Abstract
MBT domain proteins are involved in developmental processes and tumorigenesis. In vitro binding and mutagenesis studies have shown that individual MBT domains within clustered MBT repeat regions bind mono- and dimethylated histone lysine residues with little to no sequence specificity but discriminate against the tri- and unmethylated states. However, the exact function of promiscuous histone methyl-lysine binding in the biology of MBT domain proteins has not been elucidated. Here, we show that the Caenorhabditis elegans four MBT domain protein LIN-61, in contrast to other MBT repeat factors, specifically interacts with histone H3 when methylated on lysine 9, displaying a strong preference for di- and trimethylated states (H3K9me2/3). Although the fourth MBT repeat is implicated in this interaction, H3K9me2/3 binding minimally requires MBT repeats two to four. Further, mutagenesis of residues conserved with other methyl-lysine binding MBT regions in the fourth MBT repeat does not abolish interaction, implicating a distinct binding mode. In vivo, H3K9me2/3 interaction of LIN-61 is required for C. elegans vulva development within the synMuvB pathway. Mutant LIN-61 proteins deficient in H3K9me2/3 binding fail to rescue lin-61 synMuvB function. Also, previously identified point mutant synMuvB alleles are deficient in H3K9me2/3 interaction although these target residues that are outside of the fourth MBT repeat. Interestingly, lin-61 genetically interacts with two other synMuvB genes, hpl-2, an HP1 homologous H3K9me2/3 binding factor, and met-2, a SETDB1 homologous H3K9 methyl transferase (H3K9MT), in determining C. elegans vulva development and fertility. Besides identifying the first sequence specific and di-/trimethylation binding MBT domain protein, our studies imply complex multi-domain regulation of ligand interaction of MBT domains. Our results also introduce a mechanistic link between LIN-61 function and biology, and they establish interplay of the H3K9me2/3 binding proteins, LIN-61 and HPL-2, as well as the H3K9MT MET-2 in distinct developmental pathways.
Author Summary
Post-translational modifications (PTM) of histones, the proteins around which DNA is wrapped in chromatin, have been implicated in different biological processes ranging from transcriptional regulation to cell cycle progression. Many histone PTMs recruit specific proteins that translate their function into biological outcomes. Understanding the binding mode and molecular biology of these factors is key for our comprehension of epigenetic processes. In this study, we found that the Caenorhabditis elegans LIN-61 protein specifically interacts with particular PTMs on histone H3, di- and trimethylation of lysine 9 (H3K9me2/3), which are implicated in transcriptional repression. LIN-61 contains so-called malignant brain tumor repeats (MBT), which have been found to bind histone methyl-lysine residues in other proteins and model systems. However, these interactions are limited to the mono- and dimethylated states, and there is little to no sequence specificity for particular histone lysine residues. Importantly, H3K9me2/3 binding by LIN-61 is essential for the function of this factor within the synMuv pathway of C. elegans vulva cell fate determination. Besides identifying the first sequence and methylation state specific MBT protein, our studies define novel functions of LIN-61 and manifest a role of H3K9me2/3 in the synMuv pathway.
Introduction
Proteins containing MBT (malignant brain tumor) domains potentially act as tumor suppressors and might modulate gene repression in the context of chromatin. The MBT repeat is a highly conserved structural motif of ca. 100 amino acids that is found from C. elegans to humans and that exists as tandem repeats of two to four elements [1]. Three linearly arranged MBT domains were first identified in the gene corresponding to the Drosophila lethal(3)malignant brain tumor (l(3)mbt) mutant, an embryonic lethal mutation associated with malignant transformations of optic neuroblasts [2], [3]. In flies, only two other MBT domain-containing proteins exist, Sex comb on midleg (Scm, containing two MBT domains) and Sex comb with four MBT domains (Sfmbt). Both are members of Polycomb group related complexes implicated in repression of Hox genes [4], [5].
Like L(3)mbt, Sfmbt and Scm are essential for Drosophila embryonic development [5]–[7]. In mammals, there are at least nine MBT repeat proteins, each containing two (SCMH1, SCML2), three (L3MBTL1, L3MBTL3, L3MBTL4) or four (L3MBTL2, MBTD1, SFMBT1, SFMBT2) MBT repeats, respectively. Disruption of scmh1, l3mbtl1 or l3mbtl in mouse germline has only minor effects, indicating possible redundant and overlapping effects [8]–[10]. Nevertheless, L3MBTL3, L3MBTL2 and SCML2 are mutated in rare cases of medulloblastoma, indicating that MBT domain proteins may be tumor suppressors [11]. In agreement, mammalian MBT domain proteins function as transcriptional repressors in different contexts, for example when directly targeted to transcriptional reporter systems via heterologous DNA binding domains or on the cyclin E promoter [12], [13]. For this function, the MBT domains appear essential [14].
MBT domain proteins in flies and human contain additional domains besides MBT repeats, such as Zn fingers and SPM regions. In contrast, the two C. elegans MBT domain proteins, LIN-61 and MBTR-1 are composed almost completely of four MBT repeats (for review see [1]). While the function of mbtr-1 is unknown, lin-61 was recently implicated in the synMuvB pathway of C. elegans vulva development [15], [16]. Here, simultaneous mutation of individual genes within two groups, synMuvA and synMuvB, causes formation of additional vulva like protrusions on the ventral side of the worm (multivulva, Muv phenotype) (for review see [17]). While a large number of chromatin factors are classified as synMuvB, the exact molecular roles of these factors within C. elegans developmental pathways have not been defined.
Diverse post-translational modifications (PTM) of histone proteins play important roles in regulating chromatin states and thereby the use and readout of the genome. Especially, methylation of distinct histone lysine residues provides a large array of regulatory options, as these confer site (position of the modified lysine in the primary sequence) and modification stage (mono- vs. di-. vs. trimethylation) specific effects [18]. This has been shown, for example, for activating methyl-lysine marks H3 lysine 4 (H3K4me) and H3 lysine 36 (H3K36me), where different methylation states are associated with distinct roles in the transcription cycle (for review see [19]). Methylation of H3 lysine 9 (H3K9me), in contrast, has been largely studied in the context of gene silencing, especially within heterochromatin. While the di- and trimethylated states of this mark (H3K9me2/3) mainly localize to repressed regions of the genome, these have also been found associated with the body of transcribed genes [20], [21]. H3K9me1, in contrast, shows a more euchromatic distribution [22], [23].
Differential and site-specific histone lysine methylation is established by distinct histone methyl-transferases (HKMT). Di- and trimethylation of H3K9 are mediated by the Suv39h1/h2 isoenzymes, CLLD8/KMT1F as well as the ESET/SETDB1 histone methyltransferase [24]–[26]. SETDB1 is mainly found in euchromatic regions, where it participates in gene silencing [27]. In contrast, CLLD8/KMT1F and Suv39-like enzymes localize to pericentromeric heterochromatin [26]. While all putative HMTs of C. elegans were analyzed for function within the synMuv pathways, only the met-1 and met-2 genes genetically interact with synMuvA factors [28]. While MET-1 appears to methylate H3K36, met-2 encodes a SETDB1 homologous H3K9MT. Besides function in vulva cell fate determination, recent work has shown that MET-2 is required for all germline H3K9me2 [29].
A number of proteins have been identified that specifically interact with histone methyl-lysine residues. These include chromodomain-, PHD finger-, tudor domain- and ankyrin domain-containing chromatin factors. In vitro, proteins containing these domains show significant site specificity in binding to distinct histone methyl-lysine marks. Also, clear preference for either higher (me2/me3) or lower (me1/me2)methylation states are observed. For example, heterochromatin protein 1 (HP1) binds H3K9me3- and H3K9me2-containing peptides preferentially over H3K9me0 and H3K9me1 targets. Also, discrimination against other sites of lysine methylation, even when in similar sequence context such as H3K27me (both H3K9 and H3K27 reside within an “ARKS” sequence patch), is observed [30]. Interestingly, one of the two HP1 orthologs in C. elegans, HPL-2 is classified as a synMuvB factor [31]. The other ortholog, HPL-1 seems to act as an enhancer of HPL-2 in this and other developmental pathways [32]. Colocalization of binding factors with cognate histone PTMs on a global scale by immunofluorescence studies or on a local scale by chromatin immunoprecipitation (ChIP) experiments have suggested that PTM recognition might be a common targeting mechanisms of chromatin factors [33]. Also, recognition of multiple histone PTMs in the context of higher order complexes of binding factors might directly affect chromatin structure, inducing higher order chromatin folds [13], [34]. However, the exact working mechanisms of many histone modification binding proteins, their recruitment, and their downstream actions have not been fully elucidated.
