Abstract
The non-visual ß-arrestins are cytosolic proteins highly conserved across species that participate in a variety of signalling events, including plasma membrane receptor degradation, recycling, and signalling, and that can also act as scaffolding for kinases such as MAPK and Akt/PI3K. In Drosophila melanogaster, there is only a single non-visual ß-arrestin, encoded by kurtz, whose function is essential for neuronal activity. We have addressed the participation of Kurtz in signalling during the development of the imaginal discs, epithelial tissues requiring the activity of the Hedgehog, Wingless, EGFR, Notch, Insulin, and TGFβ pathways. Surprisingly, we found that the complete elimination of kurtz by genetic techniques has no major consequences in imaginal cells. In contrast, the over-expression of Kurtz in the wing disc causes a phenotype identical to the loss of Hedgehog signalling and prevents the expression of Hedgehog targets in the corresponding wing discs. The mechanism by which Kurtz antagonises Hedgehog signalling is to promote Smoothened internalization and degradation in a clathrin- and proteosomal-dependent manner. Intriguingly, the effects of Kurtz on Smoothened are independent of Gprk2 activity and of the activation state of the receptor. Our results suggest fundamental differences in the molecular mechanisms regulating receptor turnover and signalling in vertebrates and invertebrates, and they could provide important insights into divergent evolution of Hedgehog signalling in these organisms.
Author Summary
Non-visual β-arrestins are key proteins involved in plasma membrane receptor internalization, recycling, and signalling. The activity of β-arrestins is generally linked to seven-transmembrane receptors, but in vertebrates they can also participate in many other signalling pathways. Consistently, β-arrestins play important roles during vertebrate development and are implicated in a variety of human pathologies. Here we take advantage of the fruit fly model to analyse the genetic requirements of the unique fly non-visual β-arrestin (kurtz) in signalling during the development of imaginal epithelia. To our surprise, we find that the complete elimination of kurtz has no major consequences in imaginal cells. Our data suggest that insect epithelial cells have evolved arrestin-independent mechanisms to control receptor turnover and signalling, so arrestin function has become less critical. On the other hand, in contrast to previous reports in vertebrates, we find that the over-expression of Kurtz blocks Hedgehog signalling by promoting the internalization and degradation of the transductor Smoothened. We suggest that such differences are based on the specific requirement of the primary cilia for Hedgehog signalling in most vertebrates. These results could provide important insights into divergent modes of membrane receptor regulation and Hedgehog signalling in vertebrates and invertebrates.
Introduction
G-protein coupled receptors (GPCRs) are seven-transmembrane proteins that play critical roles during development and in the regulation of cellular physiology. GPCRs constitute the largest superfamily of cell membrane receptors [1]. The major GPCR regulatory pathway involves phosphorylation of agonist-activated receptors by G protein–coupled receptor kinases (GRKs), followed by binding of the cytosolic arrestin proteins [2]. This interaction prevents the receptor from activating additional G proteins in a process known as desensitization [3]. GRKs and ß-arrestins also participate in signal propagation by recruiting additional proteins to the receptor complex [4]–[6]. Thus, the GRK/ß-arrestin pathway facilitates receptor internalization from the cell surface through clathrin-coated pits, and this leads to numerous physiological outcomes, including receptor degradation, receptor recycling and the activation of distinct downstream signalling events [2], [7]–[10]. Finally, more recent evidence suggest a role for ß-arrestins in signalling by other families of cellular receptors, including receptor tyrosine kinase (RTKs), non-classical 7TMRs like Smoothened and Frizzled, Notch and TGFβ receptors, and also by downstream kinases such as MAPK and Akt/PI3K [5], [11]–[13].
The arrestin family is divided in two classes: the visual arrestins (arrestin 1 and 4), which are located almost exclusively in photoreceptor cells, and the non-visual β-arrestins 1 and 2 (also named arrestin 2 and 3, respectively), which are ubiquitously distributed [4]. These proteins are closely related and their sequence is highly conserved across species [14]. In Drosophila melanogaster there is only a single non-visual β-arrestin, encoded by kurtz (krz), which function is essential for development, survival and neural function [15]–[18]. In addition, the gene CG32683 encodes a related protein that presents some homology with β-arrestins, but lacks the clathrin-binding domain (see Figure S1). The GRK family includes seven members in humans (GRK1-7) and two components in flies (Gprk1 and Gprk2). Gprk1 modulates the amplitude of the visual response, acting as a Rhodopsin kinase, whereas Gprk2 regulates the level of cAMP during Drosophila oogenesis [19]. In addition, Gprk2 and Gprk1 play a key role in the regulation of the Hedgehog (Hh) signal transduction pathway [20]–[21], where they seem to phosphorylate and activate the seven-pass transmembrane protein Smoothened (Smo) [22]. The ß-arrestin Krz has also been involved in the regulation of Notch signalling, promoting the formation of a trimeric Notch-Deltex-Krz complex that mediates the degradation of the Notch receptor in an ubiquitination-dependent pathway [23], reminiscent of ß-arrestin-mediated ubiquitination of other canonical GPCRs [8]. More recently, Krz has also been implicated in the regulation of Smo accumulation [21] and ERK phosphorylation [24]. Because Krz is the unique ß-arrestin present in Drosophila, it is likely that the protein has additional functions in the modulation of other signalling pathways.
To address the participation of Krz in signalling events, we have analyzed its function during the development of the imaginal discs, the epithelial layers that give rise to the adult structures of the fly. Imaginal discs are very convenient model systems to study the activity of signalling pathways in vivo, because their development is under the regulation of the Hh, Wingless, EGFR, Notch, Insulin and TGFβ pathways [25]. In this manner, the response of these epithelia to the manipulation of Krz levels using genetic variants is a key diagnostic to identify the functional requirements of this protein in signalling during imaginal development. Surprisingly, considering the key roles identified for vertebrate non-visual arrestins, we find that the complete elimination of Krz in imaginal cells has no major consequences during imaginal development. Thus, and as claimed previously [15], krz mutant flies are morphologically normal. In contrast, the over-expression of Krz in the wing causes a phenotype identical to the loss of Hedgehog signalling. We find that excess of Krz inhibits Hh signalling by promoting Smo internalization and degradation in a clathrin- and proteosomal- dependent manner. Contrary to that observed in vertebrates, the effects of Krz on Smo are independent of Gprk2 activity and of the activation state of the receptor. We suggest that such differences in Hh signalling are based in the strict requirement of the primary cilia, a structure that is not present in fly epidermal cells, for Hh signalling in most vertebrates [8].
Results
Expression of Krz
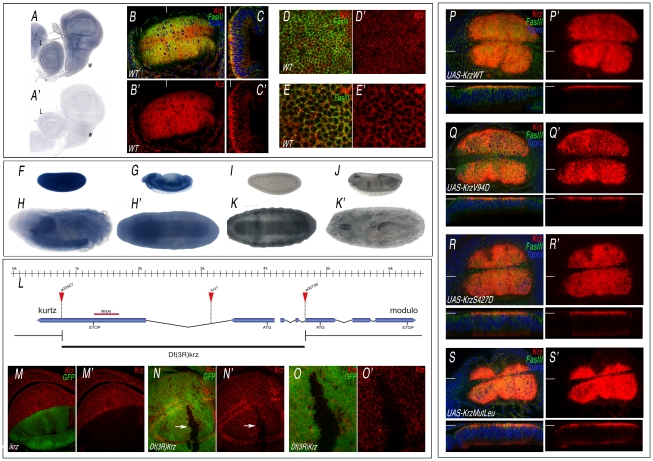
Krz is the Drosophila homologue of mammalian non-visual β-arrestins, and has all the molecular features of a canonical β-arrestin [15], [23] (Figure S1). The expression of krz occurs ubiquitously in all imaginal discs (Figure 1A and data not shown). To visualize the accumulation of the Krz protein, we generated an antibody against Krz. We found that the protein is localized in the cytoplasm of imaginal cells, being detected at higher levels close to the apical side of the epithelium (Figure 1B–1C, and 1D–1E, red). krz mRNA is also expressed in early blastoderms and during embryogenesis, mostly in the central nervous system and gut during stages 12–17, as assessed both by mRNA (Figure 1F–1H) and Krz protein (Figure 1I–1K) expression. The specificity of the antibody was confirmed by analysing the expression of Krz in loss-of-function conditions. Thus, Krz staining is lost in dorsal wing compartments expressing a krz interference RNA construct (Figure 1M–1M′), and is also absent in clones of cells homozygous for of a krz genetic deficiency (Figure 1L, 1N–1O′). The subcellular localization of the protein in wing discs over-expressing wild type Krz or different mutant forms (described below) is also in the cytoplasm, with higher levels at the apical side of the cells (Figure 1P–1S, red).
Figure 1. Expression of Krz mRNA and protein.
(A–A′) In situ hybridization with antisense (A) and sense (A′) krz probes in late third instar wing (w) and leg (L) discs. (B–C) Expression of Krz (red), FasIII (green) and Topro (Blue) in the wing pouch of a late third instar disc. (B) Apical focal plane of the wing pouch (B′ is the red channel showing the expression of Krz). (C) Longitudinal sections of the wing pouch at the place indicated in B by a white line. (C′) Red channel of C. (D–E) Transversal sections taken at the apical (D–D′) and medio-lateral (E–E′) level of the wing pouch. D′ and E′ show the expression of Krz (red). (F–H′) In situ hybridization with an antisense krz probe in the blastoderm (F), in stage 13 embryos (G) and in stage 17 embryos oriented laterally (H) or ventrally (H′), showing the prominent expression of krz in the CNS. (I–K′) Expression of Krz protein in the blastoderm (I), in stage 16 embryos (J) and in stage 17 embryos oriented ventrally (K) or dorsally (K′). (L) Representation of the krz gene indicating the intron-exon structure, the position of the ATG and Stop codons, the insertion sites of the e03507, e00739 and krz1 transposons and the extent of the krz deficiency (Df(3R)krz). The scale in Kb is indicated above. (M–M′) Loss of Krz expression (red) in ap-Gal4 UAS-GFP/UAS-ikrz wing discs. The expression of GFP is shown in green. (N–N′) Elimination of Krz expression (red) in clones of cells homozygous for the Df(3R)krz. The clone (arrow) is labelled by the absence of green. (O–O′) Higher magnification of the clone shown in N. (P–S) Expression of Krz (in red), FasIII (in green) and Topro (in blue) in wing imaginal discs expressing the following Krz-FLAG forms in the salEPv-Gal4 domain: UAS-krzWT (P–P′), UAS-krzV94D (Q–Q′), UAS-krzS427D (R–R′), UAS-krzLeu (S–S′). P′–S′ correspond to the red channel of P–S showing the expression of Krz. The planes of each transversal sections (shown below each picture) are indicated by white lines in P–S′.
