Abstract
Based on our recent work with Haloferax volcanii, ubiquitin-like (Ubl) proteins (SAMP1 and SAMP2) are known to be covalently attached to proteins in archaea. Here, we investigated the enzymes required for the formation of these Ubl-protein conjugates (SAMPylation) and whether this system is linked to sulfur transfer. Markerless in-frame deletions were generated in H. volcanii target genes. The mutants were examined for: (i) the formation of Ubl protein conjugates, (ii) growth under various conditions, including those requiring the synthesis of the sulfur-containing molybdenum cofactor (MoCo), and (iii) the thiolation of tRNA. With this approach we found that UbaA of the E1/MoeB/ThiF superfamily was required for the formation of both SAMP1- and SAMP2-protein conjugates. In addition, UbaA, SAMP1, and MoaE (a homolog of the large subunit of molybdopterin synthase) were essential for MoCo-dependent dimethyl sulfoxide reductase activity, suggesting that these proteins function in MoCo-biosynthesis. UbaA and SAMP2 were also crucial for optimal growth at high temperature and the thiolation of tRNA. Based on these results, we propose a working model for archaea in which the E1-like UbaA can activate multiple Ubl SAMPs for protein conjugation as well as for sulfur transfer. In sulfur transfer, SAMP1 and SAMP2 appear specific for MoCo biosynthesis and the thiolation of tRNA, respectively. Overall, this study provides a fundamental insight into the diverse cellular functions of the Ubl system.
Keywords: posttranslational modification, tRNA modification, proteasomes
The posttranslational conjugation of one or more molecules of ubiquitin (Ub) and ubiquitin-like (Ubl) proteins to selected proteins plays an integral role in a wide variety of functions in eukaryotic cells. Many of these processes are central to cell physiology, including the regulation of gene expression, heterochromatin formation, genome stability, protein trafficking, cell division, morphogenesis, DNA repair, autophagy, and proteasome-mediated proteolysis (1, 2).
Elaborate ATP-dependent conjugation systems covalently and reversibly attach Ub (and Ubl) proteins to their protein targets (3). In this cascade, the C terminus of Ub is adenylated by an Ub-activating E1 enzyme, which activates Ub for nucleophilic attack by the active site cysteine of E1 to generate an E1-Ub thioester. An E2 Ub-conjugating enzyme accepts this activated form of Ub on its active site cysteine to form the second thioester linkage in this pathway. From E2, Ub is transferred to the ε-amino group of a lysine residue either within the target protein or the growing poly-Ub chain, thus forming an isopeptide bond. Transfer is often assisted by an E3 Ub-ligase, either forming a transient E3-Ub thioester intermediate or facilitating the transfer of Ub from E2 directly to the substrate protein.
Ub and Ubl-proteins have a β-grasp fold that is common to a superfamily of proteins found in all three domains of life (4, 5). Proteins with this fold are diverse and mediate a variety of functions beyond the ubiquitination of proteins. For example, (i) 2[Fe-S] ferredoxins facilitate electron transfer, (ii) ThrRS, GTPase, and SpoT domains of tRNA synthetases are required for RNA-protein interactions, (iii) ThiS/MoaD proteins serve as sulfur carriers in thiamine and tungsten/molybdenum cofactor (W/MoCo) biosynthesis, and (iv) Atg8 and LC3 are conjugated to phospholipids (5–8).
β-Grasp fold-proteins that function as sulfur carriers and protein modifiers share a common chemistry. Both types of proteins are adenylated at their C terminus by an ATP-dependent E1/MoeB/ThiF-type enzyme (8, 9). This adenylation activates the β-grasp fold-protein for either the acceptance of sulfur as a C-terminal thiocarboxylate or the formation of an E1-Ubl thioester intermediate for subsequent protein modification.
Urm1, one of the most ancestral of eukaryotic Ubl-proteins, along with its E1-activating enzyme Uba4p (MOCS3) provide the only example to date of a system that functions in both protein conjugation (urmylation) (10, 11) and sulfur transfer (2-thiolation of tRNAs) (12–18). Uba4p adenylates and transfers sulfur to the C terminus of Urm1, resulting in the formation of a C-terminal thiocarboxylated form of Urm1 (14) that is required for both tRNA thiolation and urmylation (19). Although lysine residues of protein substrates and an apparent thioester intermediate are required for urmylation (19), the detailed chemistry of how Uba4p forms these covalent adducts and differentiates between protein conjugation and sulfur transfer remain to be determined.
Recently, we identified two Ubl proteins (SAMP1 and SAMP2) that are differentially conjugated to proteins in the haloarchaeon Haloferax volcanii (20). The SAMP2 C-terminal carboxylate was demonstrated to form an isopeptide bond with the ε-amino group of lysine residues on target proteins. Although related to ubiquitination, this type of posttranslational modification (SAMPylation) appears to be simplified in comparison. Like H. volcanii, all archaea encode Ubl proteins. However, only a single member of the E1/MoeB/ThiF superfamily is present and E2- and E3-like proteins are not predicted in the majority of archaeal genomes (8).
Here we demonstrate that the E1-like UbaA and Ubl SAMPs of H. volcanii are required for protein conjugation and sulfur transfer, including the thiolation of tRNA and most likely MoCo biosynthesis. Thus, archaeal Ubl-systems are highly versatile in their function and provide an example of a Ubl-system that forms not only classic isopeptide bonds with diverse protein targets, but also mediates sulfur transfer.