MBT domains constitute a separate class of histone methyl-lysine reading modules that seem not to have high selectivity for their target sites. In vitro, diverse MBT domain regions from different proteins and organisms display specificity for mono- and dimethylated lysine residues over the unmodified and trimethylated states. However, site discrimination and specificity for selected sites is very low [1], [5], [12], [35]–[39]. These findings are complemented by structural studies that have implemented three aromatic caging residues and an aspartate moiety in methyl-lysine binding. In this “cavity insertion mode,” there is little contribution of protein surface residues in histone sequence recognition [35]–[41]. Interestingly, only individual MBT domains within multiple MBT repeat-containing elements are implicated in histone methyl-lysine binding [12], [35], [36]. In this sense, the biological role and functional implications of the other MBT repeats within the linearly arranged MBT regions are unclear.
MBT domain proteins might exert higher specificity for particular histone methyl-lysine PTMs in vivo as suggested by indirect targeting experiments [12]. While it is clear that MBT domain proteins contribute to the complex organization of chromatin as readers and effectors of histone PTMs that is critical for the establishment of specific cellular differentiation states [33], [34], [42], the exact contribution of methyl-lysine binding activity to the biology of these proteins has not been worked out.
Here, we show that C. elegans LIN-61 specifically interacts with H3K9me2/3. Sequence homology and mutagenesis studies imply the fourth MBT repeat in this interaction. However, a minimum of three C-terminal MBT repeats is required for H3K9me2/3 binding and analysis of previously identified lin-61 point mutant alleles indicates complex conformational regulation of ligand interaction of LIN-61 MBT domains. We find that in vivo H3K9me2/3 binding of LIN-61 is necessary for C. elegans vulva development within the synMuvB pathway. We also demonstrate that lin-61 genetically interacts with hpl-2, an HP1 ortholog H3K9me2/3 binding factor and met-2, an H3K9 methyl transferase in determining C. elegans vulva development and fertility.
Results
LIN-61 Specifically Binds H3K9me2/3
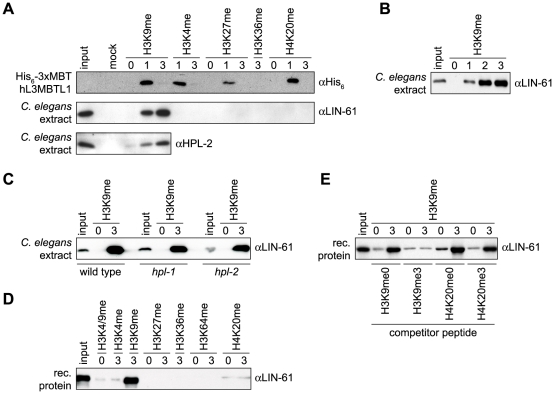
We used peptide affinity purification experiments to identify binding partners of H3K9me3 in C. elegans extracts. Besides other factors, LIN-61 and MBTR-1 were identified in MS analysis of proteins bound to H3K9me3 peptides compared to the H3K9me0 control (Figure S1A-S1C). Both proteins share the same four MBT domain overall structure and are 40% identical in sequence (Figure S1D). We raised antibodies specific for LIN-61 (Figure S2D) and confirmed this finding using a panel of lysine methylated histone peptides in affinity purification experiments. As Figure 1A shows, LIN-61 from C. elegans extracts interacted preferentially with the H3K9me3 peptide compared to the H3K9me1 peptide. No interaction with the H3K9me0 peptide was found under these conditions. Binding to H3K9me was specific as no binding to H3K4me3-, H3K27me3-, H3K36me3- or H4K20me3-containing peptides was observed. In contrast and as was previously found [12], the recombinant MBT domains of human L3MBTL1 bound to the monomethylated forms of all tested histone methyl-lysine sites with little to no sequence preference in the same assay (Figure 1A). Additional experiments showed that LIN-61 binds to H3K9me2 equally well as to H3K9me3 (Figure 1B).
Figure 1. LIN-61 specifically interacts with H3K9me2/3.
(A), (B) Affinity purification of the recombinant three MBT repeats of hL3MBTL1 or C. elegans extract using the indicated biotinylated histone tail peptides carrying different methyl-lysine marks immobilized on avidin agarose resin. (C) Affinity purification as in (A) and (B) using C. elegans extract from wild type or hpl-1 and hpl-2 mutant worms. (D) Affinity purification experiment using bacterially produced recombinant MBP-LIN-61 and the indicated immobilized peptides. (E) Affinity purification of bacterially produced recombinant MBP-LIN-61 using unmodified and H3K9me3 peptides in presence of the indicated competitor peptides (100-fold excess). Western blot analyses of the recovered material using the indicated antibodies are shown. Input, 4% (A) or 2% (B-E); mock, avidin agarose resin without peptide.
HP1 proteins in different model systems have been described as major H3K9me3 interacting chromatin factors. On the basis of immunofluorescence experiments, it was nevertheless suggested that HPL-2, one of the two C. elegans HP1 orthologous factors might not be a direct binding protein of H3K9me3 [43]. We detected HPL-2 in affinity purification experiments of C. elegans extracts using H3K9me peptides. We found slight preference for the trimethlated over the monomethylated form and discrimination against the unmodified template (Figure 1A).
To exclude indirect binding of LIN-61 to H3K9me2/3 via bridging interaction by the C. elegans HPL-1 and HPL-2 HP1 proteins, we repeated the affinity purification experiments using extracts prepared from hpl-1 and hpl-2 mutant worms. LIN-61 was recovered on H3K9me3 peptide-containing beads under these conditions comparable to the wild type situation (Figure 1C). Further, we purified recombinant MBP-LIN-61 fusion protein after expression in bacteria. In affinity purification experiments, the recombinant LIN-61 protein showed strong preference for the H3K9me3 peptide over H3K4, H3K27, H3K36, H3K64 and H4K20 trimethylated peptides and for the trimethylated H3K9 site over the monomethylated form reminiscent of the endogenous C. elegans protein (Figure 1D and Figure S1E). Also, in vitro translated LIN-61 and MBTR-1 proteins specifically bound H3K9me3 (Figure S1F and S1G). Only H3K9me3 peptide but not H3K9me0, H4K20me0 or H4K20me3 peptides was able to compete with the binding of recombinant LIN-61 to the immobilized H3K9me3 target (Figure 1E). From these experiments we conclude that the C. elegans MBT domain protein LIN-61 specifically and autonomously interacts with H3K9me. In contrast to other MBT proteins that display little to no sequence specific methyl-lysine binding and that discriminate against the tri-methylated state, LIN-61 shows preference for the di- and tri-methylated states of the H3K9 site.
LIN-61 Methyl-Lysine Binding Is Distinct from Other MBT Domain Proteins
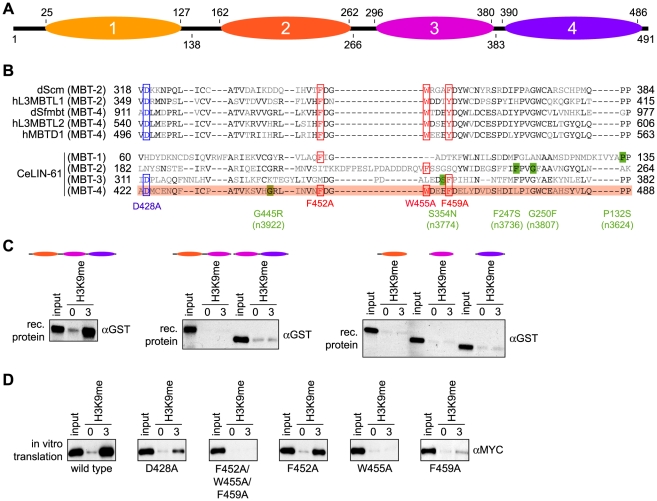
Alignment of LIN-61 with other MBT domain proteins, dScm [35], hL3MBTL1 [37], dSfmbt [5], [36], hL3MBTL2 [41] and hMBTD1 [40] indicated that only the fourth MBT repeat of LIN-61 contains all three aromatic caging residues and the aspartate moiety that have been implicated in methyl-lysine binding of other MBT repeats (Figure 2A and 2B). From the structural insights available from these MBT domain proteins, we predicted that LIN-61 MBT domain four is involved in H3K9me3 peptide interaction. To characterize the H3K9me binding of LIN-61, we first analyzed a series of mutant recombinant GST-fusion proteins where different MBT repeats were deleted (Figure 2C). In peptide affinity purification experiments we found the first MBT repeat of LIN-61 dispensable for H3K9me interaction. When additional MBT repeats were deleted, neither truncated LIN-61 protein corresponding to MBT repeats three and four nor truncated LIN-61 protein corresponding to MBT repeats two and three bound the H3K9me3 peptide. Also, none of the individual MBT repeats showed any interaction.