Functional requirement of Krz during imaginal development
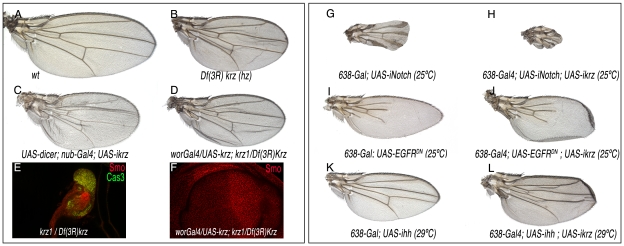
β-arrestin has widespread functions during mammalian development and cellular homeostasis [2], [8]. To identify the functional requirements of Krz during the development of the fly wing, we constructed flies with wings homozygous for a krz deficiency (Figure 1L). The elimination of Krz in the wing pouch (in 638-Gal4; FRT82 M(3)z/FRT82 Df(3R)krz; UAS-FLP/+ flies) produces a slightly folded wing of smaller than normal size but without any major defects in the pattern of veins or wing margin (Figure 2A–2B). This phenotype was also observed when the expression of krz was reduced in the entire wing blade (UAS-dicer/+; nub-Gal4/UAS-ikrz; Figure 2C). In wing discs of a similar genotype (638-Gal4/UAS-ikrz), we found a 70% reduction in krz mRNA levels (Figure S2). The very modest effects of krz elimination in the wing disc imply that the signalling pathways regulating wing patterning operate normally in the absence of Krz in imaginal cells. We also studied the consequences of krz elimination in the entire larva in Df(3R)krz homozygotes and in the Df(3R)krz/krz1 combination. These two genotypes survive until the third larval instar, where they became immobile and flaccid and form melanotic tumors, as described for krz1 homozygotes [16]. The imaginal discs of Df(3R)krz/Df(3R)krz and Df(3R)krz/krz1 larvae are very reduced in size, and express high levels of activated Caspase 3, indicating massive cell death in all imaginal tissues (Figure 2E). As expected, krz mRNA is absent in Df(3R)krz homozygous larva (Figure S2). We rescued the viability of Df(3R)krz/krz1 larvae by expressing Krz in the central nervous system in UAS-krz/wor-Gal4; Df(3R)krz/krz1 flies. The surviving flies display slightly folded wings that were very similar to those of flies where Krz levels are eliminated only in the wing (Figure 2D, compare with 2B and 2C). The corresponding wing discs have a normal appearance and the expression of signalling molecules occurs in the normal domains (Figure 2F for Smo and data not shown). These data confirm the essential function of Krz in the CNS for larval development [15]–[16] and indicate that the degeneration of imaginal tissues observed in Df(3R)krz/krz1 larvae is a consequence of the loss of krz in the CNS. Interestingly, the rescued flies, both males and females, albeit morphologically normal are sterile (data not shown), suggesting that krz is required during germ cell development.
Figure 2. Loss-of-function phenotype of krz in the wing.
(A) Wild type control wing. (B) 638-Gal4/+; UAS-FLP/+; FRT82 Df(3R)krz/FRT82 M(3)w. In this genotype all wing cells are homozygous for the Df(3R)krz, and the wings show reduced size and incorrect folding. (C) UAS-dicer/+; nub-Gal4/+; UAS-ikrz/+. In this genotype krz iRNA is expressed in the entire wing pouch, and the wing is very similar to that shown in B. (D) wor-Gal4/UAS-krz; krz1/Df(3R)krz. The expression of krz in the CNS driven by wor-Gal4 rescues the lethality of the krz1/Df(3R)krz combination, and the wings develop without krz expression. (E) Third instar wing disc of krz1/Df(3R)krz genotype, showing a reduced size and the expression of activated-Cas3 (green) throughout the disc. (F) Third instar wing disc of wor-Gal4/UAS-krz; krz1/Df(3R)krz genotype, showing a normal expression of Smo (red) and the rescue of wing disc size. (G–H) Genetic interaction between Notch and Krz. The reduction in Notch expression (638-Gal4/+; UAS-iNotch/+; G) causes the thickening of the wing veins and the elimination of the wing margin (G). This phenotype is augmented when the expression of krz is also reduced (638-Gal4/+; UAS-iNotch/UAS-ikrz; H). (I–J) Genetic interaction between EGFR and Krz. The reduction in EGFR activity (638-Gal4/+; UAS-EGFRDN/+; I) causes the loss of veins and a reduction in wing size (I). This phenotype is not modified when the expression of krz is also reduced (638-Gal4/+; UAS-EGFRDN/UAS-ikrz; H). (K–L) Genetic interaction between Hh and Krz. The reduction in Hh expression (638-Gal4/+; UAS-ihh/+; I) causes a reduction in wing size and in the distance between the veins L3 and L4 (K). This phenotype is not modified when the expression of krz is also reduced (638-Gal4/+; UAS-ihh/UAS-ikrz; L).
Krz requirements for EGFR, Notch, and Smoothened signalling
The function of β-arrestin 2 [26]–[28] and Krz [23]–[24] has been related to the regulation of EGFR, Notch or Smo signalling in different experimental systems (reviewed in [8]). Therefore, and despite of the lack of a krz mutant phenotype suggestive of an alteration in any of these signalling pathways, we searched for genetic interactions between loss of krz and genetic variants affecting the efficiency of signalling by the Notch, EGFR or Hh pathways. We used the 638-Gal4/UAS-ikrz genotype as a background condition in which the levels of Notch, EGFR or Hh components were reduced. In these combinations, we only observed a genetic interaction between Notch and Krz, as the reduction of krz increases the phenotype of wing margin loss and thicker veins caused by loss of Notch (Figure 2G–2H). No effects of loss of krz were detected upon a reduction of EGFR (Figure 2I–2J) or Hh signalling (Figure 2K–2L).
Since the activities of β-arrestins have been mostly linked to mechanisms of receptor turnover and activation, we next studied the expression and subcellular localization of the EGFR, Notch and Smoothened proteins in krz mutant cells, as a second approach to identify Krz roles during imaginal development. We first looked at the expression of EGFR, Notch and Smo proteins in wing imaginal discs where krz levels are reduced only in the dorsal compartment (ap-Gal4/+; UAS-ikrz/+), because in these discs we can compare the dorsal (mutant) with the ventral (control) compartments in the same disc (Figure 3A–3D, 3F–3G and 3I–3I′). Only in the case of Notch did we observe a slight relative increase in the accumulation of Notch at the apical side of dorsal cells (Figure 3A–3D). No differences were detected in the expression of EGFR or Smo in dorsal versus ventral cells in ap-Gal4/+; UAS-ikrz/+ wing discs (Figure 3F–3G and 3I–3I′, respectively). To extend these results to krz null mutant cells, we generated clones of cells homozygous for the krz deficiency (Df(3R)krz) and for the krz1 allele in the wing imaginal disc. In both cases the clones were grown at 25°C and at 29°C, to determinate whether there are temperature-dependent effects of the loss of Krz, as described for Gprk2 [21]. The expression of Smo and EGFR in krz null cells is normal, and the subcellular localization of these receptors remains as in wild type cells (Figure 3H–3H′ and 3J–3J′). In the case of Notch, we could only observe a subtle increase of Notch accumulation in the apical membrane of some krz mutant cells (Figure 3E–3E′). This increase is not observed in all cells of the same clone, and most clones (60%) displayed a normal expression of Notch (Figure 3E–3E′). We noticed that some of the clones contained cells shorter than wild type cells (Figure 3E–3F). In these cases, the maximal expression of Notch is detected at a different focal plane of the epithelium because of the shortening of the cells in the apico-basal axis (Figure 3E–3E′, Figure S3 and Figure S4). Otherwise no major changes in Notch accumulation were observed in transversal sections of the disc (Figure 3E–3E′, Figure S3 and Figure S4). In summary, the analysis of krz loss-of-function conditions uncovered a modest requirement of this gene for Notch signalling, which was only observed upon a reduction of Notch levels of expression, and a variable effect of loss of krz on Notch protein levels. These results are in contrast to the key requirements of vertebrate Krz homologs in several signalling pathways described in cell cultures and in vivo systems (reviewed in [8]).
Figure 3. Expression of Notch, EGFR, and Smo in krz loss-of-function conditions.
(A–D) Expression of Notch (in red) in ap-Gal4 UAS-GFP/+; UAS-ikrz/+. A and B show two different focal planes in the ventral (A) and dorsal (B) wing disc surfaces. Below each panel are transversal sections in the ventral (“V” in A′) and dorsal (“D” in B′) disc surfaces. The bottom panels are the corresponding red channels (Notch protein expression) in these transversal sections. The white lines indicate the position of the different sections. (C) Longitudinal section of the wing blade showing the dorsal (green) and ventral (lack of GFP) surfaces showing the expression of Notch (red) Topro (blue) and GFP (green). The individual red channel is shown below. (D) Projection of 10 optical sections (indicated by a white vertical line in B′ and white horizontal line in C) spanning the apical side of an ap-Gal4 UAS-GFP/+; UAS-ikrz/+ wing disc showing Notch (red) and GFP (green) expression. Note in all cases the modest difference between wild type cells (ventral) and ikrz-expressing cells (dorsal). (E) Expression of Notch in third instar wing discs bearing clones of cells homozygous for the krz deficiency. Below each focal plane are the transversal sections showing the expression of Notch (red) and GFP (green). (E′) corresponding red channel of E. Clones were induced in hsFLP1.22/+; FRT82 Df(3R)krz/FRT82 Ubi-GFP M(3)w larvae 48–72 h. AEL (in panels E, H and J). (F) Expression of EGFR (in red), GFP (in green) and Topro (in blue) in ap-Gal4 UAS-GFP/+; UAS-ikrz/+ third instar discs. Below are transversal sections in the ventral (V, middle panel) and dorsal (D, bottom panel) disc surfaces. (F′) Red channel of F showing only EGFR expression. (F″) Longitudinal section of the wing blade showing the expression of EGFR (red) Topro (blue) and GFP (green). The individual red channel is shown below. (G) Projection of 10 optical sections spanning the apical side of a ap-Gal4 UAS-GFP/+; UAS-ikrz/+ disc showing EGFR (red) and GFP (green) expression. (H) Expression of EGFR in third instar wing discs bearing clones of cells homozygous for the krz deficiency. Below and to the right are the corresponding transversal and longitudinal sections showing the expression of EGFR (red) and GFP (green). (H′) Corresponding red channel of I. (I–I′) Expression of Smo (red) in ap-Gal4 UAS-GFP/+; UAS-ikrz/+. Below are transversal sections in the ventral (V, middle panel) and dorsal (D, bottom panel) disc surfaces. To the right are longitudinal sections in the posterior (p) and anterior (a) compartments. The individual red channels are shown in I′. (J) Expression of Smo in third instar wing discs bearing clones of cells homozygous the krz deficiency. Below each focal plane are the transversal sections showing the expression of Smo (red) and GFP (green). (J′) corresponding red channel of J. Note that in all genotypes tested the reduction or loss of krz does not modify EGFR (G–I) or Smo (J–L) expression.
In Drosophila, besides the visual arrestins that are only expressed in the eye, there is another gene (CG32683) encoding a protein structurally related to Krz. It is unlikely that CG32683 is providing an arrestin-like function in the absence of Krz, because this protein lacks several conserved aminoacid motifs present in all members of the arrestin family (Figure S1). Furthermore, the expression of CG32683 is only observed during embryonic development (data not shown), and no transcripts are detected in the wing imaginal disc by RT-qPCR or in situ hybridization (Figure S2). As expected, the expression of interference RNA directed against CG32683, either alone or in combination with ikrz, does not cause any alteration in the wing (data not shown). Finally, the over-expression of CG32683-FLAG in the wing does not cause any mutant phenotype, although in this background (salEPv-Gal4/UAS-CG32383-FLAG) the protein is present at high levels in a pattern similar to that of Krz (Figure S2). In this manner, we conclude that CG32683 does not have any role during imaginal development, and that it cannot substitute for Krz in the absence of this gene.
