Abstract
When yeast cell extracts that faithfully transcribe class III genes are provided with different electrolyte ions, the pattern of transcripts changes. A transcription unit in pBR322, silent with 0.1M potassium chloride, becomes active in the presence of 0.1M potassium acetate. This pseudogene depends on transcription factors B and C and RNA polymerase III like a tRNA gene. The transcribed region contains the only sequence in pBR322 homologous to the modified B block consensus sequence GTTCRDNNC found in normal tRNA genes. The presence of a block A sequence is less evident. When a block A deleted tRNA(GLU) gene was constructed, it behaved similarly: poorly transcribed with 0.1M potassium chloride, well transcribed with 0.1M potassium acetate. In fact, the deletion of the A block promoter element from the tRNA(GLU) gene did not dramatically lower its transcription when tested with potassium acetate, while it had a strong negative effect when tested with potassium chloride. Consequently the requirement for this promoter element is not constant but is a function of the electrolyte composition.
Full text
PDF



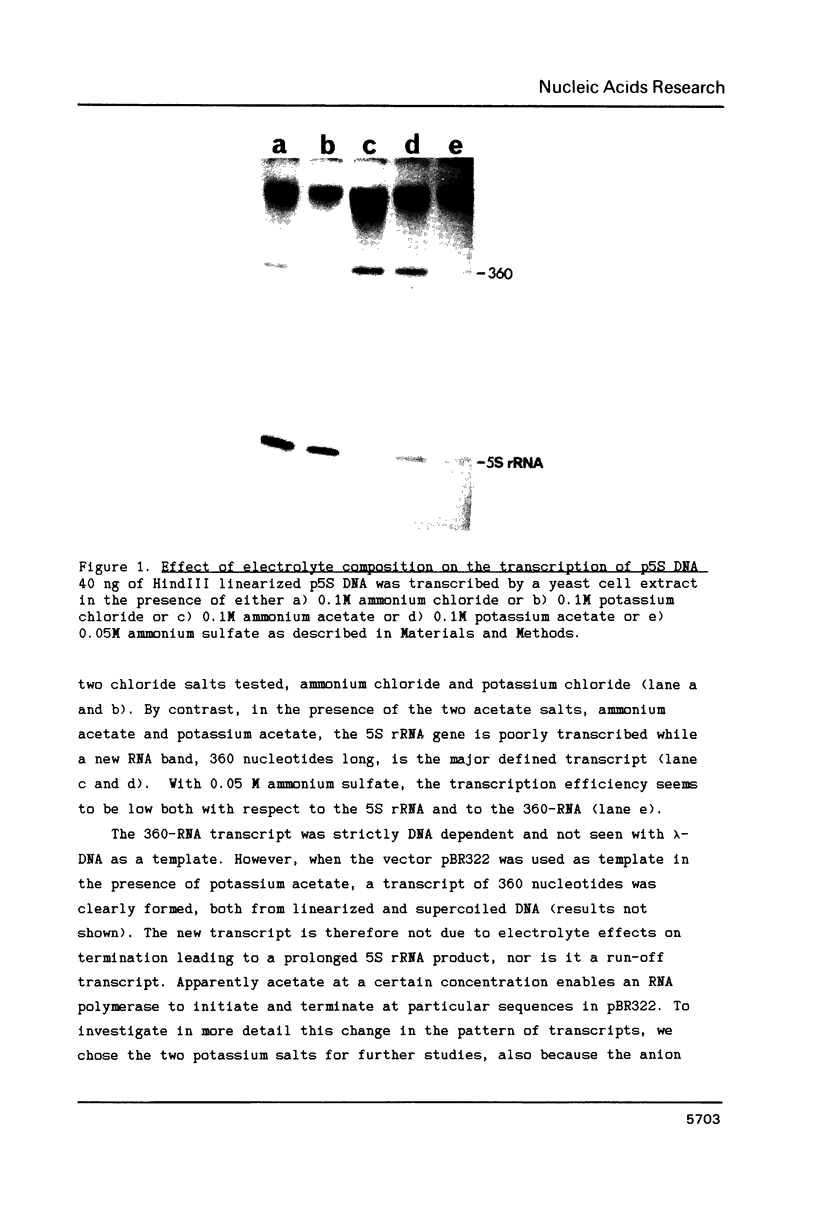










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman R., Schultz L. D., Hall B. D. Transcription in yeast: separation and properties of multiple FNA polymerases. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1702–1706. doi: 10.1073/pnas.69.7.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison D. S., Goh S. H., Hall B. D. The promoter sequence of a yeast tRNAtyr gene. Cell. 1983 Sep;34(2):655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
- Allison D. S., Hall B. D. Effects of alterations in the 3' flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985 Oct;4(10):2657–2664. doi: 10.1002/j.1460-2075.1985.tb03984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Gabrielsen O., Hall B. D. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem. 1986 Apr 25;261(12):5275–5282. [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Cortese R. Transcription by RNA polymerase III. Curr Top Dev Biol. 1983;18:59–88. doi: 10.1016/s0070-2153(08)60579-7. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Melton D. A., Cortese R. Promoter of a eukaryotic tRNAPro gene is composed of three noncontiguous regions. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1195–1199. doi: 10.1073/pnas.79.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto G., Raugei G., Costanzo F., Dente L., Cortese R. Common and interchangeable elements in the promoters of genes transcribed by RNA polymerase iii. Cell. 1983 Mar;32(3):725–733. doi: 10.1016/0092-8674(83)90058-2. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985 May 10;13(9):3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H., Olah J., Friedenreich H. Sequence of a yeast DNA fragment containing a chromosomal replicator and a tRNA Glu 3 gene. Nucleic Acids Res. 1981 Jun 25;9(12):2949–2959. doi: 10.1093/nar/9.12.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Baker C. C., Manley J. L., Ziff E. B., Sharp P. A. In vitro transcription of adenovirus. J Virol. 1981 Dec;40(3):703–719. doi: 10.1128/jvi.40.3.703-719.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk W. R., Hofstetter H. A detailed mutational analysis of the eucaryotic tRNAmet1 gene promoter. Cell. 1983 Jun;33(2):585–593. doi: 10.1016/0092-8674(83)90439-7. [DOI] [PubMed] [Google Scholar]