Figure 2. The three C-terminal MBT repeats of LIN-61 are essential for H3K9me3 interaction.
(A) Schematic representation of the LIN-61 protein indicating the amino acid position of the four (1-4) MBT repeat domains on top (according to GenBank using RPS-BLAST). Bottom, boundaries of the deletion constructs used (amino acid positions). (B) Sequence alignment of the four LIN-61 MBT core domains with MBT core domains of other MBT factors implicated in methyl-lysine binding. Amino acids identical in at least three of the sequences are in black. Residues shown to be essential for methyl-lysine interaction of dScm, hL3MBTL1 (isoform I), hL3MBTL2, dSfmbt (isoform C) and hMBTD1 are boxed in red (aromatic cage residues mediating hydrophobic and π-cation interactions) or in blue (conserved aspartate residue mediating ion pairing and hydrogen bonding to mono- and dimethylammonium moiety of lysine ε-amino group). MBT repeat four of LIN-61 containing all residues determined to be essential in methyl-lysine binding of other MBT domain proteins is highlighted in red. Amino acid positions of point mutants generated in this study (red and blue) or corresponding to lin-61 alleles identified in genetic screens (green) are indicated. (C) Affinity purification experiments of bacterially produced recombinant GST-LIN-61 proteins corresponding to the indicated MBT regions using immobilized unmodified and H3K9me3 peptides. αGST Western blot analyses of the recovered material are shown. Input, 2%. (D) Affinity purification experiments of in vitro translated wild type or point mutant MYC-LIN-61 proteins using immobilized unmodified and H3K9me3 peptides. αMYC Western blot analyses of the recovered material are shown. Input, 7.5%.
Next, we analyzed a series of LIN-61 proteins with point mutations in conserved residues in MBT domain four. In other factors, these residues were implicated in MBT repeat methyl-lysine binding. To this end, affinity purifications were carried out with in vitro translated MYC-tagged LIN-61 proteins (Figure 2D). Mutation of the highly conserved aspartate residue (LIN-61 D428A) caused reduced binding, but did not abolish interaction. This finding is in stark contrast to other MBT domain protein interaction with mono- and dimethylated histone lysine residues where the corresponding residue is absolutely required [35]–[37]. In agreement with an aromatic cage in MBT domain four being involved in H3K9me3 binding, simultaneous mutation of three conserved aromatic residues in LIN-61 MBT domain four (LIN-61 F452A/W455A/F459A) completely abolished H3K9me3 interaction. However, we found only two of these residues, W455 and F459, essential for methyl-lysine binding, whereas F452 was not necessary for interaction. We conclude that MBT domain four of LIN-61 is likely directly involved in H3K9me3 binding. The results suggest that the overall structure of the three MBT repeats two to four is essential for this interaction. In general, LIN-61 binding to methyl-lysine residues appears to be different from other MBT repeat-containing methyl-lysine binding factors.
LIN-61 H3K9me2/3 Binding Is Essential for C. elegans Vulva Development
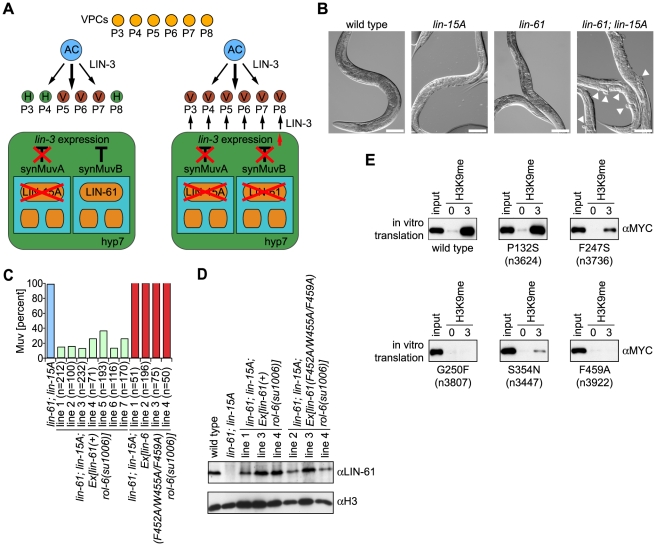
To analyze the biological role of LIN-61 H3K9me2/3 binding, we obtained a deletion mutant worm strain from the National BioResource Project. Analysis of this strain, lin-61(tm2649) indicated loss of a 672 bp fragment spanning from the middle of exon two to exon four (Figure S2A). While shortened mRNA could be detected by reverse transcription PCR analysis (Figure S2B), Western blotting using two different antibodies directed against an N- terminal region of LIN-61 or the full-length recombinant protein verified that lin-61(tm2649) is a protein null mutant allele (Figure S2D and S2E). In agreement with previous observations on lin-61 mutant C. elegans, lin-61(tm2649) did not have any apparent phenotype on the level of the organism [15]. We also did not detect profound changes on a cellular level, and the global degree of H3K9me2/3 was unchanged (data not shown). Nevertheless, congruent with the earlier studies, we detected a synMuv phenotype for lin-61 (Figure 3A and 3B). Only when individual members of two sets of genes, synMuvA and synMuvB, in the C. elegans vulva development pathway are simultaneously mutated is a multivulva (Muv) phenotype observed. Individual synMuvA or SynMuvB gene mutation does not cause a phenotype. lin-61(tm2649) in the background of lin-15A(n767), a synMuvA gene, caused extra ventral protrusions (pseudovulvae) in 78.3% of worms at 20°C and in 99.9% of worms at 24.5°C. Similarly, RNAi knockdown of the lin-15A gene product in lin-61(tm2649) C. elegans caused a Muv phenotype with a frequency of 94.0% at 24.5°C (Table 1). These finding confirm lin-61 as a synMuvB gene.
Figure 3. LIN-61–H3K9me3 binding is essential for C. elegans vulva development within the synMuvB pathway.
(A) Schematic representation of C. elegans vulva development pathways. The anchor cell (AC) secretes the EGF factor LIN-3, thereby inducing vulval cell fate determination in three out of six vulva precursor cells (VPC). Normally, LIN-3 expression and secretion in the hypodermis (hyp7) is repressed by the parallel synMuvA and synMuvB pathways. When components of each class of factors, synMuvA (e.g. lin-15A) and synMuvB (e.g. lin-61) are mutated, spurious LIN-3 signal results in induction of additional VPCs causing pseudovulvae formation (adapted from ref. [49]). (B) Representative images (DIC optics) of worms of the indicated genotypes. Arrowheads point to pseudovulvae. Scale bar represents 100 µm. (C) lin-61; lin-15A double mutant worms were injected with a genomic fragment of the lin-61 gene or the lin-61 gene encoding for a F452A/W455A/F459A triple mutant protein. rol-6(su1006) served as marker for transgenic worms. Animals with a minimum of one ectopic ventral protrusion (pseudovulva) were scored Muv. For each independently established worm line, the indicated number (n) of worms was analyzed. (D) Western blot analysis of worm extracts of the indicated transgenic worm lines using the indicated antibodies. (E) Affinity purification experiments of in vitro translated wild type or point mutant MYC-LIN-61 proteins using immobilized unmodified and H3K9me3 peptides. αMYC Western blot analyses of the recovered material are shown. Input, 7.5%.