Krz downregulates Smoothened signalling
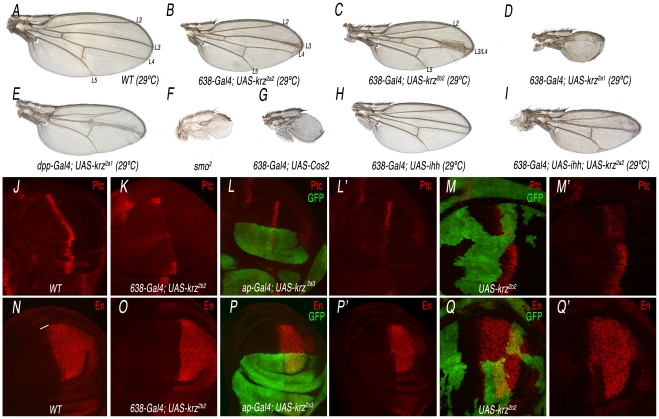
The over-expression of β-arrestin is often sufficient to promote internalization of its agonist-activated GPCRs [28], [29]. In this manner, increasing the levels of Krz might reveal other activities of the protein not uncovered by the loss-of-function approach. To this purpose, we made several constructs to express under the UAS promoter the complete krz cDNA or different modified forms of the protein. When Krz is over-expressed in the wing blade (638-Gal4/+; UAS-krz/+), we obtained a variable phenotype of reduced wing size and changes in the pattern of veins (Figure 4A–4D and Figure S5). The strength of the phenotype depended on the transgenic UAS-krz line used in these combinations, and in the most severe cases the wing was very reduced in size and all longitudinal veins failed to differentiate (Figure 4D and Figure S5). These combinations were raised at 29°C, as we did not find any major pattern defects at 25°C. This is likely a consequence of insufficient levels of ectopic expression, because when we used two copies of the UAS-krz construct at 25°C we obtained similar results than with one copy of the UAS-krz at 29°C (Figure S6B–S6D). The most obvious phenotype of gain of Krz expression is the reduction of the L3/L4 intervein territory. This phenotype is caused by Krz over-expression in the anterior compartment, because when Krz is over-expressed only in anterior cells located in the central domain of the wing blade (dpp-Gal4/+; UAS-krz/+) the fusion of the L3 and L4 veins is also observed (Figure 4E). These veins and the L3/L4 intervein correspond to the territory specified by Hh signalling [30]. In fact, the observed phenotypes are very similar to those resulting from loss of Hh signalling, detected when, for example, Smo or hh expression is reduced or when Costal2 or Patched (Ptc) are over-expressed (Figure 4F–4H and data not shown). Furthermore, wings expressing lower levels of Hh have a stronger phenotype when Krz is over-expressed (638-Gal4/+; UAS-ihh/UAS-krz; Figure 4I, compare with 4B and 4H). These results indicate a negative effect of Krz on Hh signalling when Krz levels are higher than normal.
Figure 4. Analysis of the krz gain-of-expression phenotype and its relationship with Smo signalling.
(A) Wild type control wing grown at 29°C showing the positions of the longitudinal veins L2 to L5. (B–D) Weak (B), moderate (C) and strong (D) phenotypes resulting from krz over-expression in the wing blade (638-Gal4), using three different UAS-krz lines (2a2 in B, 2b2 in C and 2a1 in D). All flies were grown at 29°C. (E) Phenotype of krz over-expression in the domain of dpp expression (dpp-Gal4/UAS-krz2a1). (F–G) Strong Hh loss-of-function phenotypes obtained in smo2 homozygous wings (F; 638-Gal4/+; FRT42 smo2/FRF42 M(2)l2; UAS-FLP/+) and in Cos2 over-expressing wings (638-Gal4/+; UAS-Cos2/+; G). (H–I) Genetic interaction between hh loss of expression and krz gain of expression. The hh loss-of-function phenotype (638-Gal4/+; UAS-ihh/+; H) is strongly increased by the over-expression of Krz (638-Gal4/+; UAS-ihh/UAS-krz; I). (J–M) Expression of the Hh-target gene ptc in different Krz over-expression conditions. (J) Control third instar wing disc showing the normal expression of Ptc (red). (K) Krz over-expression in the wing blade (638-Gal4/+; UAS-krz2b2/+) reduces Ptc expression. (L) Krz over-expression in the dorsal compartment (ap-Gal4 UAS-GFP/+; UAS-krz2a3/+) reduces Ptc expression in dorsal cells (labelled in green). L′ is the red channel of L. (M) Clones of cells over-expressing Krz (labelled in green) show a cell-autonomous reduction in Ptc expression. M′ is the red channel of M. (O–Q) Expression of the Hh-target gene en in different Krz over-expression conditions. (N) Control third instar wing disc showing the normal expression of En (red). (O) Krz over-expression in the wing blade (638-Gal4/+; UAS-krz2b2/+) eliminates En expression in anterior cells (labelled by a white line in N). (P) Krz over-expression in the dorsal compartment (ap-Gal4 UAS-GFP/+; UAS-krz2a3/+) eliminates En expression in dorsal cells (labelled in green). P′ is the red channel of L. (Q) Clones of cells over-expressing Krz (labelled in green) show a cell-autonomous elimination of En expression. M′ is the red channel of M. As expected, the expression of Ptc and En is detected in wild type cells anterior to the clones of Krz over-expressing cells (M′ and Q′).
To demonstrate that increasing the level of Krz diminishes Hh signalling, we analyzed the expression of several Hh-target genes, such as Ptc and Engrailed (En), in Krz over-expression conditions. We found that the expression of Ptc and En is strongly reduced or absent in anterior cells of wing discs over-expressing Krz in the entire wing blade (638-Gal4/+, UAS-krz/+; Figure 4J–4K and 4N–4O), the dorsal compartment (ap-Gal4/+; UAS-krz/+; Figure 4L and 4P) or in clones of cells (hsFLP1.22; act<FRT>Gal4; UAS-GFP/UAS-krz; Figure 4M and 4Q). All together, these results indicate that Krz has the potential to antagonize Hh signalling, although this antagonism is only observed upon its over-expression.
Krz affects Smoothened accumulation
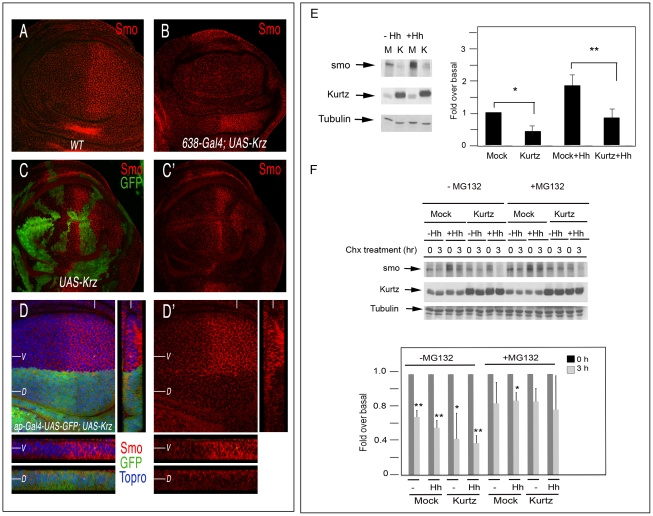
It has been described that β-arrestin 2 interacts with Smo in cell cultures and promotes Shh signalling during Zebra fish development [8], [26]–[27]. Consequently, we analyzed the possible effects of Krz in the regulation of Smo accumulation in vivo. The expression of the smo gene occurs in all wing disc cells, but the protein is only detected in the cell membrane of posterior cells and of anterior cells where Hh signalling is more active [31]–[32]. We confirmed that the expression of Smo in the membrane of posterior compartment cells and at the A/P boundary is very much reduced in wing discs over-expressing Krz in the entire wing (638-Gal4/+; UAS-krz/+; Figure 5A–5B, see also [21]). The loss of Smo is also observed in clones of cells over-expressing Krz (hsFLP1.22; act<FRT>Gal4; UAS-GFP/UAS-krz; Figure 5C–5C′) and in dorsal cells of ap-Gal4/+; UAS-krz/+ genotype (Figure 5D). The resulting levels of Smo in Krz over-expressing cells are very similar to those of the anterior compartment, suggesting that Krz promotes Smo elimination. Interestingly, loss of Smo is also observed in wing discs raised at 25°C, even though the corresponding adult wings are almost normal (Figure S6 and [21]). These results suggest that Krz mostly promotes Smo elimination, and that only above a certain level of Smo reduction, Smo signalling is compromised. In addition, the effects produced by the ectopic expression of Krz on Smo accumulation appear to be very specific, because the localization of other membrane receptors, such as EGFR and Notch, is not modified by excess of Krz. Thus, these two proteins are expressed at normal levels in dorsal and ventral cells of ap-Gal4/+; UAS-krz/+ wing discs (Figure S8).
Figure 5. Effects of krz on Smo expression.
(A–D) Expression of Smo in different genetic backgrounds in which Krz is over-expressed. (A) Wild type control third instar disc showing normal Smo expression (red). (B) Elimination of Smo expression of the wing blade of 638-Gal4/+; UAS-krz2a2/+ discs. (C–C′) Expression of Smo (red) in clones of cells over-expressing Krz (labelled in green in C). C′ is the red channel of C. (D) Expression of Smo in ap-Gal4 UAS-GFP/+; UAS-krz2a3/+. Below are transversal sections in the ventral (V, above) and dorsal (D, below) compartments and to the right a longitudinal section showing Smo expression. Note in C and D the cell-autonomous elimination of Smo by excess of Krz. (E, F) Krz modulates Smo stability and degradation by the proteasome pathway. (E) Krz over-expression decreases steady-state Smo levels. Stable Myc-Smo S2 cells were transiently transfected with either empty pUAS/actinGAL4 vector or pUAS-krz/actinGAL4 vector, and incubated in presence or absence of Hh conditioned medium, as indicated in the figure. The levels of Smo were determined in the whole cellular lysates by immunoblotting. The basal amount of Smo in the mock-transfected cells was defined as 1 and all the data were normalised by Tubulin protein levels. Data are the mean±SEM of five independent experiments. A representative gel is shown. * (P<0.05); *** (P<0.001) when compared to values of mock-transfected cells. (F) Smo turnover is inhibited by the proteosome inhibitor MG132, and the stability of Myc-Smo is increased in the presence of MG132 even when Krz is over-expressed. Smo protein levels were examined by immunoblot analysis in cells incubated with control or Hh conditioned medium upon treatment with Cycloheximide (Chx) at 0 and after two hours treatment, in the absence or presence of MG132 as indicated. The amount of Smo at 0 h was defined as 1 for each case, and data normalized by Tubulin protein levels. Data are the mean±SEM of three independent experiments. A representative gel is shown. * (P<0.05); *** (P<0.001) when compared to values at 0 h.
To further analyse the effects of Krz on Smo accumulation, we analysed the expression of Smo in S2 cells transiently transfected with Krz. To this purpose, we generated a stable cell line over-expressing myc-Smo and these cells were transfected with the pUASt-Krz and pActin-Gal4 vectors. Analysis of whole-cell lysates in Western blots revealed a decrease in Smo protein levels when Krz is over-expressed, both in the absence or presence of Hh in the medium (Figure 5E). These data are consistent with those obtained in the imaginal discs. To investigate how Krz reduces Smo levels, we performed assays in presence of the protein synthesis inhibitor cycloheximide and the proteosome-specific inhibitor MG132. The expression of Krz appears to favour Smo turnover either in the absence or presence of Hh (Figure 5E). In addition, proteosome inhibition inhibits the reduction of Smo levels, both in control (myc-Smo S2, mock transfected) and Krz over-expressing cells (myc-Smo S2, pUASt-Krz/pAct-Gal4 transfected) (Figure 5F). These results suggest that Krz enhances Smo degradation via the proteosomal pathway.