- Gabrielsen O. S., Andersen K. E., Oyen T. B. Yeast RNA polymerase III. Chromatographic, catalytic and DNA-binding properties are highly dependent on the type of anion. Eur J Biochem. 1984 Jun 1;141(2):345–350. doi: 10.1111/j.1432-1033.1984.tb08198.x. [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Hall B. D., Clarkson S. G., Tocchini-Valentini G. Transcription initiation of eucaryotic transfer RNA genes. Cell. 1982 May;29(1):3–5. doi: 10.1016/0092-8674(82)90083-6. [DOI] [PubMed] [Google Scholar]
- Hofstetter H., Kressman A., Birnstiel M. L. A split promoter for a eucaryotic tRNA gene. Cell. 1981 May;24(2):573–585. doi: 10.1016/0092-8674(81)90348-2. [DOI] [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Partial purification and characterization of the Saccharomyces cerevisiae transcription factor TFIIIB. J Biol Chem. 1986 Feb 25;261(6):2819–2827. [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Specific transcription of homologous class III genes in yeast-soluble cell-free extracts. J Biol Chem. 1982 Jul 25;257(14):8432–8441. [PubMed] [Google Scholar]
- Koski R. A., Allison D. S., Worthington M., Hall B. D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4 gene transcription. Nucleic Acids Res. 1982 Dec 20;10(24):8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. C., Roeder R. G. Transcription of adenovirus type 2 genes in a cell-free system: apparent heterogeneity of initiation at some promoters. Mol Cell Biol. 1981 Jul;1(7):635–651. doi: 10.1128/mcb.1.7.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T. M. Kinetics of protein-nucleic acid interactions: use of salt effects to probe mechanisms of interaction. CRC Crit Rev Biochem. 1986;19(3):191–245. doi: 10.3109/10409238609084656. [DOI] [PubMed] [Google Scholar]
- Marzouki N., Camier S., Ruet A., Moenne A., Sentenac A. Selective proteolysis defines two DNA binding domains in yeast transcription factor tau. Nature. 1986 Sep 11;323(6084):176–178. doi: 10.1038/323176a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Tower J., Sollner-Webb B. A complex control region of the mouse rRNA gene directs accurate initiation by RNA polymerase I. Mol Cell Biol. 1985 Mar;5(3):554–562. doi: 10.1128/mcb.5.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan V., Madden M. J., Salzman N. P. Preferential stimulation of transcription from simian virus 40 late and adeno IVa2 promoters in a HeLa cell extract. J Biol Chem. 1983 Dec 10;258(23):14652–14655. [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Pieler T., Oei S. L., Hamm J., Engelke U., Erdmann V. A. Functional domains of the Xenopus laevis 5S gene promoter. EMBO J. 1985 Dec 30;4(13B):3751–3756. doi: 10.1002/j.1460-2075.1985.tb04144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Mills P., Mossing M., Roe J. H. Ions as regulators of protein-nucleic acid interactions in vitro and in vivo. Adv Biophys. 1985;20:109–135. doi: 10.1016/0065-227x(85)90033-4. [DOI] [PubMed] [Google Scholar]
- Roe J. H., Burgess R. R., Record M. T., Jr Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985 Aug 5;184(3):441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- Roe J. H., Record M. T., Jr Regulation of the kinetics of the interaction of Escherichia coli RNA polymerase with the lambda PR promoter by salt concentration. Biochemistry. 1985 Aug 27;24(18):4721–4726. doi: 10.1021/bi00339a002. [DOI] [PubMed] [Google Scholar]