Table 1. Genetic interaction of lin-61 with other factors in causing Muv phenotype.
| 20°C | 24.5°C | |||
| Genotype | % Muv ± SD | (n) | % Muv ± SD | (n) |
| lin-15A(RNAi) | ND | 0±0 | (>750) | |
| lin-15A(n767) | 0±0 | (>100) | 0±0 | (976) |
| lin-61(tm2649) | 0.2±0.1 | (914) | 0.3±0.2 | (1197)* |
| mbtr-1(n4775) | 0±0 | (>100) | 0±0 | (>200) |
| met-2(n4256) | 0±0 | (695) | 0.4±0.2 | (875) |
| hpl-1(tm1624) | 0±0 | (>100) | 0±0 | (>100) |
| hpl-2(tm1489) | 0±0 | (1150) | 2.2±0.7 | (1068) |
| mbtr-1(n4775); lin-15A(RNAi) | ND | 0±0 | (1040) | |
| mbtr-1(n4775) lin-61(tm2649) | 0±0 | (>200) | 0±0 | (>200) |
| mbtr-1(n4775); hpl-2(tm1489) | 0±0 | (738) | 1.4±0.8 | (819) |
| lin-61(tm2649); lin-15A(RNAi) | ND | 94.0±2.1 | (783) | |
| lin-61(tm2649); lin-15A(n767) | 78.3±14 | (825) | 99.9±0.2 | (833) |
| lin-61(tm2649); met-2(n4256) | 0.8±0.7 | (482) | 12.9±3.1 | (507) |
| lin-61(tm2649); hpl-1(tm1624) | 0±0 | (>100) | 0±0 | (179) |
| lin-61(tm2649); hpl-2(tm1489) | 0.8±0.5 | (1551) | 84.4±4 | (1416) |
Three trials of independent growth per genotype were conducted. Animals were scored as Muv if one or more ectopic ventral protrusions (pseudovulva) were observed. SD, standard deviation; n, total number of worms analyzed; asterisk marks strain that was raised on control RNAi feeding plates using Ht115(DE3) bacteria containing empty L4440 plasmid. ND, not determined.
To analyze whether H3K9me3 binding is essential for LIN-61 function within the synMuv pathway, we injected a genomic lin-61 DNA fragment into lin-61(tm2649); lin-15A(n767) C. elegans (Figure 3C). In seven independent C. elegans lines, we observed significant rescue of the Muv phenotype where the individual lines displayed a residual frequency of worms containing pseudovulvaes from 12.1% to 35.8%. In contrast, injection of the same genomic DNA fragment carrying the F452A/W455A/F459A triple mutation that abolished LIN-61–H3K9me3 interaction did not rescue the Muv phenotype at all. In four independent C. elegans lines, we observed Muv phenotypes with a frequency close to 100%. Importantly, Western blot analysis of transgene expression in several of the C. elegans lines verified that the exogenous lin-61 wild type and mutant gene copies were expressed to levels comparable to those in wild type worms (Figure 3D).
A number of lin-61 point mutant alleles with synMuv phenotypes have been described [15]. While some of these mutants show significantly reduced expression levels (n3807, n3922) others have diminished (n3624, n3736) or wild type LIN-61 content (n3447) [15]. We expressed the corresponding mutant proteins in an in vitro translation system and tested their interaction with H3K9me3 peptides in affinity purification experiments (for a map of the mutants see Figure 2B). As Figure 3E shows, the G445R (within MBT domain four) and G250F (within MBT domain two) mutant proteins encoded by the lin-61(n3922) and lin-61(n3807) alleles did not bind to H3K9me3. The P132S mutant protein (within MBT domain one) encoded by the lin-61(n3624) allele, in contrast, showed H3K9me3 interaction similar to the wild type factor. The F247S mutant protein (within MBT domain two) encoded by the lin-61(n3736) allele showed somewhat reduced binding. However, the S354N mutant protein (within MBT domain three) encoded by the lin-61(n3447) allele showed significantly reduced H3K9me3 interaction. Considering the expression levels of the mutant proteins, we conclude from these experiments that H3K9me2/3 binding is essential for LIN-61 function in the synMuv pathway and that amino acid residues outside of MBT repeat four are essential for interaction of LIN-61 with H3K9me2/3. Also, additional protein interactions that map outside the MBT domains two to four, which are required for H3K9me2/3 binding, are likely involved in LIN-61 synMuv function.
LIN-61 Acts—at Least Partially—in Parallel to HPL-2 and MET-2 in Determining C. elegans Vulva Cell Fate and Fertility
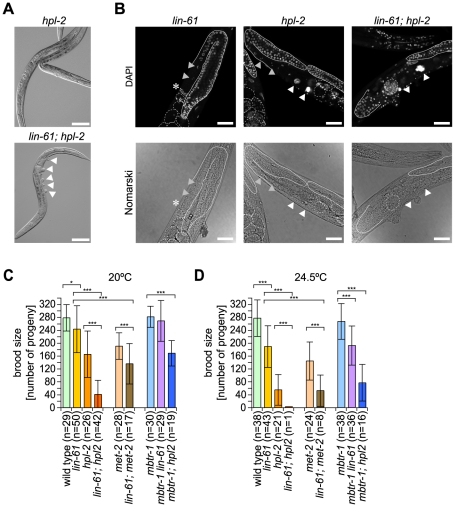
Only one of the two HP1 orthologous factors in C. elegans, hpl-2 but not hpl-1 is a synMuvB gene [31]. Since we detected both LIN-61 and HPL-2 interacting with H3K9me2/3, we asked whether these two factors act synergistically in C. elegans vulva cell fate determination. It has been suggested that such genetic enhancement studies can be used to dissect the relationship of genes within biological pathways [44]. In agreement with earlier findings, hpl-2 mutant worms did not show a phenotype at 20°C, but displayed low frequency of Muv phenotype at elevated temperature (24.5°C) (Table 1) [32], [43]. Interestingly, lin-61;hpl-2 double mutant worms showed a very high frequency of Muv phenotype under these conditions in the absence of a synMuvA mutation, indicating strong genetic interaction between these factors within this process (Figure 4A and Table 1). Importantly, mbtr-1, which itself is not a synMuvB gene [15], but whose protein product binds H3K9me2/3, did not act synergistically either with hpl-2 or lin-61 in these experiments (see also Figure S2F). Also, hpl-1, which enhances the mild Muv phenotype of hpl-2 observed at elevated temperature [32], did not synergize with lin-61. We infer that LIN-61 and HPL-2 might regulate the same target genes in the synMuv pathway, possibly via binding the same H3K9me2/3 regions of the genome.
Figure 4. lin-61 and hpl-2 act synergistically in C. elegans vulva development and fertility control.
(A) Representative images (DIC optics) of worms of the indicated mutant genotypes. Arrowheads point to pseudovulvae. Scale bar represents 100 µm. (B) Representative images of C. elegans of lin-61, hpl-2 and lin-61; hpl-2 mutant genotype raised at 24.5°C. DNA was stained with DAPI, Nomarski images were taken with DIC optics. The shape of gonad arms is outlined (solid lines). Dashed lines outline the embryos. White arrowheads mark nuclei of endomitotic cells implying failure in oocyte maturation and/or fertilization. Grey arrowheads mark oocytes. Asterisk marks spermatheca. Scale bar represents 50 µm. Mean brood size of non-sterile C. elegans of the indicated genotype at 20°C (C) and 24.5°C (D). Error bars reflect standard deviation. n, number of worms analyzed. One asterisk marks a significance interval of p<0.02 and three asterisks mark a significance interval of p<0.001 determined using a two-tailed Student's t-test.
MET-2 is the H3K9MT within the synMuvB pathway. If a linear pathway from MET-2, which establishes H3K9me2/3 modification on target regions of the genome involved in determining C. elegans vulva cell fate, to LIN-61, which binds and translates this chromatin mark, exists, then the corresponding genes should show no interaction. Similar to earlier studies [28], we did not detect worms with pseudovulvaes with considerable frequency for met-2 even at elevated temperature. In context of lin-61;met-2 we found no significant Muv phenotype at 20°C. However, at 24.5°C we found 12.9% of double mutant worms containing peseudovulvaes indicating a weak but robust genetic interaction of lin-61 and met-2. The results indicate that met-2 is not only upstream of lin-61, but also acts to some degree parallel to this gene in determining C. elegans vulva cell fate.