Results and Discussion
Identification of Putative Genes Required for SAMPylation.
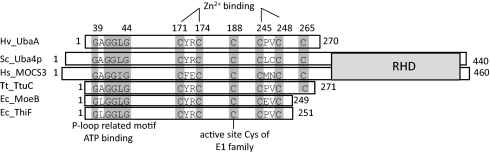
Similar to most archaea, H. volcanii encodes a single protein of the E1/MoeB/ThiF superfamily (HVO_0558), which we have termed Ubl-activating enzyme of archaea or UbaA for its potential role in activating SAMPs. Our previous work identified UbaA conjugated to SAMP1 and SAMP2 by MS-based proteomics (20). To further understand the function of UbaA, its protein sequence was compared with E1/MoeB/ThiF-type homologs by multiple amino acid sequence alignment (Fig. 1 and SI Appendix, Fig. S1). Based on this alignment, UbaA had all of the conserved residues needed to catalyze the ATP-dependent adenylation of the SAMPs, including a glycine-rich region related to the NTPase P-loop motif. In addition, UbaA C188 was analogous to the active site cysteine required for the formation of the E1-Ub thioester intermediate of ubiquitination in eukaryotes (21) and the acyldisulfide-linked ThiF-ThiS intermediate of thiamine biosynthesis in bacteria (22). UbaA also had four conserved cysteine residues predicted to coordinate Zn2+ for structural integrity similar to E1-like enzymes. However, UbaA did not have the extended C-terminal rhodanese domain (RHD) that is common to Uba4p and MOCS3. Instead, like other archaea, the H. volcanii RHDs are separately encoded by multiple genes including ubaB (HVO_0559) that is divergently transcribed from ubaA (SI Appendix, Fig. S2).
Fig. 1.
Schematic representation of the H. volcanii (Hv) UbaA with members of the MoeB/E1 superfamily. The glycine rich region related to the NTPase P-loop motif, conserved cysteine residues, and C-terminal RHD are indicated with conserved amino acid residues shaded and numbered according to UbaA (HVO_0558). Uba4p of yeast (Saccharomyces cerevisiae, Sc), MOCS3 of human (Homo sapiens, Hs), TtuC of Thermus thermophilus (Tt), and ThiF and MoeB of Escherichia coli (Ec) are presented for comparison.
In addition to UbaA, archaea encode homologs of MoaE (MOCS2B) (SI Appendix, Fig. S3), known in bacteria (and eukaryotes) to associate with the Ubl MoaD (MOCS2A) (23). This association occurs after the C terminus of MoaD is adenylated by the E1-like MoeB and thiocarboxylated by the IscS cysteine desulfurase. Molybdopterin (MPT) synthase is a heterotetramer of MoaE and thiocarboxylated MoaD (MoaD-COSH) that catalyzes the transfer of sulfur from the bound MoaD-COSH to cyclic pyranopterin monophosphate (cPMP or precursor Z). This catalyzation forms the dithiolene moiety of MPT before the insertion of molybdenum. In methanogenic and halophilic archaea, MoaE is often fused to an N-terminal P-loop NTPase MobB domain (e.g., HVO_1864 of H. volcanii, termed MoaE) (SI Appendix, Fig. S3). MobB functions downstream of MoaE in bacteria to form molybdopterin guanine dinucleotide common to enzymes of the DMSO reductase family (24, 25). The MoaE homolog of H. volcanii and other archaea is likely to be linked to Ubl protein function, based on its amino acid sequence relationship to the large subunit of MPT synthase and its association with SAMP1 as detected by MS (20).
Gene Knockout to Investigate the Relationship of Protein Conjugation and Sulfur Transfer in Archaea.
To further investigate the roles of the archaeal Ubl SAMPs and to identify enzymes catalyzing their activation and covalent modification, a series of knockout strains was generated in the H. volcanii genome using the markerless pyrE2-based deletion strategy (26, 27) (strains listed in SI Appendix, Table S1). Targets for deletion were the genes encoding SAMP1 and SAMP2, as well as genes implicated in SAMPylation based on comparative genomics (discussed above) and MS-based proteomics (20). This latter set included genes encoding UbaA (HVO_0558), MoaE (HVO_1864), and UbaB (HVO_0559). In addition, a gene (HVO_2177), encoding a third Ubl protein that has a β-grasp fold and C-terminal diglycine motif similar to SAMP1 and SAMP2, was deleted in single and triple knockout with the SAMP1 and SAMP2 genes. Mutant strains were confirmed by Southern blot, PCR and RT-qPCR (for details see SI Appendix, Tables S1–S3 and Figs. S4 and S5).
UbaA Is Required for SAMPylation.