- Ruet A., Camier S., Smagowicz W., Sentenac A., Fromageot P. Isolation of a class C transcription factor which forms a stable complex with tRNA genes. EMBO J. 1984 Feb;3(2):343–350. doi: 10.1002/j.1460-2075.1984.tb01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L. D., Hall B. D. Transcription in yeast: alpha-amanitin sensitivity and other properties which distinguish between RNA polymerases I and III. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1029–1033. doi: 10.1073/pnas.73.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J. Assembly of a yeast 5 S RNA gene transcription complex. J Biol Chem. 1986 Sep 5;261(25):11578–11584. [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Shaner S. L., Melançon P., Lee K. S., Burgess R. R., Record M. T., Jr Ion effects on the aggregation and DNA-binding reactions of Escherichia coli RNA polymerase. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):463–472. doi: 10.1101/sqb.1983.047.01.055. [DOI] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Dingermann T., Farrell P., Söll D. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6657–6661. doi: 10.1073/pnas.78.11.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., Dingermann T., Schaack J., DeFranco D., Söll D. Transcription of eukaryotic tRNA genes in vitro. I. Analysis of control regions using a competition assay. J Biol Chem. 1983 Feb 25;258(4):2440–2446. [PubMed] [Google Scholar]
- Sharp S., Dingermann T., Schaack J., Sharp J. A., Burke D. J., DeRobertis E. M., Söll D. Each element of the Drosophila tRNAArg gene split promoter directs transcription in Xenopus oocytes. Nucleic Acids Res. 1983 Dec 20;11(24):8677–8690. doi: 10.1093/nar/11.24.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of sequences of tRNA genes. Nucleic Acids Res. 1982 Jan 22;10(2):r57–r81. [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Caspers P., Geiduschek E. P. Effects of temperature and single-stranded DNA on the interaction of an RNA polymerase III transcription factor with a tRNA gene. Cell. 1985 Feb;40(2):311–317. doi: 10.1016/0092-8674(85)90145-x. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Geiduschek E. P. Differential binding of a S. cerevisiae RNA polymerase III transcription factor to two promoter segments of a tRNA gene. EMBO J. 1984 Apr;3(4):847–853. doi: 10.1002/j.1460-2075.1984.tb01895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Sivertsen A. L., Zentner P. G., Geiduschek E. P. Correlations between transcription of a yeast tRNA gene and transcription factor-DNA interactions. J Biol Chem. 1984 Jun 25;259(12):7955–7962. [PubMed] [Google Scholar]
- Taylor M. J., Segall J. Characterization of factors and DNA sequences required for accurate transcription of the Saccharomyces cerevisiae 5 S RNA gene. J Biol Chem. 1985 Apr 10;260(7):4531–4540. [PubMed] [Google Scholar]
- Traboni C., Ciliberto G., Cortese R. A novel method for site-directed mutagenesis: its application to an eukaryotic tRNAPro gene promoter. EMBO J. 1982;1(4):415–420. doi: 10.1002/j.1460-2075.1982.tb01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traboni C., Ciliberto G., Cortese R. Mutations in Box B of the promoter of a eucaryotic tRNAPro gene affect rate of transcription, processing, and stability of the transcripts. Cell. 1984 Jan;36(1):179–187. doi: 10.1016/0092-8674(84)90087-4. [DOI] [PubMed] [Google Scholar]
- Wu C. W., Tweedy N. Mechanistic aspects of promoter binding and chain initiation by RNA polymerase. Mol Cell Biochem. 1982 Sep 17;47(3):129–149. doi: 10.1007/BF00229597. [DOI] [PubMed] [Google Scholar]
- deHaseth P. L., Lohman T. M., Burgess R. R., Record M. T., Jr Nonspecific interactions of Escherichia coli RNA polymerase with native and denatured DNA: differences in the binding behavior of core and holoenzyme. Biochemistry. 1978 May 2;17(9):1612–1622. doi: 10.1021/bi00602a006. [DOI] [PubMed] [Google Scholar]