LIN-61 is globally distributed and shows chromatin association in adult worms and focal distribution in embryos [15]. Therefore, the synMuv pathway represents only part of LIN-61 biology. In addition, several synMuvB genes have been found to interact with hpl-2 [28], [31], [43]. hpl-1 also enhances the frequency of C. elegans with pseudovulvaes. As hpl-2 might therefore be a special synMuvB gene displaying pleiotropic effects [28], we wanted to test whether lin-61 and hpl-2 interact beyond the synMuv pathway. As both, HPL-2 and MET-2 have been implied in germ line development [29], [31], [32], we investigated whether LIN-61 also synergizes with these factors in determining C. elegans fertility and brood size.
In agreement with earlier findings, hpl-2 mutation caused some sterility of worms (13%) that was enhanced at higher temperature (25% at 24.5°C, Table 2) [31], [32]. For lin-61 C. elegans, we detected only very low sterility at elevated temperature (2% at 24.5°C compared to 0% at 20°C). However, lin61;hpl-2 double mutant worms showed 22% sterility at 20°C and 98% sterility at 24.5°C, indicating strong interaction. These effects were not caused by general loss of germ cells but were due to enhancement of the defects observed for hpl-2 worms [31]. While 1% of hpl-2 worms had no oocytes and 2% displayed failures in gonad elongation, 32% showed defects in oocyte maturation or fertilization (n = 88). In contrast, 35% of lin61;hpl-2 double mutant worms had no oocytes, 25% displayed failures in gonad elongation and 22% showed defects in oocyte maturation or fertilization at 24.5°C (n = 113, Figure 4B). Importantly, no synergism between mbtr-1 and lin-61 or hpl-2 in causing C. elegans sterility was observed. Similar to the results of the Muv analysis, we also detected some – albeit weaker – synergism between lin-61 and met-2, which itself brought about only a mild phenotype under the conditions tested, in causing sterility of C. elegans, (Table 2) [29].
Table 2. Fertility analysis of lin-61 in conjunction with other factors.
| 20°C | 24.5°C | |||
| Genotype | % Sterile | (n) | % Sterile | (n) |
| lin-61(tm2649) | 0 | (50) | 2 | (44) |
| mbtr-1(n4775) | 0 | (30) | 0 | (38) |
| met-2(n4256) | 7 | (30) | 8 | (26) |
| hpl-2(tm1489) | 13 | (30) | 25 | (28) |
| mbtr-1(n4775) lin-61(tm2649) | 0 | (29) | 0 | (36) |
| lin-61(tm2649); met-2(n4256) | 0 | (17) | 47 | (15) |
| lin-61(tm2649); hpl-2(tm1489) | 22 | (54) | 98 | (60) |
| mbtr-1(n4775); hpl-2(tm1489) | 5 | (20) | 16 | (19) |
Sterility of different worm strain was assayed as described in Materials and Methods. n, total number of worms analyzed.
Observations analogous to those made when evaluating C. elegans sterility were made when analyzing the brood size of mutant worms (Figure 4C and 4D). lin-61 caused mild but significant reduction in brood size at 20°C and brought about more severe reduction at 24.5°C. At both conditions tested, a clear synergism with the brood size reduction of hpl-2 was observed. Similarly, interaction of lin-61 with met-2 was detected in this assay. We conclude that within different pathways of differentiation of somatic cells such as determining C. elegans vulva cell fate and of development of germ cells such as determining C. elegans fertility, the H3K9me2/3 binding proteins LIN-61 and HPL-2 have overlapping functionality by at least partially working in parallel pathways. Similarly, lin-61 and met-2 show some redundancy.
Discussion
Our results have two main implications. On one side, they are important for the understanding of MBT protein histone methyl-lysine binding and on the other side they provide molecular insights into the synMuv pathway in C. elegans. Why is LIN-61 (and MBTR-1) binding to H3K9me2/3 highly specific whereas MBT containing proteins of Drosophila and human origin show promiscuous interaction with methyl-lysine residues embedded in different sequence context? The specific ligands for other MBT domain proteins might not yet have been identified and the analyzed histone methyl-lysine target sites might only be surrogate interaction partners. Yet, in the available structures of MBT domain–ligand complexes, there is very little specific interaction of MBT surfaces with histone sequences. Binding appears largely dependent on unspecific contact of charged interfaces [35]–[41]. Alternatively, interaction might be more specific in cellular context. To this point, binding studies investigating MBT domain behavior have been undertaken with recombinant proteins. In vivo, the MBT proteins of higher eukaryotes might need to be modified or interacting with other factors to allow for sequence specific histone methyl-lysine binding. Along this line, H4K20me1, but not H3K9me1/2, recruits hL3MBTL1 in cellular target assays to chromatin, despite the fact that peptides containing these histone marks bind to the recombinant MBT repeats with similar affinities in vitro [12]. Lastly, the methyl-lysine binding mode of LIN-61 might be completely different from other MBT proteins. Mutagenesis of two of three conserved putatively methyl-group caging aromatic residues abolishes LIN-61–H3K9me3 interaction. However, mutagenesis of the third aromatic residue, F452 does not have an effect. Further, mutation of D428, a highly conserved, charged residue implicated in essential salt bridging and hydrogen bonding of the mono- and dimethylammonium moieties of the methylated lysines in other MBT domain–ligand complexes did not abolish interaction [35]–[37]. A different binding mode is clearly inferred from the fact that LIN-61 prefers the di-and trimethylated states of its target H3K9me residue to the unmodified and mono-methylated forms, whereas other MBT-proteins discriminate against the fully methylated states.
Structural studies of Drosophila and human factors have revealed interdigitation of the N-terminal arms of predicted MBT repeats with β-barrel folds of adjacent MBT domains [35]–[41], [45]. While the exact functional consequence of this overall folding principle of MBT proteins is unclear, expression of the individual MBT repeats of hSCML2 was not possible [46]. We could investigate the individual MBT repeats of LIN-61, but found them incapable of mediating H3K9me2/3 interaction. We also note that proteins containing only two MBT repeats of the SCM type display about 50- to 100-fold weaker binding to methyl-lysine residues compared to three or four MBT repeat containing factors [5], [12], [35]–[39]. Interestingly, four MBT domains of dSfmbt, hL3MBTL2 and hMBTD1 fold into three propeller blade like structures similar to three MBT domain proteins (hL3MBTL1 [37], [38], [45] with the fourth MBT domain stacked atop the planar arrangement [36], [40], [41]. Our results on LIN-61 indeed indicate that the first MBT repeat is dispensable for H3K9me2/3 binding.
What is it about three MBT domain structures that mediate stable interactions with methyl-lysine ligands? In the ring form with each domain making contact to two adjacent domains, the individual MBT repeats and especially the single ligand binding domains might be structurally stabilized. Indeed, almost all mutations that cause the malignant phenotype in Drosophila l(3)mbt or the polycomb phenotype of Scm map to the MBT repeats but not necessarily to the domain that has been implicated in methyl-lysine interaction [4], [47]. While the corresponding mutant proteins have not yet been explicitly tested in in vitro binding experiments, we show that several single amino acid exchanges in LIN-61 that map outside of MBT repeat four interfere with H3K9me2/3 interaction. Some of these mutations, G445R (n3922) and G250F (n3807) obviously affect overall protein stability as they are detected only at very low levels in mutant C. elegans [15]. However, other mutant LIN-61 proteins, which are expressed to significant (F247S (n3736)) or even wild type (S354N (n3447)) levels, nevertheless interfere with H3K9me2/3 interaction. While these findings might indicate a composite LIN-61 binding surface with elements of different MBT repeats, they could also point to conformational linkage within the three MBT repeat structures [1]. Slight differences of MBT domain architecture and especially the methyl-lysine binding pockets crystallized under different conditions or in the absence and presence of ligand have been observed [39], [45]. It remains open whether additional ligands regulate the cross-talk of MBT domains or whether other mechanisms of MBT domain interaction and regulation exist. Structural insights of LIN-61–H3K9me2/3 binding and of mutant LIN-61 proteins are required to better understand not only LIN-61 but also general MBT domain function.