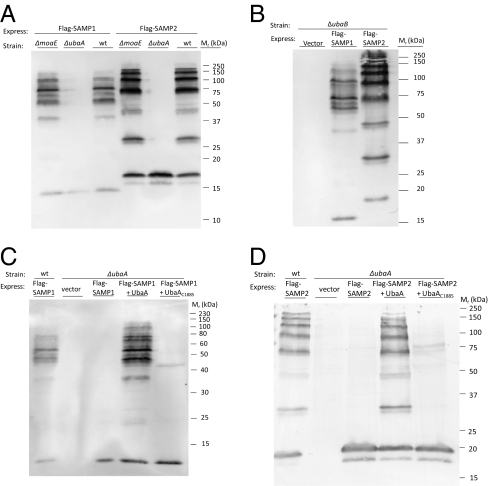
To examine whether ubaA, moaE, or ubaB are required for SAMPylation, parent and mutant strains were transformed with plasmids expressing Flag-SAMP1 and Flag-SAMP2 and grown under conditions known to enhance protein-conjugate levels (aerobically with N-limitation) (20). Cellular protein was separated by reducing SDS/PAGE and analyzed by α-Flag immunoblot. With this approach, we found that the moaE and ubaB genes were not required for the formation of SAMP protein-conjugates (Fig. 2 A and B). In contrast, the ubaA mutant was deficient in SAMPylation with unconjugated Flag-SAMP1 and Flag-SAMP2 proteins detected at 15 to 20 kDa (Fig. 2A), in a pattern similar to previously reported for the SAMPs synthesized with C-terminal diglycine motif deletions (ΔGG) (20). The reason for the doublet of SAMP2-specific bands (at 16 and 17 kDa) in the ubaA mutant is unclear, yet comparable to cells expressing SAMP2ΔGG (20). The ubaA knockout was complemented by providing a wild-type copy, but not a C188S variant of ubaA in trans (Fig. 2 C and D), with production of these UbaA proteins confirmed by immunoblot (SI Appendix, Fig. S7). Thus, UbaA Cys188 appears to function as an active site residue in SAMPylation similar to E1-type enzymes.
Fig. 2.
UbaA is required for SAMPylation. Deletion of ubaA reduces the level of SAMP1- and SAMP2- protein conjugates in the cell (A), moaE and ubaB are not required for formation of these conjugates (A and B), and the ubaA mutation is complemented by providing a wild-type copy of ubaA in trans (C and D). H. volcanii strains (indicated above, where wild-type represents parent H26) were grown aerobically to stationary phase in N-limiting medium. Cell lysate was separated by reducing SDS/PAGE. Flag-SAMP proteins and conjugates were detected by α-Flag immunoblot. Equivalent protein loading was confirmed by staining parallel gels for total protein with Coomassie blue (SI Appendix, Fig. S6). Molecular mass standards are indicated on the right.
UbaA, MoaE, and SAMP1 Appear Crucial for MoCo Biosynthesis.
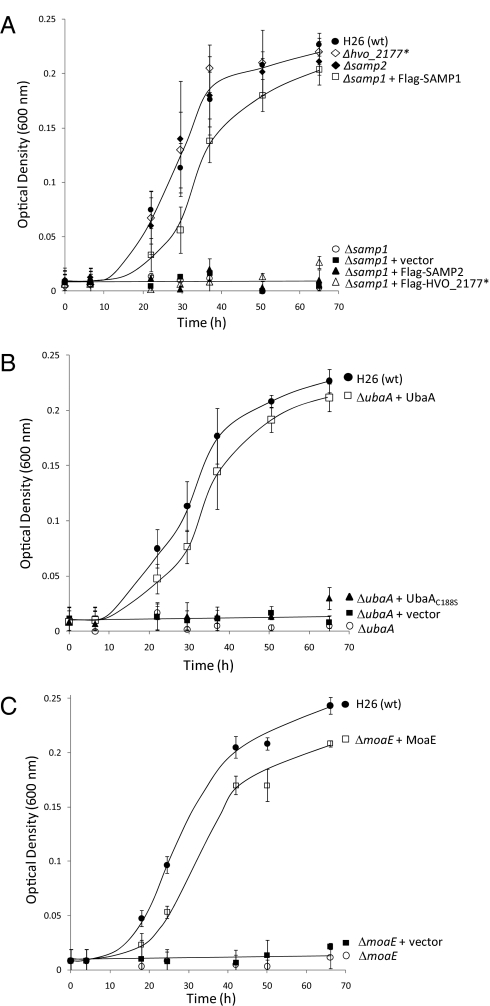
Growth of mutant strains, including single knockouts of ubaA, moaE, and ubaB, as well as single and triple knockouts of the Ubl genes (samp1, samp2, and hvo_2177), was investigated in rich media at optimal growth temperature (42 °C) in the presence of either oxygen or DMSO as the terminal electron acceptor. In the presence of oxygen, growth was relatively similar to wild-type for all mutant strains examined (SI Appendix, Fig. S8). Likewise, growth rates were similar to wild-type for the samp2, hvo_2177, and ubaB single knockouts under anaerobic conditions with DMSO (Fig. 3A and SI Appendix, Fig. S9). In contrast, mutants with deletions in ubaA, moaE, or samp1 did not grow anaerobically with DMSO but could be complemented by providing a wild-type copy of the corresponding gene in trans (Fig. 3). The gene encoding UbaA C188S did not complement the ubaA knockout (Fig. 3B), revealing that Cys188 is likely to be important for the catalytic function of UbaA under these conditions. Furthermore, plasmids expressing Flag-SAMP2 and Flag-HVO_2177 did not complement the samp1 mutation (Fig. 3A), suggesting the role of SAMP1 is distinct from these two other Ubl β-grasp fold-proteins during anaerobic growth on DMSO.
Fig. 3.