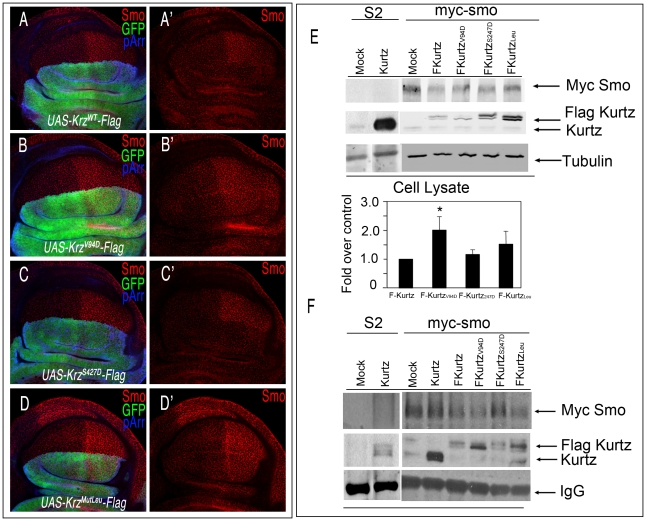
We identified in Krz several conserved aminoacids (Val94, Leu440/IsoLeu441/Leu443 and Ser427; see Figure S1) whose human counterparts are implicated in the targeting of GPCR to clathrin-coated pits without affecting receptor signalling (Val94) [33]–[34], in the binding of β-arrestin to clathrin (Leu440/IsoLeu441/Leu443) [34]–[35], or that reduce its ability to promote internalization of the β2-adrenergic receptor (Ser427) [36]–[37]. To explore whether the function of these residues is conserved in Krz, we made several constructs with mutant forms of Krz fused to the Flag tag (UAS-krzV94D-Flag, UAS-krzS427D-Flag, and UAS-krzLeu-Flag). As a control construct we used wild type Krz fused to Flag (UAS-krzWT-Flag). The expression of Krz-Flag in the dorsal compartment eliminates Smo from the cell membranes of dorsal cells (Figure 6A). In contrast, neither the over-expression of KrzV94D-Flag nor of KrzLeu-Flag affects the localization of Smo (Figure 6B and 6D). These results show that Val94 and the Leu440/IsoLeu441/Leu443 domain are conserved regions essential to the function of Krz, and suggest that Krz binds to Smo and internalizes it via clathrin-coated vesicles. The over-expression of KrzS427D-Flag causes the same reduction in Smo levels as the wild type form (Figure 6C), suggesting that modulation at Ser427 is not functional in Drosophila. Over-expression experiments using the wild-type and the mutant Flag-Krz forms in myc-Smo S2 cells were consistent with the in vivo data (Figure 6E). Co-immunoprecipitation studies showed that all mutant Krz forms interact with Smo (Figure 6F). Interestingly, KrzV94D and KrzLeu mutants co-immunoprecipitated higher levels of Smo (3 and 4 times over wild type Krz, respectively), consistent with an altered Smo internalization and degradation in such conditions. Overall, these results suggest that Krz promotes Smo internalization via clathrin vesicles, and that this step is relevant for its enhancing effect on Smo degradation.
Figure 6. Interactions between Krz mutant forms and Smo.
(A–D) Effects of different Krz-Flag mutant forms expressed in the dorsal compartment on Smo expression. (A) Control ap-Gal4 UAS-GFP/+; UAS-krz-Flag/+ showing Flag (blue) and GFP (green), and the elimination of Smo (red) in the dorsal compartment. (B) ap-Gal4 UAS-GFP/+; UAS-krzV94D-Flag/+. The mutant protein KrzV94D is over-expressed (blue staining in B), but it does not affect Smo expression (red in B′). (C) ap-Gal4 UAS-GFP/+; UAS-krzS427D-Flag/+. The mutant protein KrzS427D is over-expressed (blue staining in C), and eliminates Smo expression from dorsal cells (red in C′). (D) ap-Gal4 UAS-GFP/+; UAS-krzLeu-Flag/+. The mutant protein KrzLeu is over-expressed (blue staining in D), but it does not affect Smo expression (red in D′). A′–D′ corresponds to the single red channels showing Smo expression. (E, F) Interactions between Krz and Smo in S2 cells. Stable Myc-Smo S2 or S2 control cells were transiently transfected with either empty pUAS/actinGAL4 vector (mock lanes), pUAS-krz/actinGAL4 (Kurtz lanes) or pAWF-krz mutant constructs (pAWF-krzwt, pAWF-krzV94D, pAWF-krzS247D or pAWF-krzLeu; F-Kurtz construct lanes) as indicated in the figure, in presence of Hh conditioned medium for 18–24 hrs. Cell extracts were immunoprecipitated with anti-Myc affinity gel. Immunoprecipitates (F) and whole-cell lysates (E) were immunoblotted with antibodies against Krz (detecting both endogenous Kurtz, lower band and the Flag-Kurtz protein, upper band), Myc (detecting Myc-Smo protein) and Tubulin. In control IPs carried out in Smo-Myc non-expressing cells transfected with pAWF-krzwt, pAWF-krzV94D, pAWF-krzS247D or pAWF-krzLeu, we did not detect any Krz or FLAG-Krz protein (data not shown). The levels of Smo were determined in the whole cellular lysates by immunoblotting. The basal amount of Smo in the Krzwt transfected cells was defined as 1 and all data were normalised by tubulin protein levels. Data are the mean ±SEM of three independent experiments. A representative gel is shown. * (P<0.05); *** (P<0.001) *, P<0.05; ***, when compared to values of Krzwt transfection.
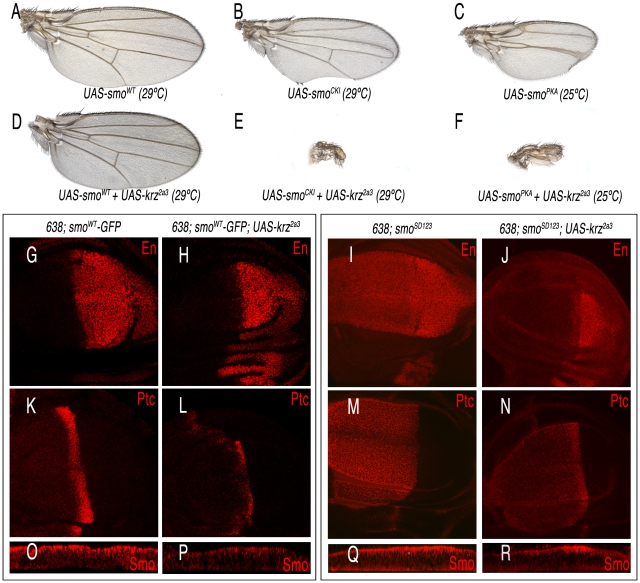
The characteristic phenotype of krz over-expression, and the effects of Krz on Smo accumulation prompted us to study the possible interactions between Krz and Smo in wing discs. We found that the phenotype of reduced L3–L4 intervein in wings over-expressing Krz is rescued by the simultaneous over-expression of Smo, resulting in the formation of normal wings (Figure 7A and 7D). As expected, the corresponding wing imaginal discs express normal levels of Ptc and En (638-Gal4/+; UAS-smoWT-GFP/UAS-krz; Figure 7G–7H and 7K–7L), indicating normal Hh signalling. In this genotype, the over-expression of Krz reduces the level of ectopic Smo-GFP (Figure 7O–7P). The interaction of vertebrate β-arrestin-2 and Smo depends on Smo activation [26], [28]. Drosophila Smo is activated by phosphorylation [38]–[39], and consequently we studied the effects of Krz over-expression in the background of Smo mutant forms affecting its phosphorylation sites. The expression of Smo mutant forms lacking the CK1 and PKA phosphorylation sites (SmoCK1 and SmoPKA) causes a weak Hh loss-of-function phenotype (Figure 7B–7C), and the co-expression of Krz in these backgrounds (638-Gal4/+; UAS-smoCK1/UAS-krz and 638-Gal4/+; UAS-smoPKA/UAS-krz) strongly enhances these phenotypes, resulting in the loss of the entire wing (Figure 7E–7F). We also combined Krz with a Smo variant that mimics phosphorylation in the PKA and CK1 sites (SmoSD123; [38]). Discs expressing SmoSD123 are overgrown and show ectopic expression of Ptc and En in the anterior compartment (638-Gal4/+; UAS-SmoSD123/+; Figure 7I, 7M). The co-expression of Krz in this background reduces the levels of ectopic En and Ptc, and also the accumulation of SmoSD123 in the anterior compartment (Figure 7J and 7N and 7Q–7R). These results indicate that Krz is able to eliminate Smo independently of its phosphorylation state by the kinases CK1 and PKA. In agreement, the SmoPKA form is also eliminated from the cell surface by the over-expression of Krz (Figure S7).
Figure 7. Interactions between Krz and different Smo mutant forms.
(A–C) Control phenotypes caused by the over-expression of wild type Smo (638-Gal4/+; UAS-smo/+; A), and phosphorylation-defective forms in the CK1 (638-Gal4/+; UAS-smoCK1/+; B) and PKA (638-Gal4/+; UAS-smoPKA/+; C) sites. (D) Rescue of the Krz over-expression phenotype by increased levels of Smo (638-Gal4/+; UAS-smo/UAS-krz2a3). (E–F) Synergistic effects of increased Krz expression in smo mutant backgrounds (638-Gal4/+; UAS- smoCK1/UAS-krz in E and 638-Gal4/+; UAS-smoPKA/UAS-krz2a3 in F). (G–J) Expression of Engrailed in Smo-Krz combinations. (G) 638-Gal4/+; UAS-smo/+. (H) 638-Gal4/+; UAS-smo/UAS-krz2a3. (I) 638-Gal4/+; UAS-smoSD123/+. (J) 638-Gal4/+; UAS-smoSD123/UAS-krz2a3. (K–N) Expression of Ptc in Smo-Krz combinations. (K) 638-Gal4/+; UAS-smo/+. (L) 638-Gal4/+; UAS-smo/UAS-krz2a3. (M) 638-Gal4/+; UAS-smoSD123/+. (N) 638-Gal4/+; UAS-smoSD123/UAS-krz2a3. Expression of Smoothened in Smo-Krz combinations. Transversal sections along the dorso-ventral boundary of 638-Gal4/+; UAS-smo/+ (O); 638-Gal4/+; UAS-smo/UAS-krz2a3 (P); 638-Gal4/+; UAS-smoSD123/+ (Q) and 638-Gal4/+; UAS-smoSD123/UAS-krz2a3 (R) third instar discs.
Krz promotes Smo degradation independently of Gprk2 activity
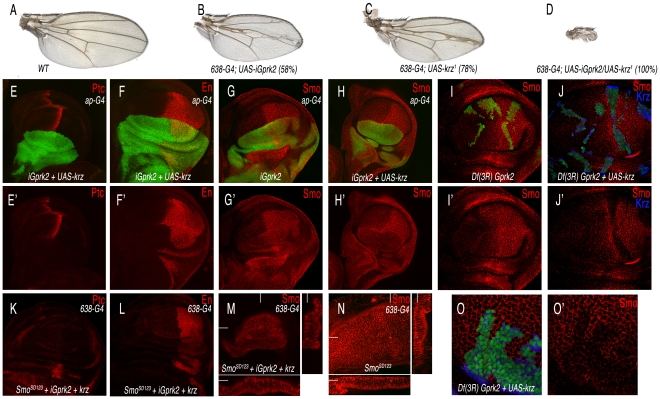
Gprk2 has a positive role in Hh signalling [20]–[21], and its vertebrate homologues directly regulate Smo by phosphorylation, triggering ß-arrestin recruitment [26]. Although the loss of Gprk2 and the gain of Krz diminish Hh signalling, their effects on Smo accumulation are entirely different. Thus, the reduction in Gprk2 stabilizes inactive Smo in the cell membrane of anterior cells, whereas the over-expression of Krz induces internalization and degradation of Smo in both anterior and posterior cells, preventing its activity. To analyse the relationships between Gprk2 and Krz in the wing disc, we expressed simultaneously a Gprk2 interference RNA (iGprk2) and the UAS-krz in the same cells. In this background the formation of the wing fails entirely (Figure 8A–8D), suggesting that Hh signalling is severely compromised. When the miss-expression is directed to the dorsal compartment (ap-Gal4/UAS-iGprk2; UAS-GFP/UAS-krz), we find that the expression of Ptc and En is lost in anterior-dorsal cells, confirming a complete loss of Smo activity (Figure 8E–8F). Interestingly, the expression of Smo in this genetic background is absent in anterior and posterior dorsal cells (Figure 8H, compare to 8G). The same loss of Smo was obtained in cells homozygous for a Gprk2 deficiency (Df(3R)Gprk2) that simultaneously over-expressed Krz (Figure 8J–8J′, compare with 8I–8I′ and 8O). In this manner, it appears that Krz can internalize Smo independently of the activity of Gprk2. The efficiency of Krz to down-regulate Smo independently of Gprk2 activity is not modified when we over-expressed the phosphomimic form of Smo (SmoSD123). Thus, reducing Gprk2 together with an over-expression of Krz also eliminates SmoSD123 accumulation (Figure 8M–8N), and abolishes the ectopic expression of Ptc and En in the anterior compartment (Figure 8K–8L). Taken together, these results show that Krz promotes Smo degradation independently of the Smo phosphorylation state and of Gprk2 activity.