Besides the possible implications for the biochemistry of MBT domain-containing proteins, our studies on LIN-61 also involve and further emphasize a significant role for H3K9me in the synMuv pathway of C. elegans vulva cell fate determination. They also stress a role of H3K9me2/3 in germline development. We confirm here that the HP1 homolog synMuvB factor HPL-2 recognizes H3K9 methylation and introduce LIN-61 as novel H3K9me2/3 reader. The high degree of synergy between lin-61 and hpl-2 in vulva cell fate determination and germline development might indicate that these are the only reader proteins in these pathways acting in parallel to translate the effects of H3K9me2/3. While both factors might act redundantly, binding H3K9me2/3 independently and targeting similar downstream machinery to the same regions of the genome, we also consider a direct interplay between these factors. LIN-61 and HPL-2 might be part of the same multiprotein complex(es). These associations might be transient in the nucleoplasm, as no stable interaction of LIN-61 and HPL-2 was observed in immunoprecipitation experiments [15]. However, these might be stabilized by additional components on the target chromatin regions. While in many instances the H3K9me2/3 binding activity of either HPL-2 or LIN-61 might be sufficient, under conditions of stress (such as elevated temperature) both proteins might be necessary to stably anchor effector machinery at certain sites of the genome. It has been proposed that such multivalency binding modes could be crucial to the stable readout of patterns of histone modifications [34].
Earlier work has implied enzymatic activity of MET-2 in the synMuv pathway [28]. It has also been found that this protein in the germline of C. elegans exclusively establishes H3K9me2 [29]. We detect mild but significant genetic interaction of met-2 with lin-61 in determining C. elegans vulva cell fate and in germline development. Such interaction would not be expected in a strictly singular linear writer (H3K9MT) – reader (H3K9me2/3 binding protein) relationship. Obviously, met-2 is not essential in both pathways of worm development. Other HKMTs can substitute at least partially for MET-2 function. One candidate is met-1, which has been shown to affect global H3K9me3 to some degree and which is also a synMuvB factor [28]. Another candidate is MES-2, which in the germline mediates H3K9me3 but does not affect H3K9me2 [29]. Genetic interaction of met-2 and lin-61 might then be due to possible self-reinforcing loops, where H3K9MTs redundant to MET-2 have to be brought to and maintained at target regions via anchoring activity of LIN-61 as an H3K9me binding protein. HPL-2 might only partially be able to substitute for this function, for example if it does not directly interact with the H3K9MTs. Indeed, direct interactions between HKMTs and proteins binding the corresponding histone modifications have been observed in several instances, for example in the case of Suv-39/HP1 [48] and PR-SET7/L3MBTL1 [12].
What are the targets of the MET-2/LIN-61/HPL-2/H3K9me2/3 system? Penetrance of the Muv phenotype has directly been linked to aberrant, increased expression levels of lin-3 EGF in the hyp7 syncytium, which is secreted and induces abnormal differentiation of additional vulva precursor cells (VPC) (see Figure 3A) [44], [49]. Indeed, all three components, lin-61, hpl-2 and met-2, have been shown to cause elevated LIN-3 levels in a lin-15A background [28], [44]. However, the available genome wide mapping data of H3K9me3 do not show enrichment of H3K9me3 at the lin-3 locus [50]. Also, we have failed so far to detect H3K9me2/3 at this region using direct ChIP approaches. Therefore, the mechanisms by which MET-2/LIN-61/HPL-2/H3K9me2/3 regulate lin-3 might be indirect, involving intermediate factors. Since no synMuvA mutation is needed for inducing a Muv phenotype in lin-61;hpl-2 and lin-61;met-2 (this study) as well as met-2 hpl-2 [28] double mutant C. elegans, we infer nevertheless that H3K9me2/3 is central in vulva cell fate determination. The results also support a more operational definition of synMuv genes using genetic enhancement tests [44] compared to the original classification where synMuvB mutants are supposed to result in a Muv phenotype only in combination with mutations in the synMuvA pathway, and where factors of one class do not genetically interact [51].
Since LIN-61 is globally distributed in adult worms and embryos [15], it is clear that this factor has other functionality outside of vulva cell fate determination and germline development. A role in genomic stability has been suggested using an RNAi screening strategy [52]. It remains to be seen whether this and putative other functions of LIN-61 also depend on H3K9me2/3 binding. Since mutation of P132 (corresponding to the n3624 allele of lin-61) does not interfere with H3K9me2/3 interaction and as the n3447 allele is not a very strong synMuv mutant [15], additional biochemical functions of LIN-61 clearly await discovery.
Materials and Methods
DNA Constructs
Cosmid R06C7 was obtained from the Sanger Institute (UK). Plasmids R06C7.7 pL4440-Dest-containing cDNA corresponding to LIN-61 and ZK678.1 pL4440-Dest-containing cDNA corresponding to LIN-15A were obtained from OpenBiosystems. l3mbtl1pCMV-SPORT6 plasmid containing the cDNA of human L3MBTL1 (GenBank BC039829.1) was obtained from Geneservice. MBTR-1 cDNA containing the full ORF was amplified from total C. elegans mRNA after reverse transcription. cDNAs were subcloned into the following vectors using standard PCR and cloning procedures: pETM-40 (gift of G. Stier, EMBL Heidelberg) for generating MBP-tagged LIN-61; pcDNA3.1 to generate MYC-tagged LIN-61 and MYC-tagged MBTR-1. hL3MBTL1 cDNA corresponding to amino acid residues 197-526 were cloned into pET16b (Merck) to generate His6-3xMBT-L3MBTL1. cDNAs corresponding to the following amino acid residues of LIN-61 were cloned into pGEX-4T-3: GST-MBT(2-4)-LIN61, aa 138-491; GST-MBT(2-3)-LIN61, aa 138-383; GST-MBT(3-4)-LIN61, aa 266-491; GST-MBT(2)-LIN61, aa 138-265, GST-MBT(3)-LIN61, aa 266-383; GST-MBT(4)-LIN61, aa 384-491. Side directed mutagenesis (LIN61 D248A; F452A/W455A/F459A; F452A; W455A; F459A; P132S; F247S; G250F; S354N; G445R) was carried out according to the QuickChange protocol (Stratagene). A 4387 bp fragment of C. elegans genomic DNA generated by StuI/SacII restriction digest of cosmid R06C7 was cloned into pBluescript SK (-) (Stratagene) and used for microinjection experiments [15]. Further details of cloning procedures are available upon request.
Peptide Affinity Purification
Experiments were performed as described with slight modification [53]. 1 ml C. elegans extract (5 mg/ml), 17 µl TNT in vitro translation reaction or 25 µg recombinant protein with 30 µl avidin agarose resin (Thermo Scientific) were used per reaction. PD150 buffer was supplemented with 0.5% (w/v) low fat dry milk as blocking reagent. Peptides carried a biotinylated lysine residue at the C-terminus for affinity purifications: H3K4me1/3: ARTK(me1/3)QTARKSTGGKAPRKQLK-biotin; H3K9me0: ARTKQTARKSTGGKAPRKQLK-biotin; H3K9me1/2/3: ARTKQTARK(me1/2/3)STGGKAPRKQLK-biotin; H3K27me0: QLATKAARKSAPATGGVKKPHK-biotin; H3K27me1/3: QLATKAARK(me1/3)SAPATGGVKKPHK-biotin; H3K36me3: SAPATGGVK(me3)KPHRYRP-biotin; H3K64me0: STELLIRKLPFQRLVREI-biotin; H3K64me3: STELLIRK(me3)LPFQRLVREI-biotin; H4K20me0: KGGAKRHRKVLRDNIQ-biotin; H4K20me1/3: KGGAKRHRK(me1/3)VLRDNIQ-biotin.
Mass Spectrometry
SDS PAGE gels were stained with Coomassie Blue and entire gel lanes were cut into 23 slices of equal size. Proteins within the slices were digested according to Shevchenko et al. [54]. Peptides were extracted and analyzed by LC-coupled tandem MS on an Orbitrap Xl mass spectrometer (Thermo Fisher Scientific). CID fragment spectra were searched against NCBInr database using MASCOT (taxonomy filter C. elegans) as search engine.