SAMP1, UbaA, and MoaE are required for anaerobic growth on DMSO (A, B, and C, respectively). H. volcanii strains (indicated on the right) were grown on rich medium (YPC) with glucose supplementation and DMSO as the external electron acceptor, as described in Materials and Methods. Growth was monitored over time by an increase in OD600. Similar results were observed during growth on ATCC 974 or YPC media with glycerol and DMSO supplementation, and growth was not observed for any strains under anaerobic conditions in the absence of DMSO.
One possibility for the lack of growth of the ubaA, moaE, and samp1 mutants on DMSO is that these genes are required for sulfur transfer to cPMP to form the dithiolene intermediate in MoCo biosynthesis. This possibility would be in analogy to the Escherichia coli moeB, moaE, and moaD genes required to generate MoCo for the catalytic subunit of DMSO reductase (DmsA). To further investigate this, the ubaA, moaE, and samp1 mutant and wild-type strains were grown aerobically to log-phase and then incubated under anaerobic conditions with DMSO. Cell lysate was assayed for DMSO reductase activity, and total RNA was analyzed for dmsA-specific transcript levels by RT-PCR. Although DMSO reductase activity was readily detected in the parent and complemented strains, it was not detected in the ubaA, moaE, and samp1 mutants (Table 1). The dmsA-specific transcript, however, was present in all strains examined (SI Appendix, Fig. S10), suggesting UbaA, SAMP1, and MoaE are required for MoCo biosynthesis and, ultimately, maturation of the DmsA apoprotein into the MoCo-holoprotein. Although we do not rule out the possibility that SAMP1 modulates DMSO reductase through protein conjugation mechanisms, we propose a working model that UbaA can activate SAMP1 to function as a sulfur carrier with MoaE for MPT synthesis.
Table 1.
UbaA, SAMP1, and MoaE are required for DMSO reductase activity of H. volcanii cells
| Strain-plasmid | Description | DMSO reductase |
| Specific activity (U·mg protein−1) | ||
| H26 | Parent | 0.240 ± 0.029 |
| HM1052 | ΔubaA | UD |
| HM1052-pJAM202c | ΔubaA + vector | UD |
| HM1052-pJAM957 | ΔubaA + UbaA | 0.358 ± 0.032 |
| HM1052-pJAM1116 | ΔubaA + UbaA C188S | UD |
| HM1041 | Δsamp1 | UD |
| HM1041-pJAM202c | Δsamp1 + vector | UD |
| HM1041- pJAM947 | Δsamp1 + SAMP1 | 0.255 ± 0.024 |
| HM1053 | ΔmoaE | UD |
| HM1053-pJAM202c | ΔmoaE + vector | UD |
| HM1053-pJAM1119 | ΔmoaE + MoaE | 0.227 ± 0.026 |
UD, undetectable. Cell growth and assay of DMSO reductase activity are described in Materials and Methods. UbaA, UbaA C188S, and MoaE are synthesized as C-terminal StrepII tag fusions. SAMP1 is synthesized as a fusion with an N-terminal Flag-tag.
To bolster our proposal, we analyzed the levels of SAMP1 and SAMP2 protein-conjugates formed after cells were grown on rich medium in the presence of oxygen or DMSO (SI Appendix, Fig. S11). Similar to our previous work (20), the levels of both types of SAMP protein-conjugates appeared low when cells were grown on rich medium in the presence of oxygen, with the majority of Flag-SAMP proteins at 15 to 20 kDa in the unconjugated form. Anaerobic growth with DMSO as the terminal electron acceptor did not enhance the levels of SAMP1 protein-conjugates, suggesting SAMP1ylation is not a major mechanism of posttranslational protein modification under these conditions. Instead, the levels of SAMP2 protein-conjugates were relatively high under anaerobic conditions with DMSO and reached levels comparable to that previously observed under N-limiting conditions in the presence of oxygen (20). For cells grown in the presence of oxygen, addition of glucose to the complex medium had a stimulatory effect on SAMP2ylation. However, these levels were modest compared with the robust levels of SAMP2ylation observed during anaerobic growth with DMSO, and may be because of the reduced levels of oxygen likely to occur from the enhanced growth observed upon glucose supplementation.
UbaA and SAMP2 Are Needed for Optimal Growth at High Temperature.
To investigate whether UbaA, SAMP, and other SAMP-associated proteins are needed for optimal growth at high temperature, the aerobic growth of H. volcanii parent and single knockout strains was monitored on rich medium at temperatures equal to and above the 42 to 45 °C optimum (28). Although all strains grew similar to wild-type at 42 °C, ubaA and samp2 mutants were retarded in growth at 50 °C (SI Appendix, Fig. S12). Although many factors may be responsible for this phenotype, such as deficiencies in proteasome function (29), it is interesting to note that the samp1 and other associated mutant strains that we examined were not hypersensitive to growth at high temperature. Based on our earlier work, it is the SAMP1 (and not SAMP2) protein-conjugates that accumulate in proteasomal mutant strains, whereas SAMP2 modifies homologs of the Urm1 pathway including Tum1 (Yor251cp) and Ncs6p, both of which are important in the thiolation of tRNA (20). Interestingly, mutant strains deficient in the ability to thiolate tRNA are often temperature-sensitive (e.g., ref. 30). Thus, the temperature-sensitive phenotype of ubaA and samp2 mutants may be because of reduced thiolation of tRNAs.
UbaA and SAMP2 Are Required for the Thiolation of tRNA.