Figure 8. Interactions between Gprk2 and Krz in Smo signalling.
(A) Wild type control wing. (B) Gprk2 loss-of-function phenotype resulting from iGprk2 over-expression in the wing blade (638-Gal4/+; UAS-iGprk2/+). (C) Krz gain-of-function phenotype resulting from UAS-krz over-expression in the wing blade (638-Gal4). (D) Genetic interaction between Gprk2 loss of expression and krz gain of expression. Over-expression of Krz when Gprk2 levels are reduced cause a strong Hh los-of-function (638-Gal4/+; UAS-iGprk2/+; UAS-krz/+). (E–F) Third instar wing discs over-expressing Krz and reducing Gprk2 levels in the dorsal compartment (ap-Gal4 UAS-GFP/+; UAS-iGprk2/+; UAS-krz/+) have a complete loss of Ptc (red in E) and En (red in F) expression in anterior-dorsal cells. E′–F′ correspond to the single red channels showing Ptc (E′) and En (F′) expression. (G–G′) Expression of Smo (in red) in wing discs reducing Gprk2 expression in the dorsal compartment (ap-Gal4/UAS-GFP; UAS-iGprk2/+; GFP in green). (H–H′) Expression of Smo (in red) in wing discs over-expressing Krz and reducing Gprk2 levels in the dorsal compartment (ap-Gal4 UAS-GFP/+; UAS-iGprk2/+; UAS-krz/+). Note the difference in Smo expression between G′ and H′. (I–J and O) Expression of Smo in third instar wing discs bearing clones of cells homozygous for the Gprk2 deficiency (I–I′) and for the Gprk2 deficiency in cells that also over-express Krz (J–J′). Panels O–O′ correspond to the same genotype as J–J′ at higher magnification. Clones were induced in hsFLP1.22 actin-Gal4 UAS-GFP/+; FRT82 Df(3R)Gprk2/FRT82 tub-Gal80 (I–I′) and in hsFLP1.22 actin-Gal4 UAS-GFP/+; FRT82 Df(3R)Gprk2/FRT82 tub-Gal80; UAS-krz/+ (J–J′ and O–O′). (K–M) Expression of Ptc (K), En (L) and Smo (M) in wing imaginal disc over- expressing an active form of Smo (SmoSD123) and Krz and reducing Gprk2 expression in the wing blade (638-Gal4/SmoSD123; UAS-iGprk2/+; UAS-krz/+). (N) Expression of Smo in wing imaginal disc over-expressing an active form of Smo (SmoSD123) in the wing blade (638-Gal4). Note the reduction in Smo levels in M compared to N. Below are transversal sections and to the right longitudinal sections showing Smo expression.
Discussion
In this work we have analysed the requirement of krz during the development of the Drosophila wing disc. The wing disc is an epithelial tissue, and its patterning and growth depends on the activity of several conserved signalling pathways [25]. We therefore reasoned that any requirement of Krz in the regulation of these pathways should be uncovered by the phenotype of the complete genetic loss of krz in the disc. Surprisingly, we find that wing discs (and all other imaginal discs) can develop in an almost entirely normal manner in the total absence of Krz function (see also [15]). This finding implies that any role of Krz during normal development is dispensable for the regulation of the signalling pathways operating in the wing disc. We must emphasize that even small changes in the levels or domains of signalling by the Notch, EGFR and Hh/Smo pathways result in very characteristic and distinct phenotypes in the wing, and consequently we have to conclude that these pathways operate normally in the absence of Krz in the discs.
The function of Krz has been linked in imaginal discs with the regulation of Notch protein stability [23] and of MAPK phosphorylation [24]. These conclusions are base on sound biochemical data taken from cell culture experiments, and also on the analysis of genetic interactions evaluating the ability of krz mutations in heterozygosity to modify the phenotypes caused by Notch pathway components and MAPK alleles [23]–[24]. We also find that krz reduction enhances the phenotype of a Notch loss-of-function condition, but we never found any Notch-related phenotype in krz mutant wings. Furthermore, we only found changes in Notch accumulation in a small fraction of krz1 and Df(3R)krz mutant clones, in contrast to [23]. In this context, it is interesting to note that we were able to detect a robust accumulation of Notch when krz mutant cells over-express the Notch ligand Delta (data not shown), suggesting that the function of krz becomes critical to promote Notch turnover upon Notch-Delta interactions. In this manner, the implication of our analysis and of previous works is that Krz might be required to optimise some aspects of Notch degradation or MAPK phosphorylation, but that these processes can occur normally in the absence of Krz. It might well be that only upon particular alterations of Notch levels, or in sensitized genetic backgrounds, such as over-expressing a non-dephosphorylable form of MAPK, these fine-tuning aspects of Krz are manifested in phenotypic modifications. It is unlikely that the paucity of krz requirements during imaginal development was due to functional redundancy with other arrestin proteins, because the only Drosophila candidate, CG32683, is not expressed in imaginal discs and does not affect imaginal development when over-expressed.
The lack of a krz mutant phenotype in the discs is also surprising considering the multitude of roles assigned to its vertebrate counterparts in the Wnt, IGF, Notch, Smo and TGFβ signalling pathways and in ERK activation promoted by many GPCRs (reviewed in [8]). These roles rely both on the regulation by β-arrestins of receptor internalization and subcellular localization, and also on their functions as scaffold for a variety of proteins involved in cellular signalling. We have to postulate that insect epithelial cells have evolved arrestin-independent mechanisms to control receptor turnover and signalling, and consequently that arrestin function has become less relevant in these cells. This proposal is compatible with Krz retaining the capability to molecularly interact with similar proteins as its vertebrate counterparts, as Krz possesses both amino- and carboxy-terminal arrestin domains and is 72% similar to the mammalian ß-arrestin 2 and 74% similar to ß-arrestin 1 [15].
Implications of Krz in Smo biology and signalling
In contrast to the loss-of-function analysis of krz, the study of its over-expression offers clear-cut indications of its implication in regulating Smo internalization. Thus, over-expression of Krz causes a very specific phenotype of loss-of-Hh signalling, manifested in defects localised in the central part of the wing that in extreme cases lead to the total failure of wing development. These phenotypes are associated to the loss of expression of Hh target genes, confirming that they are caused by reduced Hh signalling. As previously described, increased levels of Krz are extremely effective in reducing Smo accumulation in the cell membrane ([21] and this work). This effect is observed with wild type forms of Smo, with Smo mutated in its phosphorylation sites and with a phospho-mimic Smo protein that is constitutively activated. The elimination of Smo is also observed in posterior cells, indicating that Krz promotes Smo elimination independently of Ptc, and also in anterior cells localised away from the source of Hh, suggesting that Krz affects Smo turnover in the absence of ligand. Finally, the elimination of Smo by excess of Krz is independent of Gprk2 activity, because it is still observed in cells deficient for the Gprk2 gene. Gprk2 is required for the transduction of Smo signal, and when Gprk2 levels are lowered, inactive Smo accumulates at the cell membrane [20]–[21]. In the double combination (excess of Krz plus loss of Gprk2), Smo is eliminated, suggesting that Smo unmodified by Gprk2 is still capable to interacting with Krz and being removed. The resulting flies show extreme hh loss-of-function phenotypes, likely the result of both loss of Gprk2-dependent Smo activation and increased, Krz-promoted, Smo turnover.
The ability of Krz to interact with Smo in the Drosophila wing is very specific, as we did not observe any other alterations in the localization and activity of other receptors, such as Notch or EGFR. In this context, it is intriguing that the function of vertebrate β-arrestins has also been linked to Smo signalling in several experimental settings. First, β-arrestin 2 promotes Smo signalling by translocating this protein to the primary cilium in mouse NIH-3T3 cells [28], [40]. Second, β-arrestin 2 promotes, upon GRK phosphorylation, the internalization of activated Smo in human embryonic kidney 293 cells [26]. Finally, β-arrestin 2 promotes Smo signalling in zebrafish embryos, and this seems to be a physiological function because it is detected in loss-of-function conditions [27]. In contrast, we only observe a clear antagonism of Krz on Smo signalling caused by Smo internalization and degradation promoted by excess of Krz, and this effect of Krz is independent of the Smo phosphorylation state and of Gprk2 activity.
One of the main differences in the Smo signalling pathway between vertebrates and Drosophila is the localization in vertebrates of active Smo to the primary cilium, a structure that is only present in the fly in sensory neurons [41]–[42]. We can only speculate that the necessity to translocate Smo complexes associated with the type II kinesin motor Kif3A to the cilium [28], a structure not present in fly epidermal cells, imposes a requirement for β-arrestins that is not observed in the fly. Nonetheless, our results show that the capability of Krz to interact with Smo is retained in Drosophila, and this is revealed upon the over-expression of Krz. Once Krz is bound to Smo it would trigger the formation of clathrin-coated pits that targets Smo for degradation in the proteasome, leading to the insufficiency of Hh signalling we observe. In this way, we propose that Krz has retained some of the molecular targets typical of vertebrate β-arrestins, but that these interactions might not occur at physiological levels of expression, or being redundant with other mechanisms of receptor trafficking and signalling.
Materials and Methods
Genetic strains
We used the krz allele krz1 [15], and made a deficiency for the gene Df(3R)krz (see below). We also used the smo2 null mutation, the Gal4 lines 638-Gal4, nub-Gal4, ap-Gal4, wor-Gal4 and salEPv-Gal4 [43], and the UAS lines, UAS-EGFRDN, UAS-Nintra, UAS-iGprk2 [20], UAS-cos2, UAS-smoWT, UAS-smoCKI, UAS-smoPKA, UAS-smoSD123 [38], UAS-FLP and UAS-GFP. We generated the following lines: UAS-krz, UAS-krzWT-Flag, UAS-krzV94D-Flag, UAS-krzS427D-Flag, UAS-krzLeu-Flag, UAS-iCG32683, UAS-CG32683-Flag and UAS-ikrz (see below). We also used the RNA interference line 4637R2 (UAS-ihh from NIG-Fly, Japan). Lines not described in the text can be found in Flybase.
Generation of a krz deficiency (Df(3R)krz)
We used the Exelixis insertions e03507 and e00739, which are separated by 3.8 Kb of DNA including krz and the 5′ untranslated end of modulo (mod). Flipase (FLP)-induced recombination was induced by a daily 1 hour heat shock at 37°C to the progeny of hsFLP1.22/+; e03507/e00739 females and w; TM2/TM6b males. Thirty putative w; e03507-e00739/TM2 offspring males were individually crossed to w; TM2/TM6b females and after 3 days were used to extract genomic DNA to determinate by PCR the existence of FLP recombination. The position of the Exelixis flanking insertions e03507 and e00739 and the extent of the krz deficiency are described in Figure 1L.
