An age-induced switch to a hyper-recombinational state. Science. 2003;301:1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- 12.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baruffini E, Lodi T, Dallabona C, Foury F. A single nucleotide polymorphism in the DNA polymerase gamma gene of Saccharomyces cerevisiae laboratory strains is responsible for increased mitochondrial DNA mutability. Genetics. 2007;177:1227–1231. doi: 10.1534/genetics.107.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrov LN, Brem RB, Kruglyak L, Gottschling DE. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics. 2009;183:365–383. doi: 10.1534/genetics.109.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hess DC, Myers CL, Huttenhower C, Hibbs MA, Hayes AP, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindstrom DL, Gottschling DE. The mother enrichment program: a genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413–422, 411SI-413SI. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen MP, Nelson ZW, Hetrick ED, Gottschling DE. A genetic screen for increased loss of heterozygosity in Saccharomyces cerevisiae. Genetics. 2008;179:1179–1195. doi: 10.1534/genetics.108.089250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cost GJ, Boeke JD. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast. 1996;12:939–941. doi: 10.1002/(SICI)1097-0061(199608)12:10%3C939::AID-YEA988%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.Johnston JR. Genetic analysis of spontaneous half-sectored colonies of Saccharomyces cerevisiae. Genet Res. 1971;18:179–184. doi: 10.1017/s0016672300012581. [DOI] [PubMed] [Google Scholar]
- 20.Zimmermann FK. A yeast strain for visual screening for the two reciprocal products of mitotic crossing over. Mutat Res. 1973;21:263–269. [PubMed] [Google Scholar]
- 21.Petes TD. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci U S A. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol. 2003;23:9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsang E, Carr AM. Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair (Amst) 2008;7:1613–1623. doi: 10.1016/j.dnarep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 27.Belli G, Gari E, Piedrafita L, Aldea M, Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Covo S, Westmoreland JW, Gordenin DA, Resnick MA. Cohesin is limiting for the suppression of DNA damage-induced recombination between homologous chromosomes. PLoS Genet. 2010;6:e1001006. doi: 10.1371/journal.pgen.1001006. doi: 10.1371/journal.pgen.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Brito IL, Villen J, Gygi SP, Amon A, et al. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 34.Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- 36.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith JS, Brachmann CB, Pillus L, Boeke JD. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmes SG, Rose AB, Steuerle K, Saez E, Sayegh S, et al. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics. 1997;145:605–614. doi: 10.1093/genetics/145.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinclair DA, Guarente L. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 40.Murray AW, Szostak JW. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- 41.Ganley AR, Ide S, Saka K, Kobayashi T. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol Cell. 2009;35:683–693. doi: 10.1016/j.molcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, et al. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 43.Taylor AF, Smith GR. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol. 1985;185:431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- 44.Burkhalter MD, Sogo JM. rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell. 2004;15:409–421. doi: 10.1016/j.molcel.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 45.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 46.Veatch JR. Seattle: University of Washington; 2008. A Crisis Caused by the Loss of the Mitochondrial Genome in Saccharomyces cerevisiae. [Google Scholar]
- 47.Teh MT, Blaydon D, Chaplin T, Foot NJ, Skoulakis S, et al. Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res. 2005;65:8597–8603. doi: 10.1158/0008-5472.CAN-05-0842. [DOI] [PubMed] [Google Scholar]
- 48.Andersen CL, Wiuf C, Kruhoffer M, Korsgaard M, Laurberg S, et al. Frequent occurrence of uniparental disomy in colorectal cancer. Carcinogenesis. 2007;28:38–48. doi: 10.1093/carcin/bgl086. [DOI] [PubMed] [Google Scholar]
- 49.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- 51.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev. 2005;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 52.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 53.Gottschling DE. Summary: epigenetics–from phenomenon to field. Cold Spring Harb Symp Quant Biol. 2004;69:507–519. doi: 10.1101/sqb.2004.69.507. [DOI] [PubMed] [Google Scholar]
- 54.Michel AH, Kornmann B, Dubrana K, Shore D. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 2005;19:1199–1210. doi: 10.1101/gad.340205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benguria A, Hernandez P, Krimer DB, Schvartzman JB. Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:893–898. doi: 10.1093/nar/gkg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kennedy BK, Austriaco NR, Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weitao T, Budd M, Hoopes LL, Campbell JL. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J Biol Chem. 2003;278:22513–22522. doi: 10.1074/jbc.M301610200. [DOI] [PubMed] [Google Scholar]
- 58.Weitao T, Budd M, Campbell JL. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat Res. 2003;532:157–172. doi: 10.1016/j.mrfmmm.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 59.Casper AM, Mieczkowski PA, Gawel M, Petes TD. Low levels of DNA polymerase alpha induce mitotic and meiotic instability in the ribosomal DNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000105. doi: 10.1371/journal.pgen.1000105. doi: 10.1371/journal.pgen.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keil RL, McWilliams AD. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics. 1993;135:711–718. doi: 10.1093/genetics/135.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burhans WC, Weinberger M. DNA replication stress, genome instability and aging. Nucleic Acids Res. 2007;35:7545–7556. doi: 10.1093/nar/gkm1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 64.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA. Identification of a small molecule inhibitor of Sir2p. Proc Natl Acad Sci U S A. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Riezman H, Hase T, van Loon AP, Grivell LA, Suda K, et al. Import of proteins into mitochondria: a 70 kilodalton outer membrane protein with a large carboxy-terminal deletion is still transported to the outer membrane. EMBO J. 1983;2:2161–2168. doi: 10.1002/j.1460-2075.1983.tb01717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johzuka K, Horiuchi T. Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells. 2002;7:99–113. doi: 10.1046/j.1356-9597.2001.00508.x. [DOI] [PubMed] [Google Scholar]
- 68.Drysdale CM, Duenas E, Jackson BM, Reusser U, Braus GH, et al. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Loss of respiratory function is less frequent in MEP strains. A) Examples of inheritance of respiratory function during pedigree analysis of UCC5185. 68% of mothers generate pedigrees with all daughters maintaining respiratory function (upper panel). 32% of mothers lose the ability to produce daughters with respiratory function (small colonies; lower panel). Respiration competence of daughter cells could be inferred from their growth rate and was confirmed by replica plating to media containing a non-fermentable carbon source (data not shown). The upper panel was photographed 2 days after placement of last daughter, while the lower panel was photographed 10 days after placement of last daughter. B) A comparison of the age of onset for the subpopulation of mother cells that lose the ability to produce daughters with respiratory function.
(TIF)
Preparation and normalization of ERC southern blotting. ERC Southern blot of total genomic DNA isolated from wild type cells and digested with BamHI and with or without RecBCD. Arrow indicates the linear rDNA array.
(EPS)
Yeast strains used in this study.
(DOC)
Oligonucleotides used in this study.
(DOC)
Plasmids used in this study.
(DOC)