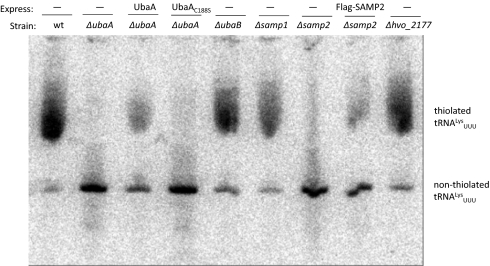
To investigate the role of the SAMPs and UbaA in the thiolation of tRNA, total RNA isolated from mutant, parent, and complemented strains was: (i) separated by [(N-acryloylamino)phenyl]mercuric chloride (APM) gel electrophoresis and (ii) hybridized to a probe specific for lysine tRNAs with anticodon UUU (tRNALysUUU) (Fig. 4). This probe was selected based on the widespread distribution of 2-thiouridine derivatives in the first position or “wobble base” in the anticodon of lysine tRNAs (in addition to glutamine and glutamate tRNAs) (31). The majority of the tRNALysUUU pool of parent H26 and samp1, hvo_2177, and ubaB mutant strains was found to be thiolated (Fig. 4), based on its retarded migration in APM gels (32). In contrast, the tRNALysUUU of ubaA and samp2 mutant strains appeared nonthiolated with this defect restored in part by expression of the corresponding genes in trans. The ubaA mutant was not complemented by the gene encoding UbaA C188S, suggesting this cysteine residue is important for UbaA function in the tRNA thiolation pathway.
Fig. 4.
UbaA and SAMP2 are required for the thiolation of tRNALysUUU. Total RNA was isolated from H. volcanii strains (indicated above), separated by APM gel electrophoresis, and hybridized with a probe complementary to tRNALysUUU, as described in Materials and Methods. Thiolated tRNALysUUU is based on its retardation in APM gels compared with the nonthiolated form, which migrates faster.
Working Model for Ubl Proteins in Archaea.
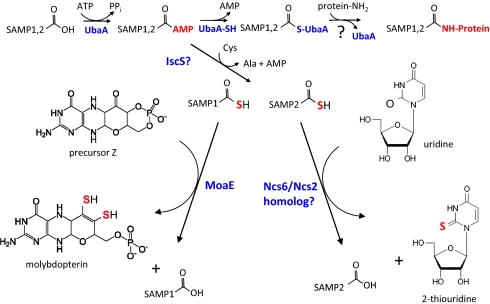
Based on our current and previous findings, we propose a working model for archaea in which the E1-like UbaA and Ubl SAMP proteins function in both protein conjugation and sulfur transfer (Fig. 5). In this model, UbaA catalyzes the adenylation of the C-terminal glycine of the SAMPs for their activation in protein conjugation. This adenylation would also activate SAMP1 and SAMP2 for their acceptance of sulfur as a C-terminal thiocarboxylate to serve as a sulfur carrier in MoCo biosynthesis and tRNA thiolation, respectively.
Fig. 5.
Working model in which the E1-like UbaA and Ubl SAMP proteins function in both protein conjugation and sulfur transfer pathways in archaea (see text for details).
During protein conjugation, a thioester intermediate is suggested to be formed between the active site Cys188 of UbaA and the C-terminal carboxyl group of the SAMPs. This prediction is based on: (i) the requirement of UbaA Cys188 for protein-conjugate formation, (ii) the conservation of UbaA Cys188 with the active site cysteine of E1-type enzymes known to form an E1-Ub thioester, and (iii) the detection of isopeptide (and not persulfide) bonds between the C-terminal carboxyl group of SAMP2 and the ε-amino group of lysine residues of target proteins. However, further studies are needed to demonstrate this intermediate.
It is not clear whether UbaA forms a covalent intermediate with the SAMPs after their adenylation in the sulfur transfer pathways and what provides the activated source of sulfur for this putative thiocarboxylation reaction. The requirement of UbaA Cys188 for anaerobic growth on DMSO and tRNA thiolation suggests this residue is important in sulfur-transfer pathways and may form either a thioester or acyldisulfide intermediate with the SAMPs. This latter intermediate would be analogous to the E1-like ThiF of E. coli that forms an acyldisulfide conjugate with the Ubl ThiS in thiamine biosynthesis (22), but contrasts with the E1-like MoeB of E. coli that appears to form only noncovalent bonds with the Ubl MoaD during MoCo biosynthesis (33). Ultimately, C-terminal thiocarboxylated forms of the SAMPs are proposed to be generated after their activation by UbaA. In many archaea, the source of sulfur for this thiocarboxylation is likely to be derived from cysteine through a persulfide intermediate of a NifS/IscS-type cysteine desulfurase. However, not all archaea generate free pools of cysteine (e.g., Methanococcus maripaludis) (34, 35), and instead may use other forms of sulfur for this thiocarboxylation reaction or not require SAMPs as sulfur carrier proteins. Whether RHD proteins provide a sulfur relay between NifS/IscS-type proteins during the thiocarboxylation of the SAMPs similar to the RHD Tum1 (YOR251c) (12) of tRNA thiolation in yeast is also unclear. In H. volcanii, the RHD protein UbaB is not required for protein conjugation, MoCo biosynthesis, or the generation of thiolated tRNALys. However, other RHD proteins may function in this sulfur relay or compensate for the loss of UbaB function in the ubaB mutant.