Generation of FLIP recombination clones
We induced clones of cells expressing krz by a 12-min heat shock in larvae of hsFLP1.22; actin<FRT>Gal4/+; UAS-GFP/UAS-krz genotype. The elimination of the FRT cassette by FLP-mediated recombination allows the expression of Gal4 under the actin promoter. Clones were indentified by the expression of GFP. Wings homozygous for smo2 were generated in 638-Gal4/+; FRT42 smo2/FRT42 M(2)l2; UAS-FLP/+. Homozygous Df(3R)krz M+ clones and krz1 clones were induced in larvae of the following genotypes: hsFLP1.22; FRT82 Df(3R)krz/FRT82 M(3)w UbiGFP and hsFLP1.22; FRT82 krz1/FRT82 M(3)w UbiGFP, respectively. Homozygous Df(3R)krz or krz1 cells were recognized in the wing disc by the absence of GFP expression. Homozygous Df(3R)Gprk2 clones and homozygous Df(3R)Gprk2 clones over-expressing Krz were induced in larvae of the following genotypes: hsFLP1.22 actin-Gal4 UAS-GFP; FRT82 Df(3R)Gprk2/FRT82 tub-Gal80 and hsFLP1.22 actin-Gal4 UAS-GFP; FRT82 Df(3R)Gprk2/FRT82 tub-Gal80; UAS-krz/+.
Generation of krz and CG32683 constructs
UAS-ikrz and UAS-iCG32683
The EST LD31082 was used as a template to amplify a 515 pb krz fragment using the following primers: 5′GCGCTCTAGAGCAAATAATAAGGATAAA3′ and 5′GCGCTCTAGAGCATGCGCCGAAAATAATAGTAGT3′. The EST RH70434 was used as a template to amplify a 697 pb CG32683 fragment using the following primers: 5′GCGCTCTAGAGCATGCAGCCAGTAAACCCACAGA3′ and 5′GCGCTCTAGACCAAAATCGGAGAGAAAG3′. In both cases, the amplified fragment was digested with the restriction enzyme XbaI (underlined sequence in the primers) and cloned into the pWIZ vector previously digested with AvrII. The resulting plasmid was digested with NheI to clone the krz PCR fragment digested with XbaI. The orientation of both XbaI fragments cloned into pWIZ was checked to confirm an inverted position.
UAS-krz
An EcoRV-XhoI fragment purified from the EST LD31082 was first cloned into pBluescriptKS, and a NotI-KpnI fragment from this construct was cloned in the pUASt vector.
krz mutant constructs
pBluescript-Krz was used as template for the generation of Krz mutant proteins. Mutations were generated with the QuikChange site-directed mutagenesis kit (Stratagene) using the following primers (altered nucleotides, for aminoacid change, are indicated as underlined sequence): Val 94 (V94D) 5′GTAAAGGACCGTAAGGATTTTGGCCAGGTGCTTGC3′ (foward) and 5′ GCAAGCACCTGGCCAAAATCCTTACGGTCCTTTAC (reverse); Ser427 (S427D) 5′GACGGAGAAACTGAAGAGGCTACTGGCGGGC3′ (foward) and 5′GCCCGCCAGTAGCCTCTTCAGTTTCTCCGTC3′ (reverse); and Clathrin interaction domain Leu440-Ile441-Leu443 to Ala mutation (Leu Mutant), 5′GTGCCAACGACAACAAATGCCGCTCAGGCGGACGACGACGAGGCAC3′ (forward) and 5′GTGCCTCGTCGTCGTCCGCCTGAGCGGCATTTGTTGTCGTTGGCAC3′ (reverse). The presence of these mutations was confirmed by sequencing the constructs.
Krz-Flag and Gateway vector constructs
in order to generate epitope tagged Krz/mutant proteins we first amplified the Krz cDNAs (wild type and mutants) using the next primer-pair for PCR: 5′ CACCATGAACGGTGGTGGTGG3′ (foward primer) and 5′GGCCTCTGTTTCAGCGCCTTTTAG3′ (reverse primer). These PCR products were directionally subcloned into pENTR/D-TOPO (Invitrogen). For generating the C-terminal-Flag-tagged fusion protein, we used the LR Clonase II reaction of Krz (wild type and mutants) - pENTR/D-TOPO clones and the pAWF (3 Flag-tag at the C-terminal) vector for tissue culture protein expression under actin promoter (pAWF-Krz, pAWF-KrzV94D, pAWF-KrzS427D or pAWF-KrzLeu), and pTWF (3 Flag-tag at the C-terminal) vector for expression in vivo in GAL4-expressing cells (pTWF-Krz, pTWF-KrzV94D, pTWF-KrzS427D or pTWF-KrzLeu), following the instructions from Invitrogen.
UAS-CG32683-FLAG
The EST RH70434 was used as a template to amplify the coding sequence of CG32683 using the following primers: 5′CACCATGTCGGACAAGCAGCAGGAAAAGG3′ (forward) and 5′CACATTCGATGACTTGGGGACT3′ (reverse). This PCR product was directionally subcloned into pENTR/D-TOPO (Invitrogen). We used the LR Clonase II reaction of CG32683-pENTR/D-TOPO and pTWF (3XFlag-tag at the C-terminal) to generate CG32683 C-terminal-Flag-tagged fusion protein following the instructions from Invitrogen.
RNA isolation and quantitative real-time RT-PCR
Total RNA was prepared from a pool of 50 wing imaginal discs (both wild type and 638-Gal4/UAS-ikrz) and a pool of 30 larvae (both wild type and homozygous Df(3R)krz) using the TRIzol reagent protocol following Life Technologies (Grand Island, NY) instructions. Total RNA (0.7 µg) was used for a first round of reverse transcription employing the Gene Amp RNA PCR kit (Applied Biosystems). Quantitative PCR analysis was performed in a APRI PRISM 7900HT SDS (Applied Biosystems) using the TaqMan probes from Universal Probe Library (Roche) for Krz and CG32683. To normalize the results of the qPCR in the ikrz and CG32683 experiments we used probes for the genes Act42A, Tub84A and RPL32 and to normalize the results of the qPCR in the Df(3R)krz experiment we used a RNApol-II probe. Three independent experiments were done and the quantification of cDNA reduction was performed using Student's t-test. A p-value≤0.05 was considered to be statistically significant.
Generation of a Krz antiserum
Protein expression and purification
Fusion protein containing aminoacids 125–470 of Krz was generated using pBluescript-Krz as template and the following primer pair: 5′GGGGATCCATTAAAAAGCTGGGGCCG 3′ (foward primer) and 5′CCCGAATTCTAGGCTC TGTTTCAG3′ (reverse primer), containing BamHI and EcoRI restriction sites respectively (underlined sequence). The amplified fragment was digested with the restriction enzymes BamHI and EcoRI, cloned in the BamHI-EcoRI site of the gluthatione-S-transferase (GST) gene fusion vector pGEX-2T (promega) vector and transformed in E.coli BL21 DE3. Selected clones were verified by sequencing.
Antibody generation
After induction, The GST-Krz125–470 protein was purified using the Profinia Protein Purification System (BioRad), and used for antibody generation in guinea pig following conventional procedures.
Inmunohistochemistry
We used rabbit anti-activated Cas3 (Cell signalling) and anti-panArrestin (BD transduction) and rat anti-EGFR (a gift from B. Shilo), we also utilized anti-En, anti-Ptc, anti-Smo anti-Nintra and anti-FasIII mouse monoclonal antibodies from the Hybridoma Bank at University of Iowa (Iowa City, IA) and anti-FlagM2 mouse from SIGMA. Secondary antibodies (used at 1∶200 dilution) were from Jackson ImmunoResearch (West Grove, PA). To stain the nuclei we used TOPRO (Invitrogen). Imaginal wing discs and embryos were dissected, fixed, and stained as described in [44]. Confocal images were taken in a LSM510 confocal microscope (Zeiss). In situ hybridization with krz and CG32683 RNA probes were carried out as described [44]. We used the ESTs LD31082 and RH70434 as templates to synthesize krz and CG32683 probes, respectively.
Cell culture and experimental treatments
Culture conditions
S2 cells were cultured at 25°C or lower temperature in Insect-XPress media (BioWhittaker) supplemented with 10% FCS, 100 units/ml penicillin, and 100 µg/ml streptomycin. Stocks were splitted every 3 days.
Stable cell lines generation
Transfection was carried out using the Cellfectin Reagent Kit (Invitrogen). The HhN inducible vector was provided by Stephen M. Cohen (European Molecular Biology Laboratory, Heidelberg, Germany). A Myc-Smo inducible vector was generated by cloning 6Myc-tagged smo (a gift from Jianhag Jia) in pRmHa-puro. Stable cell lines over-expressing either HhN or Myc-Smo were generated by puromycin treatment (25 µg/ml). Hh S2-conditioned medium was obtained by incubation with 0.7 mM CuSO4 for 24–36 h as described [20]. Myc-Smo expression was induced by incubation with 0.7 mM CuSO4 for 24–36 h prior to the experimental assays.
Transient transfections
Myc-Smo stable cells were plated at a density of 5×104 cells/cm2, 2–3 days before transfection. 107 cells were transfected using the Nucleofector Kit V (Lonza) with 8 µg of plasmid DNA (5 µg pUAS-Krz together with 3 µg of an actin-GAL4 driver plasmid (a gift from M. González Gaitan), or 8 µg of one of the following FLAG-Krz vectors pAWF-Krz, pAWF-KrzV94D, pAWF-KrzS427D or pAWF-KrzLeu, following manufacturer's instructions. Empty vector was added to keep the total amount of DNA per dish constant. 12 h after transfection Myc-Smo expression was induced by incubation with 0.7 mM CuSO4. 24 h after transient transfection cells were grown with S2 (control) or HhN-S2-conditioned media (Hh induction) for 12 h before cell treatment.
Treatments
The Myc-Smo S2 stable cell line transiently transfected with Krz constructs was induced with CuSO4 and treated with Hh or control medium, and incubated with 60 µM of the protein synthesis inhibitor cycloheximide (Calbiochem) 2 h before the experimental assay. A 60 mM cycloheximide stock solution was prepared in DMSO. Cells were also incubated when indicated with 40 µM of the proteasome inhibitor MG132 (Biomol) for the desired period of time. A 20 mM stock solution was freshly prepared in DMSO.
Western blot and immunoprecipitation
Cellular lysate
Cells were collected by centrifugation, washed with PBS, suspended in 100–200 µl of ice-cold immunoprecipitation (IP) buffer (40 mM Tris-HCl, 200 mM NaCl, 1% Chapso, 0,5% NP40, 2 mM EGTA, 2 mM EDTA, 10 mM NaF, 0.1 µM orthovanadate, 100 µM PMSF, 1 mM Benzamidine, 16 mU/ml Aprotinin, 5 mM DTT) and incubated for 1–2 h at 4°C. Lysates were clarified by centrifugation. Protein concentration in cellular lysates was determined using the Lowry-Peterson protocol [45].
Immunoprecipitation
10 µl of cellular lysate was used to assess protein expression levels. The immunoprecipitation reactions were performed by incubating the cellular lysates with 1 mg/ml BSA and specific antibodies for Myc-Smo (15 µl of anti-Myc agarose, Santa Cruz Biotechnology) at 4°C for 4–16 h, followed by incubation with protein-A sepharose when the Krz antibody was used. A pre-immune serum (diluted 1∶100) was used as a negative control.
Immunoblotting
Whole cell lysates or immunoprecipitated complexes were resolved by 6–7% SDS-PAGE and proteins transferred to nitrocellulose membranes using a wet-blotting apparatus (BioRad). Smo and Krz proteins were detected by incubating with anti-Myc monoclonal antibody (c-Myc 9E10, Santa Cruz Biotechnology) and anti-Krz guinea pig serum, respectively. Blots were also analyzed with anti-Tubulin (Sigma) as a loading control. Immunoblots were developed and quantified using IR680 and 800 labelled antibodies (Licor) with the Odyssey Infrared Imaging System (Li-Cor). When required, the amount of co-immunoprecipitated protein was normalized by the amount of the IgG protein, as assessed by specific antibodies. When the whole-cell lysates were used, the level of Myc-Smo protein was normalized with the amount of tubulin. Data are expressed as a mean value ± SEM. Specific measurements were compared using Student's t-test. A p-value≤0.05 was considered to be statistically significant.