Western Blotting
For Western blot analysis, primary antibodies were used as follows: anti-LIN-61 (rabbit), 1∶4000; anti-LIN-61 (guinea pig), 1∶1000; anti-HPL2 (gift from F. Palladino, ENS de Lyon), 1∶1000; anti-His6 (Santa Cruz), 1∶1000; anti-H3 (Abcam), 1∶10000; anti-Myc (Millipore), 1∶1000.
Recombinant Proteins
MBP-, His6- and GST-fusion proteins were expressed in E. coli strain BL21-CodonPlus (DE3)-RIL (Stratagene) using auto-inducing medium. Cells were harvested and frozen in lysis buffer: MBP lysis buffer (50 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 mM Benzamidine, pH 7.4); His6 lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, 2 mM PMSF, 2 mM benzamidine, 10 mM 2-mercaptoethanol, pH 8.0); GST lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 2 mM PMSF, 2 mM benzamidine, pH 7.4). After thawing, bacterial suspensions were passed multiple times through an EmulsiFlex-C5 homogenizer (Avestin). After clarification of extracts (40,000×g, 15 min, 4°C), protein purification was carried out on amylose (MBP-tagged proteins, NEB), Ni-NTA (His6-tagged proteins, Qiagen) or GSH resins (GST-tagged proteins, GE Healthcare), according to the manufacturers' protocols.
General C. elegans Work
C. elegans was cultivated on NGM plates seeded with E. coli OP50-1 or in liquid S medium containing E. coli HB101 as described [55]–[57]. N2 was the wild type strain. C. elegans strains with the following genotypes were used: LGI: mbtr-1(n4775) [15], lin-61(tm2649) (National BioResource project); LGIII: met-2(n4256) [28], hpl-2(tm1489) [43]; LGX: hpl-1(tm1624) [32], lin-15A(n767) [58]. Single worm PCR was carried out to analyze genotypes (details of PCR reaction conditions and primers are available upon request).
Microinjection of C. elegans was performed as described with some modifications [59]. pRF4 plasmid expressing the rol-6(su1006) allele was used as injection marker at a ratio of 4∶1 compared to linearized (SacII – PspOMI restriction digest of the pBluescript R06C7 plasmid) genomic lin-61 DNA fragment. F1 progeny displaying a roller phenotype were singled. Plates were incubated for at least one week until offspring were starved. Individuals with a roller phenotype were transferred to new plates and progeny was analyzed.
RNAi feeding experiments were performed as described [60].
C. elegans Extracts
C. elegans protein extract was prepared according to Cheeseman et al. with some modifications [61]. ∼ 5 g of frozen C. elegans were grounded using a pre-chilled mortar and pestle in liquid N2. The grounded worm pellet was supplemented with an equal volume of 2x extraction buffer (50 mM HEPES-KOH, 2 mM EGTA, 2 mM MgCl2, 200 mM KCl, 20% glycerol, 0.1% NP-40, 2x EDTA-free Complete Protease Inhibitor Cocktail (Roche), pH 7.4) while grounding and then thawed on ice. Grounded worms were sonicated (Branson) and extract was clarified by centrifugation (50,000×g, 60 min, 4°C). Protein concentrations were determined using Coomassie Plus (Thermo Scientific).
For Western blot analysis, a minimum of 50 worms were transferred to an Eppendorf tube and washed with H2O. An equal amount (w/v) of nematode solubilization buffer (0.3% (v/v) ethanolamine, 2 mM EDTA, 1 mM PMSF, 5 mM DTT) was added. The sample was sonicated for 20 min in a Bioruptor using 30 s intervals (Diagenode). An equal volume of 2x protein sample buffer was added. After boiling for 5 min, the extract was clarified by centrifugation (16,000×g, 10 min, 4°C).
Fertility and Brood Size Assay
Individual hermaphrodites at larvae stage (L3 – L4) were picked to NGM plates and transferred to fresh NGM plates at successive days. Progeny were counted at larval stage. The mean brood size and standard deviation of a particular strain was calculated and corresponds to the average brood size of all assayed hermaphrodites that were not sterile. Percentage of sterile hermaphrodites (without offspring) of particular strains was determined in parallel.
Supporting Information
Identification of LIN-61 and MBTR-1 as H3K9me3 binding proteins. (A) Affinity purification of C. elegans extract using immobilized unmodified and H3K9me3 peptides. SDS PAGE gel of recovered material stained with Coomassie Blue is shown. Molecular weight markers are indicated on the left. Protein sequences of LIN-61 (B) and MBTR-1 (C). Peptides identified by MS/MS analysis from the affinity purification reactions in (A) are highlighted in red. (D) Protein sequence alignment of LIN-61 and MBTR-1. Identical amino acids are highlighted in yellow. Colored bars indicate MBT repeats of LIN-61. (E) Affinity purification of bacterially expressed recombinant LIN-61 using immobilized unmodified, H3K9me1 and H3K9me3 peptides. Western blot analysis of the recovered material is shown. Input, 2%. Affinity purification experiments of in vitro translated MYC-tagged LIN-61 (F) and MBTR-1 (G) using immobilized unmodified and H3K9me3 peptides. Western blot analyses of the recovered material are shown. Input, 2%.
(EPS)
Characterization of the lin-61(tm2649) C. elegans strain. (A) Schematic representation of the exon-intron structure of the lin-61 gene. The bar indicates the region deleted in the lin-61(tm2649) mutant. (B) PCR analysis of cDNA corresponding to mRNA isolated from wild type and lin-61(tm2649) mutant worms. Agarose gel stained with ethidium bromide is shown. The length of DNA species amplified with lin-61 specific primers is indicated on the left. (C) Protein sequence of a hypothetical LIN-61 protein originating from the tm2649 allele. The boxed sequence corresponds to the peptide used for generating the αLIN-61 (guinea pig) antibodies. The underlined sequence indicates parts of MBT repeat one. The sequence after the dashes corresponds to non-natural LIN-61 originating from the frame shift introduced by the tm2649 mutation. Western blot analyses of C. elegans extract from wild type and lin-61(tm2649) mutant worms using antisera generated in rabbit (D) against the denatured full length protein or generated in guinea pig (E) against the peptide sequence indicated in (C). Western blot against histone H3 served as loading control. (F) Representative images (DIC optics) of worms of the indicated mutant genotypes. Scale bar represents 100 µm.