The interaction partners that facilitate sulfur transfer from the thiocarboxylated SAMP proteins to the biosynthetic intermediates are likely to be specific for each of the different sulfur transfer pathways. In MoCo biosynthesis, MoaE is proposed to form a complex with the thiocarboxylated form of SAMP1 and facilitate sulfur transfer to precursor Z. This proposal is based on: (i) the detection of MoaE with SAMP1 (and not SAMP2) as a complex by MS-based proteomics (20), (ii) the requirement of MoaE for DMSO reductase activity and not protein-conjugation, and (iii) the similarity of MoaE and SAMP1 to the subunits of the MPT synthase complex of E. coli and humans (36–40). In tRNA thiolation, the ATPase PP-loop superfamily member HVO_0580 is proposed to facilitate sulfur transfer from the SAMP2 thiocarboxyl to position 2 of the wobble uridine (U34) present in some tRNA species. HVO_0580 is homologous to tRNA modification enzymes, such as TtcA important in cytidine 2-thiolation of tRNA in bacteria (39), and Ncs6p/Ncs2p, implicated in uridine 2-thiolation of tRNA in eukaryotes (12, 15, 18, 40). HVO_0580 also associates with SAMP2 (and not SAMP1) as determined by MS (20).
Although 26S proteasomes and the enzymes mediating ubiquitination are essential for eukaryotic cell division (reviewed in refs. 41 and 42), we observe differences in archaea. Like eukaryotes, 20S proteasomal core particles are required for archaeal cell division based on the inability of H. volcanii to grow when core particles (the single β-type or both α-type subunits of these complexes) are conditionally depleted from cells (29). However, in contrast to the core particles, neither the proteasome activating nucleotidase proteins (PANs, homologs of the regulatory particle triple-A or AAA ATPase particles of eukaryal 26S proteasomes) nor the SAMPylation system are essential for archaeal cell growth. This latter conclusion is based on the robust aerobic growth observed for H. volcanii cells even after knockout of both Rpt-like genes (panA and panB) (29), all three of the Ubl genes (samp1, samp2, and hvo_2177), or depletion of ubaA gene function. This discrepancy, between the core particles being essential and the AAA-ATPase PAN and SAMPylation pathway being dispensable for growth under aerobic conditions presents a few scenarios. First, it is likely that in archaea, not all proteins degraded by core particles are targeted by the Ubl-protein conjugation pathway (SAMPylation). It is also possible that not all proteasomal substrates require the Rpt-like PANs for entry into the core particle proteolytic chamber (e.g., homologs of Cdc48-like AAA ATPases common among archaea and associated with core particle function in eukaryotes may serve this alternative role). Finally, the core particles may mediate protein degradation in the absence of AAA ATPases and facilitate other cellular processes that are essential for cell division but independent of proteasome-mediated proteolysis.
Materials and Methods
Materials.
Biochemicals and analytical-grade inorganic chemicals were purchased from Fisher Scientific, Bio-Rad, and Sigma-Aldrich. Desalted oligonucleotides were from Integrated DNA Technologies. DNA polymerases and modifying enzymes were from New England Biolabs. Hi-Lo DNA standards were from Minnesota Molecular, Inc.. APM was synthesized according to Igloi et al. (32).
Strains, Media, and Plasmids.
The strains, primers and plasmids used in this study are summarized in SI Appendix, Tables S1 and S2. Site-directed mutagenesis was performed using a QuikChange Lightning kit according to the supplier (Stratagene). E. coli strains, DH5α used for routine recombinant experiments and GM2163 used for isolation of plasmid DNA for transformation of H. volcanii (43), were grown at 37 °C in Luria-Bertani medium. H. volcanii strains were grown at 42 °C (or 50 °C where indicated). Growth media included complex media (YPC and ATCC974) and glycerol minimal medium with alanine as the nitrogen source (GMM alanine), as previously described (20, 43). Ampicillin (0.1 mg·mL−1), novobiocin (0.1 μg·mL−1), and agar (1.5% wt/vol) were included as needed. For anaerobic growth, H. volcanii strains were twice grown aerobically on complex media to log-phase (2 mL in 13 × 100-mm tubes; 200 rpm) and inoculated at 1% (vol/vol) for anaerobic growth in 10-mL screw-cap tubes on complex media supplemented with 100 mM DMSO and 2% (wt/vol) glucose (or 20 mM glycerol where indicated). For analysis of DMSO reductase activity and dsmA-specific transcripts, H. volcanii strains were first grown aerobically in YPC medium (2 mL in 13 × 100-mm tubes, 200 rpm) and inoculated (1% vol/vol) into fresh YPC medium (3 × 50 mL in 250 mL baffled flasks, 200 rpm) to late-log (OD600 of 1.4–1.5 units). Cultures were pooled, supplemented with 100 mM DMSO and 2% (wt/vol) glucose and transferred to Wheaton bottles (150 mL). Bottles were sealed with butanol rubber stoppers and incubated overnight at 42 °C. Growth was monitored at OD600.
Generation of Knockout Strains.
Target genes were deleted from the H. volcanii chromosome using an established pyrE2-based pop-in/pop-out method (26, 27). Mutants were confirmed by PCR, DNA sequencing and Southern blot as previously described (29).
Immunoblot.