Supporting Information
Amino acid sequence alignment of ß-arrestin 1 (human, accession number NP001032), ß-arrestin 2 (human, accession number NP001304), Krz (Drosophila, accession number AF221066) and CG32683 (Drosophila, accession number AAF46586). The putative Valine that prevents GPCR targeting to clathrin-coated pits (Fergurson et al 1996, science) is indicated by green asterisks. Those residues that could be phosphorylated by ERK (present in both ß-arrestin 1 and Drosophila CG32683 protein) are underlined in green. The residues that bind AP2 are boxed and the amino acid necessary for clathrin binding are underlined in red.
(1.33 MB TIF)
(A) Expression of krz mRNA in wild-type (control) and 638-Gal4/UAS-ikrz wing discs. (B) Expression of krz mRNA in wild type (control) and Df(3R)krz homozygous larvae. (C) Expression of CG32683 and RpL32 (control) in wild-type wing discs. RT-qPCR was performed as described in Material and Methods. It is represented the mean of three biological replicates. The results were normalized with the gene Tub84A in A and C panels and with the gene RNApol-II in B. (D–E) In situ hybridization with an antisense (D) and sense (E) CG32683 probes in late third imaginal wing discs. (F) Wing imaginal disc expressing the fusion protein CG32683-FLAG in the of salEPv-Gal4 domain (FLAG in red and TOPRO in blue). Below of the focal plane is a transversal section. (G) Wild type phenotype in wings expressing the fusion protein CG32683-FLAg in the 638-Gal4 domain. In F and G the crosses were grown at 29°C.
(4.33 MB TIF)
Expression of Notch in krz loss-of-function conditions. (A–C and E–F) Two examples of wing discs with clones of homozygous krz1 cells (marked by absence of GFP) grown at 25°C, showing three different apical focal planes of the same disc: most apical (A and D), medium apical (B and E), less apical (C and F) with Notch expression in red. (G–I and J–L) Two examples of wing discs with clones of homozygous Df(3R)krz cells (marked by absence of GFP) grown at 25°C (G and J–I) or at 29°C (J–L), showing three different apical focal planes of the same disc: most apical (G and J), medium apical (H and K), less apical (I and L) with Notch expression in red. Below of each focal plane are transversal sections and to the right longitudinal sections showing the expression of Notch (red) and GFP (green) in the panels A–L and the expression of Notch (red) in the panels A′–L′. Clones were induced in hsFLP1.22/+; FRT82 krz1/FRT82 Ubi-GFP M(3)w (in panels A–F) and in hsFLP1.22/+; FRT82 Df(3R)krz/FRT82 Ubi-GFP M(3)w (in panels G–L).
(15.78 MB TIF)
Expression of Notch and Notch activity in Krz loss-of-function conditions. (A) Wing disc with clones of homozygous Df(3R)krz cells (marked by absence of GFP). To the right of the focal plane are longitudinal sections of wild type cells (blue line) and mutant cells (yellow line) showing the expression of Notch (red) and GFP (green). (B–C) Wing discs expressing ikrz in the posterior compartment (marked by GFP in green) showing the expression of Krz (B–B′, Krz in red) and Cut (C–C′, Cut in red) along the dorso-ventral boundary. Cut expression is normal in posterior cells with low levels of Krz, indicating that Notch signalling is normal in loss of Krz conditions.
(2.10 MB TIF)
Over-expression phenotype of Krz. (A) Representative adult wings of 638-Gal4 combinations with different lines of the UAS-krz construct. The crosses were grown at 29°C. Wings are placed in the order of the phenotype grade. (B) Quantification of the krz over-expression phenotype in different UAS-krz/638-Gal4 combinations grown at 29°C. Phenotypic classes: (1) phenotype similar to wings 2b1 and 2c1, (2) phenotype similar to wings 2a2 and 2a3, (3) phenotype similar to wings 3 and 2a, (4) phenotype similar to wings 1 and 2b2, and (5) phenotype similar to wings 2c2 and 2a1. Note that in each combination of the different UAS-krz line with 638-Gal4, there is a predominant phenotypic class.
(4.56 MB TIF)
Effects of different levels of Krz over-expression on Smo expression. (A–A′) Wild type control wing grown at 25°C (A) and its corresponding wing disc showing the normal expression of Smo (A′, red). (B–B′) Krz over-expression phenotype in the wing (nub-Gal4/+; UAS-krz/+) at 25°C, consisting in a weak thickening of veins and a reduction of the wing size (B), and its corresponding wing disc showing a strong reduction in Smo levels (B′). (C–C′) Krz over-expression phenotype in the wing with two copies of the UAS-krz (nub-Gal4/+; UAS-krz/UAS-krz) at 25°C (C) with stronger thickening of the L3 vein, and its corresponding wing disc showing the absence of Smo (C′). (D–D′) Krz over-expression phenotype in the wing (nub-Gal4/+; UAS-krz/+) at 29°C, consisting in fusion of L3 and L4 veins and reduction of the wing size (D), and its corresponding wing disc showing the absence of Smo (D′). (E–E′) Krz loss-of-function phenotype in the wing (nub-Gal4/+; UAS-ikrz/+) at 29°C, consisting in a slightly folding of the wing and a reduction of the wing size (E), and its corresponding wing disc showing a normal expression of Smo (E′). (F–F′) Both loss- and gain- of function phenotypes are rescued in the combination nub-Gal4/+; UAS-krz/UAS-ikrz grown at 29°C in the adult wings (F) and the loss of Smo expression is rescued to normal levels in the corresponding wing discs (F′).
(3.34 MB TIF)
Effects of Krz on the expression of Smo mutant forms. (A) Wing imaginal disc of 638-Gal4/+, UAS-smoPKA/+, showing the Smo accumulation in all cells of the wing blade (red). (B) Wing imaginal disc of 638-Gal4/+, UAS-smoPKA/UAS-krz2a3, showing a reduction in the Smo accumulation (red). Below and at the right of each focal plane are the transversal sections showing the expression of Smo (red).
(0.84 MB TIF)
Expression of Notch and EGFR in krz gain-of-function conditions. (A–B) Expression of Notch (red), GFP (green) and TOPRO (blue) in ap-Gal4 UAS-GFP/+; UAS-krz2a1 wing discs. A and B show two different focal planes. (A′ and B′) Red channel (Notch protein expression) of the A and B panels. (C–D) Expression of EGFR (red), GFP (green) and TOPRO (blue) in ap-Gal4 UAS-GFP/+; UAS-krz2a1. C and D show two different focal planes in the wing disc. (C′ and D′) Red channel (EGFR protein expression) of the C and D panels. Below each panel are transversal sections in the ventral (V) and dorsal (D) compartment. Right to each panel are transversal sections. The white lines show the position of the different sections.
(7.64 MB TIF)
Acknowledgments
We are grateful to A. López-Varea, E. Caminero, M. Casado, and the Genomic service of the CBMSO for their skilful technical help. We thank the Hybridome bank at Iowa University, NIG in Japan, Bloomington Stock Center, and several colleagues for providing the tools necessary for the fly work.
Footnotes
The authors have declared that no competing interests exist.
We acknowledge grants BFU2009-09403 and CSD2007-00008 to JFdC, SAF2008-00552 to FM, and an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Premont RT, Gainetdinov RR. Physiological roles of G protein-coupled receptor kinases and arrestins. Annu Rew Physiol. 2007;69:511–534. doi: 10.1146/annurev.physiol.69.022405.154731. [DOI] [PubMed] [Google Scholar]
- 3.Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci. 2004;27:107–144. doi: 10.1146/annurev.neuro.27.070203.144206. [DOI] [PubMed] [Google Scholar]
- 4.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 5.Luttrell LM, Gesty-Palmer D. Beyond desensitization: physiological relevance of arrestin-dependent signaling. Pharmacol Rev. 2010;62:305–330. doi: 10.1124/pr.109.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Penela P, Murga C, Ribas C, Lafarga V, Mayor F., Jr The complex G protein-coupled receptor kinase 2 (GRK2) interactome unveils new physiopathological targets. British Journal of Pharmacology. 2010;160:821–832. doi: 10.1111/j.1476-5381.2010.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penela P, Murga C, Ribas C, Salcedo A, Jurado-Pueyo M, et al. G protein-coupled receptor kinase 2 (GRK2) in migration and inflammation. Archives of Physiology and Biochemistry. 2008;114:195–200. doi: 10.1080/13813450802181039. [DOI] [PubMed] [Google Scholar]
- 8.Kovacs JJ, Hara MR, Davenport CL, Kim J, Lefkowitz RJ. Arrestin development: emerging roles for beta-arrestins in developmental signaling pathways. Dev Cell. 2009;17:443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribas C, Penela P, Murga C, Salcedo A, Garcia-Hoz C, et al. The G protein-coupled receptor kinase (GRK) interactome: role of GRKs in GPCR regulation and signaling. Biochimica et Biophysica Acta. 2007;1768:913–922. doi: 10.1016/j.bbamem.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 10.Patel PA, Tilley DG, Rockman HA. Physiologic and cardiac roles of beta-arrestins. J Mol Cell Cardiol. 2009;46:300–308. doi: 10.1016/j.yjmcc.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Shenoy SK, Drake MT, Nelson CD, Houtz DA, Xiao K, et al. beta -arrestin-dependent, G protein-independent ERK1/2 activation by the beta 2 adrenergic receptor. The Journal of Biological Chemistry. 2006;281:1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 12.Spiegel A. Cell signaling. beta-arrestin–not just for G protein-coupled receptors. Science. 2003;5638:1338–1339. doi: 10.1126/science.1089552. [DOI] [PubMed] [Google Scholar]
- 13.Witherow DS, Garrison TR, Miller WE, Lefkowitz RJ. beta-Arrestin inhibits NF-kappaB activity by means of its interaction with the NF-kappaB inhibitor IkappaBalpha. PNAS. 2004;101:8603–8607. doi: 10.1073/pnas.0402851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Roman G, He J, Davis RL. kurtz, a novel nonvisual arrestin, is an essential neural gene in Drosophila. Genetics. 2000;155:1281–1295. doi: 10.1093/genetics/155.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge H, Krishnan P, Liu L, Krishnan B, Davis RL, et al. A Drosophila nonvisual arrestin is required for the maintenance of olfactory sensitivity. Chemical Senses. 2006;31:49–62. doi: 10.1093/chemse/bjj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L, Davis RL, Roman G. Exploratory activity in Drosophila requires the kurtz nonvisual arrestin. Genetics. 2007;175:1197–1212. doi: 10.1534/genetics.106.068411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson EC, Tift FW, McCauley A, Liu L, Roman G. Functional characterization of kurtz, a Drosophila non-visual arrestin, reveals conservation of GPCR desensitization mechanisms. Insect Biochemistry and Molecular Biology. 2008;38:1016–1022. doi: 10.1016/j.ibmb.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Lannutti BJ, Schneider LE. Gprk2 controls cAMP levels in Drosophila development. Dev Biol. 2001;233:174–185. doi: 10.1006/dbio.2001.0219. [DOI] [PubMed] [Google Scholar]