(EPS)
Acknowledgments
Monika Jedrusik-Bode made the initial observation of LIN-61–H3K9me2/3 interaction and provided valuable comments on the progress of this work as well as on the content of the manuscript. We thank the Proteomics group at the Max Planck Institute for Biophysical Chemistry of Henning Urlaub for excellent mass spectrometry analysis. We are grateful to Francesca Palladino and Robert Horvitz for C. elegans strains and reagents. The National BioResource Project made the lin-61(tm2649) C. elegans strain. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. We thank Christoph Biesemann and Alexandra Stützer for help with cloning expression constructs and Martina Wirth and Henriette Franz for establishing C. elegans and peptide affinity purification procedures. We further acknowledge Szabolcs Sörös and Adrian Schomburg for help with protein purification, as well as Kerstin Mosch for valuable comments on the progress of this study.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the Max Planck Society and the European Union (FP6, NoE “The Epigenome”). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bonasio R, Lecona E, Reinberg D. MBT domain proteins in development and disease. Semin Cell Dev Biol. 2010;21:221–230. doi: 10.1016/j.semcdb.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wismar J, Loffler T, Habtemichael N, Vef O, Geissen M, et al. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech Dev. 1995;53:141–154. doi: 10.1016/0925-4773(95)00431-9. [DOI] [PubMed] [Google Scholar]
- 3.Gateff E, Loffler T, Wismar J. A temperature-sensitive brain tumor suppressor mutation of Drosophila melanogaster: developmental studies and molecular localization of the gene. Mech Dev. 1993;41:15–31. doi: 10.1016/0925-4773(93)90052-y. [DOI] [PubMed] [Google Scholar]
- 4.Bornemann D, Miller E, Simon J. Expression and properties of wild-type and mutant forms of the Drosophila sex comb on midleg (SCM) repressor protein. Genetics. 1998;150:675–686. doi: 10.1093/genetics/150.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bornemann D, Miller E, Simon J. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development. 1996;122:1621–1630. doi: 10.1242/dev.122.5.1621. [DOI] [PubMed] [Google Scholar]
- 7.Breen TR, Duncan IM. Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev Biol. 1986;118:442–456. doi: 10.1016/0012-1606(86)90015-1. [DOI] [PubMed] [Google Scholar]
- 8.Takada Y, Isono K, Shinga J, Turner JM, Kitamura H, et al. Mammalian Polycomb Scmh1 mediates exclusion of Polycomb complexes from the XY body in the pachytene spermatocytes. Development. 2007;134:579–590. doi: 10.1242/dev.02747. [DOI] [PubMed] [Google Scholar]
- 9.Arai S, Miyazaki T. Impaired maturation of myeloid progenitors in mice lacking novel Polycomb group protein MBT-1. EMBO J. 2005;24:1863–1873. doi: 10.1038/sj.emboj.7600654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, Van Buren D, Huang HS, Zhong L, Mostoslavsky R, et al. Chromatin protein L3mbtl1 is dispensable for development and tumor suppression in mice. J Biol Chem. 2010;285:27767–27775. doi: 10.1074/jbc.M110.115410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27:4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 14.Boccuni P, MacGrogan D, Scandura JM, Nimer SD. The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6). J Biol Chem. 2003;278:15412–15420. doi: 10.1074/jbc.M300592200. [DOI] [PubMed] [Google Scholar]
- 15.Harrison MM, Lu X, Horvitz HR. LIN-61, one of two Caenorhabditis elegans malignant-brain-tumor-repeat-containing proteins, acts with the DRM and NuRD-like protein complexes in vulval development but not in certain other biological processes. Genetics. 2007;176:255–271. doi: 10.1534/genetics.106.069633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 2005;24:2613–2623. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fay DS, Yochem J. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev Biol. 2007;306:1–9. doi: 10.1016/j.ydbio.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 19.Hublitz P, Albert M, Peters AH. Mechanisms of transcriptional repression by histone lysine methylation. Int J Dev Biol. 2009;53:335–354. doi: 10.1387/ijdb.082717ph. [DOI] [PubMed] [Google Scholar]
- 20.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 21.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 23.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, An W, Cao R, Xia L, Erdjument-Bromage H, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Falandry C, Fourel G, Galy V, Ristriani T, Horard B, et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J Biol Chem. 2010;285:20234–20241. doi: 10.1074/jbc.M109.052399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development. 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- 29.Bessler JB, Andersen EC, Villeneuve AM. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 2010;6:e1000830. doi: 10.1371/journal.pgen.1000830. doi: 10.1371/journal.pgen.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couteau F, Guerry F, Muller F, Palladino F. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 2002;3:235–241. doi: 10.1093/embo-reports/kvf051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schott S, Coustham V, Simonet T, Bedet C, Palladino F. Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev Biol. 2006;298:176–187. doi: 10.1016/j.ydbio.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 33.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimm C, de Ayala Alonso AG, Rybin V, Steuerwald U, Ly-Hartig N, et al. Structural and functional analyses of methyl-lysine binding by the malignant brain tumour repeat protein Sex comb on midleg. EMBO Rep. 2007;8:1031–1037. doi: 10.1038/sj.embor.7401085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, et al. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 2009;28:1965–1977. doi: 10.1038/emboj.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Fischle W, Wang W, Duncan EM, Liang L, et al. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, et al. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14:1229–1230. doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- 39.Santiveri CM, Lechtenberg BC, Allen MD, Sathyamurthy A, Jaulent AM, et al. The malignant brain tumor repeats of human SCML2 bind to peptides containing monomethylated lysine. J Mol Biol. 2008;382:1107–1112. doi: 10.1016/j.jmb.2008.07.081. [DOI] [PubMed] [Google Scholar]
- 40.Eryilmaz J, Pan P, Amaya MF, Allali-Hassani A, Dong A, et al. Structural studies of a four-MBT repeat protein MBTD1. PLoS ONE. 2009;4:e7274. doi: 10.1371/journal.pone.0007274. doi: 10.1371/journal.pone.0007274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Y, Nady N, Qi C, Allali-Hassani A, Zhu H, et al. Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2. Nucleic Acids Res. 2009;37:2204–2210. doi: 10.1093/nar/gkp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 43.Coustham V, Bedet C, Monier K, Schott S, Karali M, et al. The C. elegans HP1 homologue HPL-2 and the LIN-13 zinc finger protein form a complex implicated in vulval development. Dev Biol. 2006;297:308–322. doi: 10.1016/j.ydbio.2006.04.474. [DOI] [PubMed] [Google Scholar]
- 44.Andersen EC, Saffer AM, Horvitz HR. Multiple levels of redundant processes inhibit Caenorhabditis elegans vulval cell fates. Genetics. 2008;179:2001–2012. doi: 10.1534/genetics.108.092197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang WK, Tereshko V, Boccuni P, MacGrogan D, Nimer SD, et al. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure. 2003;11:775–789. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sathyamurthy A, Allen MD, Murzin AG, Bycroft M. Crystal structure of the malignant brain tumor (MBT) repeats in Sex Comb on Midleg-like 2 (SCML2). J Biol Chem. 2003;278:46968–46973. doi: 10.1074/jbc.M306469200. [DOI] [PubMed] [Google Scholar]
- 47.Yohn CB, Pusateri L, Barbosa V, Lehmann R. l(3)malignant brain tumor and three novel genes are required for Drosophila germ-cell formation. Genetics. 2003;165:1889–1900. doi: 10.1093/genetics/165.4.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBOJ. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui M, Chen J, Myers TR, Hwang BJ, Sternberg PW, et al. SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev Cell. 2006;10:667–672. doi: 10.1016/j.devcel.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferguson EL, Horvitz HR. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989;123:109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, et al. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franz H, Mosch K, Soeroes S, Urlaub H, Fischle W. Multimerization and H3K9me3 binding are required for CDYL1b heterochromatin association. J Biol Chem. 2009;284:35049–35059. doi: 10.1074/jbc.M109.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 55.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- 57.Stiernagle T. WormBook; 2006. Maintenance of C. elegans. pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferguson EL, Horvitz HR. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110:17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 60.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 61.Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, et al. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of LIN-61 and MBTR-1 as H3K9me3 binding proteins. (A) Affinity purification of C. elegans extract using immobilized unmodified and H3K9me3 peptides. SDS PAGE gel of recovered material stained with Coomassie Blue is shown. Molecular weight markers are indicated on the left. Protein sequences of LIN-61 (B) and MBTR-1 (C). Peptides identified by MS/MS analysis from the affinity purification reactions in (A) are highlighted in red. (D) Protein sequence alignment of LIN-61 and MBTR-1. Identical amino acids are highlighted in yellow. Colored bars indicate MBT repeats of LIN-61. (E) Affinity purification of bacterially expressed recombinant LIN-61 using immobilized unmodified, H3K9me1 and H3K9me3 peptides. Western blot analysis of the recovered material is shown. Input, 2%. Affinity purification experiments of in vitro translated MYC-tagged LIN-61 (F) and MBTR-1 (G) using immobilized unmodified and H3K9me3 peptides. Western blot analyses of the recovered material are shown. Input, 2%.
(EPS)
Characterization of the lin-61(tm2649) C. elegans strain. (A) Schematic representation of the exon-intron structure of the lin-61 gene. The bar indicates the region deleted in the lin-61(tm2649) mutant. (B) PCR analysis of cDNA corresponding to mRNA isolated from wild type and lin-61(tm2649) mutant worms. Agarose gel stained with ethidium bromide is shown. The length of DNA species amplified with lin-61 specific primers is indicated on the left. (C) Protein sequence of a hypothetical LIN-61 protein originating from the tm2649 allele. The boxed sequence corresponds to the peptide used for generating the αLIN-61 (guinea pig) antibodies. The underlined sequence indicates parts of MBT repeat one. The sequence after the dashes corresponds to non-natural LIN-61 originating from the frame shift introduced by the tm2649 mutation. Western blot analyses of C. elegans extract from wild type and lin-61(tm2649) mutant worms using antisera generated in rabbit (D) against the denatured full length protein or generated in guinea pig (E) against the peptide sequence indicated in (C). Western blot against histone H3 served as loading control. (F) Representative images (DIC optics) of worms of the indicated mutant genotypes. Scale bar represents 100 µm.
(EPS)