Cells were harvested by centrifugation (14,000 × g, 10 min, 25 °C), resuspended in SDS loading buffer [100 mM Tris-Cl buffer at pH 6.8 with 2% (wt/vol) SDS, 10% (vol/vol) glycerol, 0.6 mg·mL−1 bromophenol blue, and 2.5% (vol/vol) β-mercaptoethanol] and boiled for 20 to 30 min. Proteins were separated by SDS-PAGE (10 or 12%) and electroblotted onto PVDF membranes (Amersham). Equivalent protein loading was determined by OD600 of cell culture (0.065 units per lane) and confirmed by staining parallel gels with Coomassie blue. Epitope-tagged proteins were detected by immunoblot using alkaline phosphatase-linked anti-Flag M2 monoclonal antibody (Sigma) or rabbit anti-StrepII polyclonal antibody (GenScript) combined with goat anti-rabbit IgG (H+L)-alkaline phosphatase-linked antibody (SouthernBiotech). Alkaline phosphatase activity was detected colorimetrically using nitroblue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and by chemiluminescence using CDP-Star (Applied Biosystems) with X-ray film (Hyperfilm; Amersham Biosciences).
DMSO Reductase Activity Assay.
Cells (15 mL culture) were harvested by centrifugation (6,000 × g, 20 min at 4 °C), washed in 15 mL buffer A (50 mM Tris pH 7.5, 1 mM EDTA pH 8.0, 2 M NaCl), resuspended in 1 mL buffer A, and lysed by sonication (4 × 20 s at 140 W). Cell lysate was clarified by centrifugation (14,000 × g, 20 min at 4 °C), and protein concentration was determined using the Bradford assay with BSA as a standard (BioRad). DMSO reductase activity was monitored at A600 nm (15-s intervals for 3.5 min) with nitrogen as the headspace. Assays (4 mL) included cell lysate (1–1.5 mg protein) and 0.3 mM methyl viologen in buffer A. The mixture was titrated with fresh 20 mM sodium dithionite (Na2S2O4) in 20 mM sodium bicarbonate (NaHCO3) to 1 to 1.2 A600 nm units before addition of 10 mM DMSO. One unit (U) of enzyme activity is defined as 1-μmol substrate consumed per minute at room temperature with an extinction coefficient A600 nm of 13.6 (mM−1·cm−1) for methyl viologen. All assays were performed in biological triplicate with the means ± SDs calculated.
RNA Isolation.
Total RNA was isolated for RT-(q)PCR (see SI Appendix, Figs. S5 and S10 for details) using the RNeasy Mini Kit (Qiagen) with a typical yield of 10 to 15 μg RNA per 1 mL culture. RNA samples were treated with RNase-free DNase (Qiagen) to remove genomic DNA contamination as confirmed by PCR. For tRNA thiolation assays, total RNA was extracted, as previously described (44), from log-phase cells grown in ATCC 974 medium (37.5 mL of 100 mL culture in a 500-mL flask; 42 °C at 200 rpm). RNA was further purified by extraction with equal volume of acidic-phenol (pH 5.0): chloroform: isoamyl alcohol (25:24:1) followed by chloroform: isoamyl alcohol (24:1). RNA was precipitated in 0.25 M sodium acetate (pH 5.0) with two volumes of 95% ethanol (−70 °C, 15 min) and washed with 70% ethanol. The air-dried RNA pellet was resuspended in 30 μL DEPC-treated water with a typical yield of 100 to 150 μg RNA. RNA quality was assessed by agarose gel electrophoresis, and RNA concentration was determined by A260 nm.
Assay for tRNA Thiolation.
APM-gel retardation analysis of tRNA was performed as follows. Total RNA (10 μg per lane) was separated by electrophoresis using 12% urea-PAGE gels supplemented with 30 μg APM per milliliter. RNA was transferred to a Hybond N+ nylon membrane (GE Healthcare) and immobilized by UV-crosslinking before hybridization. The oligonucleotide probe (see SI Appendix, Table S1 for details) was 5′ end-labeled using T4 polynucleotide kinase and [γ-32P]ATP. Excess [γ-32P]ATP was removed by passing the reaction through a MicroSpin G-25 column (GE Healthcare). The membrane was prehybridized in ULTRAhyb-Oligo buffer (Ambion) for 30 min at 42 °C before addition of the end-labeled oligonucleotide (~106 cpm per milliliter). After hybridization at 42 °C for 14 h, the membrane was washed twice with buffer consisting of 2× SSC and 0.5% SDS (for 30 min each time at 42 °C). The blot was exposed to an imaging plate (FujiFilm) and scanned on a Molecular Dynamics Storm 860 Phosphoimager (GE Healthcare).
Supplementary Material
Acknowledgments
Thanks to T. Allers for Haloferax volcanii strain H26 and plasmid pTA131; S. Shanker and the staff at the University of Florida Interdisciplinary Center for Biotechnology Research for DNA sequencing. This work was funded in part by National Institutes of Health Grants GM57498 (to J.A.M.-F.) and GM22854 (to D. Söll), and the Department of Energy Office of Basic Energy Sciences Grants DE-FG02-05ER15650 (to J.A.M.-F.) and DE-FG02-98ER20311 (to D. Söll).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018151108/-/DCSupplemental.