- 20.Molnar C, Holguin H, Mayor F, Jr, Ruiz-Gomez A, de Celis JF. The G protein-coupled receptor regulatory kinase GPRK2 participates in Hedgehog signaling in Drosophila. PNAS. 2007;104:7963–7968. doi: 10.1073/pnas.0702374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng S, Maier D, Neubueser D, Hipfner DR. Regulation of Smoothened by Drosophila G-protein-coupled receptor kinases. Dev Biol. 2010;337:99–109. doi: 10.1016/j.ydbio.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meloni AR, Fralish GB, Kelly P, Salahpour A, Chen JK, et al. Smoothened signal transduction is promoted by G protein-coupled receptor kinase 2. Mol Cell Biol. 2006;26:7550–7560. doi: 10.1128/MCB.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, et al. Regulation of Notch signalling by non-visual beta-arrestin. Nature Cell Biol. 2005;7:1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- 24.Tipping M, Kim Y, Kyriakakis P, Tong M, Shvartsman SY, et al. beta-arrestin Kurtz inhibits MAPK and Toll signalling in Drosophila development. The EMBO J. 2010;29:3222–3235. doi: 10.1038/emboj.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Celis JF. Pattern formation in the Drosophila wing: the development of the veins. Bio Essays. 2003;25:443–451. doi: 10.1002/bies.10258. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Ren XR, Nelson CD, Barak LS, Chen JK, et al. Activity-dependent internalization of Smoothened mediated by beta-arrestin 2 and GRK2. Science. 2004;306:2257–2260. doi: 10.1126/science.1104135. [DOI] [PubMed] [Google Scholar]
- 27.Wilbanks AM, Fralish GB, Kirby ML, Barak LS, Li YX, et al. Beta-arrestin 2 regulates zebrafish development through the hedgehog signaling pathway. Science. 2004;306:2264–2267. doi: 10.1126/science.1104193. [DOI] [PubMed] [Google Scholar]
- 28.Kovacs JJ, Whalen EJ, Lui R, Xiao K, Kim J, Chen M, Wang J, Chen W, Lefkowitz β-arrestin mediated localization of Smoothened to the primary cilium. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Z-L, Zhao J, Sun Y, Hu W, Wu Y-L, Cen B, Wu G-X, Pei G. ß- Arrestin differentially regulates the chemokine receptor CXCR4-mediated signaling and receptor internalization, and this implicates multiple interaction sites between ß-Arrestin and CXCR4. J Biol Chem. 2000;275:2479–2485. doi: 10.1074/jbc.275.4.2479. [DOI] [PubMed] [Google Scholar]
- 30.Crozatier M, Glise B, Vincent A. Connecting Hh, Dpp and EGF signalling in patterning of the Drosophila wing; the pivotal role of collier/knot in the AP organiser. Development. 2002;129:4261–4269. doi: 10.1242/dev.129.18.4261. [DOI] [PubMed] [Google Scholar]
- 31.Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of Patched and Smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 32.Zhu AJ, Zheng L, Suyama K, Scott MP. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev. 2003;17:1240–1252. doi: 10.1101/gad.1080803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson SS, Downey WE, 3rd, Colapietro AM, Barak LS, Menard L, et al. Role of beta-arrestin in mediating agonist-promoted G protein-coupled receptor internalization. Science. 1996;271:363–366. doi: 10.1126/science.271.5247.363. [DOI] [PubMed] [Google Scholar]
- 34.Goodman OB, Jr, Krupnick JG, Gurevich VV, Benovic JL, Keen JH. Arrestin/clathrin interaction. Localization of the arrestin binding locus to the clathrin terminal domain. J Biol Chem. 1997;272:15017–15022. doi: 10.1074/jbc.272.23.15017. [DOI] [PubMed] [Google Scholar]
- 35.Krupnick JG, Goodman OB, Jr, Keen JH, Benovic JL. Arrestin/clathrin interaction. Localization of the clathrin binding domain of nonvisual arrestins to the carboxy terminus. J Biol Chem. 1997;272:15011–15016. doi: 10.1074/jbc.272.23.15011. [DOI] [PubMed] [Google Scholar]
- 36.Lin FT, Chen W, Shenoy S, Cong M, Exum ST, Lefkowitz RJ. Phosphorylation of beta-arrestin2 regulates its function in internalization of beta(2)-adrenergic receptors. Biochemistry. 2002;41:10692–10699. doi: 10.1021/bi025705n. [DOI] [PubMed] [Google Scholar]
- 37.Kim YM, Barak LS, Caron MG, Benovic JL. Regulation of arrestin-3 phosphorylation by casein kinase II. J Biol Chem. 2002;277:16837–16846. doi: 10.1074/jbc.M201379200. [DOI] [PubMed] [Google Scholar]
- 38.Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;7020:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 39.Apionishev S, Katanayeva NM, Marks SA, Kalderon D, Tomlinson A. Drosophila Smoothened phosphorylation sites essential for Hedgehog signal transduction. Nature Cell Biol. 2005;7:86–92. doi: 10.1038/ncb1210. [DOI] [PubMed] [Google Scholar]
- 40.Ayers KL, Thérond PP. Evaluating Smoothened as a G-protein-coupled receptor for Hedgehog signalling. Trends Cell Biol. 2010;20:287–298. doi: 10.1016/j.tcb.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Han YG, Kwok BH, Kernan MJ. Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr Biol. 2003;13:1679–1686. doi: 10.1016/j.cub.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 42.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, et al. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- 43.Cruz C, Glavic A, Casado M, de Celis JF. A Gain of Function Screen Identifying Genes Required for Growth and Pattern Formation of the Drosophila melanogaster Wing. Genetics. 2009;183:1005–1023. doi: 10.1534/genetics.109.107748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Celis JF. Expression and function of decapentaplegic and thick veins in the differentiation of the veins in the Drosophila wing. Development. 1997;124:1007–1018. doi: 10.1242/dev.124.5.1007. [DOI] [PubMed] [Google Scholar]
- 45.Peterson GL. Determination of total protein. Methods Enzymol. 1983;91:95–121. doi: 10.1016/s0076-6879(83)91014-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence alignment of ß-arrestin 1 (human, accession number NP001032), ß-arrestin 2 (human, accession number NP001304), Krz (Drosophila, accession number AF221066) and CG32683 (Drosophila, accession number AAF46586). The putative Valine that prevents GPCR targeting to clathrin-coated pits (Fergurson et al 1996, science) is indicated by green asterisks. Those residues that could be phosphorylated by ERK (present in both ß-arrestin 1 and Drosophila CG32683 protein) are underlined in green. The residues that bind AP2 are boxed and the amino acid necessary for clathrin binding are underlined in red.
(1.33 MB TIF)
(A) Expression of krz mRNA in wild-type (control) and 638-Gal4/UAS-ikrz wing discs. (B) Expression of krz mRNA in wild type (control) and Df(3R)krz homozygous larvae. (C) Expression of CG32683 and RpL32 (control) in wild-type wing discs. RT-qPCR was performed as described in Material and Methods. It is represented the mean of three biological replicates. The results were normalized with the gene Tub84A in A and C panels and with the gene RNApol-II in B. (D–E) In situ hybridization with an antisense (D) and sense (E) CG32683 probes in late third imaginal wing discs. (F) Wing imaginal disc expressing the fusion protein CG32683-FLAG in the of salEPv-Gal4 domain (FLAG in red and TOPRO in blue). Below of the focal plane is a transversal section. (G) Wild type phenotype in wings expressing the fusion protein CG32683-FLAg in the 638-Gal4 domain. In F and G the crosses were grown at 29°C.
(4.33 MB TIF)
Expression of Notch in krz loss-of-function conditions. (A–C and E–F) Two examples of wing discs with clones of homozygous krz1 cells (marked by absence of GFP) grown at 25°C, showing three different apical focal planes of the same disc: most apical (A and D), medium apical (B and E), less apical (C and F) with Notch expression in red. (G–I and J–L) Two examples of wing discs with clones of homozygous Df(3R)krz cells (marked by absence of GFP) grown at 25°C (G and J–I) or at 29°C (J–L), showing three different apical focal planes of the same disc: most apical (G and J), medium apical (H and K), less apical (I and L) with Notch expression in red. Below of each focal plane are transversal sections and to the right longitudinal sections showing the expression of Notch (red) and GFP (green) in the panels A–L and the expression of Notch (red) in the panels A′–L′. Clones were induced in hsFLP1.22/+; FRT82 krz1/FRT82 Ubi-GFP M(3)w (in panels A–F) and in hsFLP1.22/+; FRT82 Df(3R)krz/FRT82 Ubi-GFP M(3)w (in panels G–L).
(15.78 MB TIF)
Expression of Notch and Notch activity in Krz loss-of-function conditions. (A) Wing disc with clones of homozygous Df(3R)krz cells (marked by absence of GFP). To the right of the focal plane are longitudinal sections of wild type cells (blue line) and mutant cells (yellow line) showing the expression of Notch (red) and GFP (green). (B–C) Wing discs expressing ikrz in the posterior compartment (marked by GFP in green) showing the expression of Krz (B–B′, Krz in red) and Cut (C–C′, Cut in red) along the dorso-ventral boundary. Cut expression is normal in posterior cells with low levels of Krz, indicating that Notch signalling is normal in loss of Krz conditions.
(2.10 MB TIF)
Over-expression phenotype of Krz. (A) Representative adult wings of 638-Gal4 combinations with different lines of the UAS-krz construct. The crosses were grown at 29°C. Wings are placed in the order of the phenotype grade. (B) Quantification of the krz over-expression phenotype in different UAS-krz/638-Gal4 combinations grown at 29°C. Phenotypic classes: (1) phenotype similar to wings 2b1 and 2c1, (2) phenotype similar to wings 2a2 and 2a3, (3) phenotype similar to wings 3 and 2a, (4) phenotype similar to wings 1 and 2b2, and (5) phenotype similar to wings 2c2 and 2a1. Note that in each combination of the different UAS-krz line with 638-Gal4, there is a predominant phenotypic class.
(4.56 MB TIF)
Effects of different levels of Krz over-expression on Smo expression. (A–A′) Wild type control wing grown at 25°C (A) and its corresponding wing disc showing the normal expression of Smo (A′, red). (B–B′) Krz over-expression phenotype in the wing (nub-Gal4/+; UAS-krz/+) at 25°C, consisting in a weak thickening of veins and a reduction of the wing size (B), and its corresponding wing disc showing a strong reduction in Smo levels (B′). (C–C′) Krz over-expression phenotype in the wing with two copies of the UAS-krz (nub-Gal4/+; UAS-krz/UAS-krz) at 25°C (C) with stronger thickening of the L3 vein, and its corresponding wing disc showing the absence of Smo (C′). (D–D′) Krz over-expression phenotype in the wing (nub-Gal4/+; UAS-krz/+) at 29°C, consisting in fusion of L3 and L4 veins and reduction of the wing size (D), and its corresponding wing disc showing the absence of Smo (D′). (E–E′) Krz loss-of-function phenotype in the wing (nub-Gal4/+; UAS-ikrz/+) at 29°C, consisting in a slightly folding of the wing and a reduction of the wing size (E), and its corresponding wing disc showing a normal expression of Smo (E′). (F–F′) Both loss- and gain- of function phenotypes are rescued in the combination nub-Gal4/+; UAS-krz/UAS-ikrz grown at 29°C in the adult wings (F) and the loss of Smo expression is rescued to normal levels in the corresponding wing discs (F′).
(3.34 MB TIF)
Effects of Krz on the expression of Smo mutant forms. (A) Wing imaginal disc of 638-Gal4/+, UAS-smoPKA/+, showing the Smo accumulation in all cells of the wing blade (red). (B) Wing imaginal disc of 638-Gal4/+, UAS-smoPKA/UAS-krz2a3, showing a reduction in the Smo accumulation (red). Below and at the right of each focal plane are the transversal sections showing the expression of Smo (red).
(0.84 MB TIF)
Expression of Notch and EGFR in krz gain-of-function conditions. (A–B) Expression of Notch (red), GFP (green) and TOPRO (blue) in ap-Gal4 UAS-GFP/+; UAS-krz2a1 wing discs. A and B show two different focal planes. (A′ and B′) Red channel (Notch protein expression) of the A and B panels. (C–D) Expression of EGFR (red), GFP (green) and TOPRO (blue) in ap-Gal4 UAS-GFP/+; UAS-krz2a1. C and D show two different focal planes in the wing disc. (C′ and D′) Red channel (EGFR protein expression) of the C and D panels. Below each panel are transversal sections in the ventral (V) and dorsal (D) compartment. Right to each panel are transversal sections. The white lines show the position of the different sections.
(7.64 MB TIF)