References
- 1.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shukla A, Chaurasia P, Bhaumik SR. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol Life Sci. 2009;66:1419–1433. doi: 10.1007/s00018-008-8605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: The extraordinary functional and structural diversity of the β-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like β-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatogawa H, Oh-oka K, Ohsumi Y. Lipidation of Atg8: How is substrate specificity determined without a canonical E3 enzyme? Autophagy. 2008;4:911–913. doi: 10.4161/auto.6646. [DOI] [PubMed] [Google Scholar]
- 7.Burroughs AM, Balaji S, Iyer LM, Aravind L. A novel superfamily containing the β-grasp fold involved in binding diverse soluble ligands. Biol Direct. 2007;2:4. doi: 10.1186/1745-6150-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: Implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessler D. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol Rev. 2006;30:825–840. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 10.Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem. 2000;275:7462–7465. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 11.Goehring AS, Rivers DM, Sprague GF., Jr Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell. 2003;2:930–936. doi: 10.1128/EC.2.5.930-936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci USA. 2008;105:18255–18260. doi: 10.1073/pnas.0808756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz J, et al. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47:6479–6489. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- 15.Leidel S, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 16.Huang B, Lu J, Byström AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedrioli PG, Leidel S, Hofmann K. Urm1 at the crossroad of modifications. ‘Protein Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep. 2008;9:1196–1202. doi: 10.1038/embor.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem. 2008;283:27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 19.Van der Veen AG, et al. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci USA. 2011;108:1763–1770. doi: 10.1073/pnas.1014402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humbard MA, et al. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang DT, et al. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xi J, Ge Y, Kinsland C, McLafferty FW, Begley TP. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: Identification of an acyldisulfide-linked protein-protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proc Natl Acad Sci USA. 2001;98:8513–8518. doi: 10.1073/pnas.141226698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- 24.McLuskey K, Harrison JA, Schuttelkopf AW, Boxer DH, Hunter WN. Insight into the role of Escherichia coli MobB in molybdenum cofactor biosynthesis based on the high resolution crystal structure. J Biol Chem. 2003;278:23706–23713. doi: 10.1074/jbc.M301485200. [DOI] [PubMed] [Google Scholar]
- 25.Johnson JL, Bastian NR, Rajagopalan KV. Molybdopterin guanine dinucleotide: A modified form of molybdopterin identified in the molybdenum cofactor of dimethyl sulfoxide reductase from Rhodobacter sphaeroides forma specialis denitrificans. Proc Natl Acad Sci USA. 1990;87:3190–3194. doi: 10.1073/pnas.87.8.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bitan-Banin G, Ortenberg R, Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J Bacteriol. 2003;185:772–778. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson JL, et al. Growth kinetics of extremely halophilic archaea (family halobacteriaceae) as revealed by arrhenius plots. J Bacteriol. 2005;187:923–929. doi: 10.1128/JB.187.3.923-929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou G, Kowalczyk D, Humbard MA, Rohatgi S, Maupin-Furlow JA. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:8096–8105. doi: 10.1128/JB.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shigi N, Sakaguchi Y, Suzuki T, Watanabe K. Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J Biol Chem. 2006;281:14296–14306. doi: 10.1074/jbc.M511675200. [DOI] [PubMed] [Google Scholar]
- 31.Rogers KC, Crescenzo AT, Söll D. Aminoacylation of transfer RNAs with 2-thiouridine derivatives in the wobble position of the anticodon. Biochimie. 1995;77:66–74. doi: 10.1016/0300-9084(96)88106-5. [DOI] [PubMed] [Google Scholar]
- 32.Igloi GL. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry. 1988;27:3842–3849. doi: 10.1021/bi00410a048. [DOI] [PubMed] [Google Scholar]
- 33.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414:325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 34.Sauerwald A, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Sieprawska-Lupa M, Whitman WB, White RH. Cysteine is not the sulfur source for iron-sulfur cluster and methionine biosynthesis in the methanogenic archaeon Methanococcus maripaludis. J Biol Chem. 2010;285:31923–31929. doi: 10.1074/jbc.M110.152447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudolph MJ, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat Struct Biol. 2001;8:42–46. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 37.Rudolph MJ, Wuebbens MM, Turque O, Rajagopalan KV, Schindelin H. Structural studies of molybdopterin synthase provide insights into its catalytic mechanism. J Biol Chem. 2003;278:14514–14522. doi: 10.1074/jbc.M300449200. [DOI] [PubMed] [Google Scholar]
- 38.Daniels JN, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal structure of a molybdopterin synthase-precursor Z complex: Insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency. Biochemistry. 2008;47:615–626. doi: 10.1021/bi701734g. [DOI] [PubMed] [Google Scholar]
- 39.Jäger G, Leipuviene R, Pollard MG, Qian Q, Björk GR. The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica serovar Typhimurium. J Bacteriol. 2004;186:750–757. doi: 10.1128/JB.186.3.750-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dewez M, et al. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci USA. 2008;105:5459–5464. doi: 10.1073/pnas.0709404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura Y, Tanaka K. Regulatory mechanisms involved in the control of ubiquitin homeostasis. J Biochem. 2010;147:793–798. doi: 10.1093/jb/mvq044. [DOI] [PubMed] [Google Scholar]
- 42.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 43.Dyall-Smith M. The Halohandbook: Protocols for Haloarchaeal Genetics. 2008 Available at http://www.haloarchaea.com/resources/halohandbook/Halohandbook_2008_v7.pdf. [Google Scholar]
- 44.Nieuwlandt DT, et al. Archaea: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 161–162. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.