Abstract
One in four proteins in Plasmodium falciparum contains asparagine repeats. We probed the function of one such 28-residue asparagine repeat present in the P. falciparum proteasome lid subunit 6, Rpn6. To aid our efforts, we developed a regulatable, fluorescent affinity (RFA) tag that allows cellular localization, manipulation of cellular levels, and affinity isolation of a chosen protein in P. falciparum. The tag comprises a degradation domain derived from Escherichia coli dihydrofolate reductase together with GFP. The expression of RFA-tagged proteins is regulated by the simple folate analog trimethoprim (TMP). Parasite lines were generated in which full-length Rpn6 and an asparagine repeat-deletion mutant of Rpn6 were fused to the RFA tag. The knockdown of Rpn6 upon removal of TMP revealed that this protein is essential for ubiquitinated protein degradation and for parasite survival, but the asparagine repeat is dispensable for protein expression, stability, and function. The data point to a genomic mechanism for repeat perpetuation rather than a positive cellular role. The RFA tag should facilitate study of the role of essential genes in parasite biology.
Keywords: malaria, conditional expression, polyasparagine, amino acid repeat, trinucleotide repeat
Malaria is a parasitic disease that causes high mortality and morbidity in tropical and subtropical regions of the world (1). The causative agents of malaria in humans are intracellular parasites of the genus Plasmodium. The most deadly of these parasites is Plasmodium falciparum. The proteome of P. falciparum is replete with amino acid repeats; about 25% of all proteins in P. falciparum contain asparagine repeats (2). These repeats are found in proteins with diverse functions and structures (2). Proteins containing asparagine repeats are prone to forming amyloid fibrils (3); however, despite their abundance, there is no evidence that P. falciparum proteins form these tangles in vivo. Many hypotheses have been proposed to explain the function of these repeats and how they could have arisen (4, 5), but there is a paucity of experimental evidence to support any of the suggestions. It has been proposed that asparagine repeats in the P. falciparum proteome are evolving under positive selection (6). Alternatively, it has been suggested that the propagation and evolution of these repeats is occurring primarily through neutral mechanisms (4). We set out to determine what role, if any, these repeats play in protein function and other aspects of cellular biology within the parasite.
Ideally, one would want to study a protein that has a single, large asparagine repeat, a fairly small size, and a predicted function that provides some clues as to its biological role. One protein that fulfills these criteria is the proteasome lid regulatory subunit 6, Rpn6 (PlasmoDB ID: PF14_0025). The Rpn6 protein has a single, large 28-residue asparagine repeat that is present in a Plasmodium-specific insertion. Rpn6 is a nonenzymatic component of the proteasome. It is thought to be important for the assembly of the proteasome in yeast (7). In Drosophila melanogaster (8) and Trypanosoma brucei (9) this protein was found to be essential for the survival of the organism. It has a clear predicted function, although the homology with Rpn6 proteins from other organisms is low (Fig. S1). Its function in P. falciparum is unknown.
The tools available to study the function of genes, especially essential genes, in P. falciparum are limited. Because of the haploid nature of the parasite, it often is not feasible or informative to use knockout technology to study essential genes, even though this technology is well established (10). Conditional expression systems such as the tetracycline-repressor system (11) have not been generally useful. FK506-binding protein degradation domain (ddFKBP)-based conditional expression was reported recently to work in the parasite (12–14). However, the small molecule that is used to stabilize the ddFKBP, Shield-1, is expensive and is somewhat toxic to the parasite at the concentrations required for protein stabilization (13).
A degradation domain based on the Escherichia coli dihydrofolate reductase (DHFR) enzyme was reported recently (15). The DHFR degradation domain (DDD) is stabilized by inexpensive folate analogs such as trimethoprim (TMP) (15). We combined the DDD with GFP and HA sequences to make a regulatable fluorescent affinity (RFA) tag (Fig. 1A) that allows us to probe aspects of protein biology such as its localization and interacting partners.
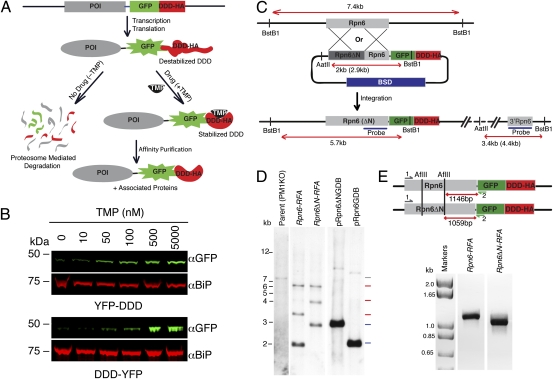
Fig. 1.
Utilizing the DDD and the RFA tag in P. falciparum. (A) Outline of the RFA tag scheme. The protein of interest (POI) is RFA-tagged (GFP+DDD+HA), allowing protein knockdown, live fluorescence microscopy, and affinity purification of associated proteins. (B) Stabilization of DDD by TMP. Synchronized ring-stage parasites transfected with YFP-DDD (Upper) or DDD-YFP (Lower) were incubated for 24 h with different TMP concentrations (as indicated). Western blots of whole-cell protein extracts were probed with anti-GFP antibody. BiP was the loading control. (C) Scheme showing the strategy used to incorporate the RFA tag at the 3′ end of the endogenous locus by single-crossover homologous integration. Plasmid (pRpn6GDB or pRpn6ΔNGDB) was transfected into the parent strain, PM1KO. The plasmids contained a BSD cassette for positive drug selection. BstBI and AatII restriction sites and the probe used to detect integration along with the expected sizes are indicated (in brackets for Rpn6ΔN). (D) Southern blots of genomic DNA and plasmid DNA digested with BstBI and AatII. Bands expected from a single-crossover recombination event were seen in Rpn6-RFA and Rpn6ΔN-RFA integrant clones (red lines). The plasmid bands also were seen in Rpn6-RFA and Rpn6ΔN-RFA integrant clones (blue lines), suggesting that a plasmid concatamer integrated into the gene, a common occurrence in P. falciparum. A single band was seen for the parental strain (PM1KO, gray line) that was absent in the integrant clones. This lane was exposed for a longer time than the other lanes to visualize the band. The expected bands also were seen for the plasmid controls pRpn6GDB and pRpn6ΔNGDB (blue lines). (E) PCR products from Rpn6-RFA and Rpn6ΔN-RFA genomic DNA, generated by the primers 1 and 2. (Upper) Map showing AflII restriction sites and the expected size of restriction fragments. (Lower) Restriction fragments generated after AflII digest of PCR products. The 87-bp difference between Rpn6-RFA and Rpn6ΔN-RFA indicates deletion of the asparagine repeat.
In this study, we report the use of the DDD in P. falciparum, the development of the RFA tag for studying Rpn6 function, and the role of the Rpn6 asparagine repeat.
Results
Validation of the DDD in P. falciparum.
We assessed episomal expression of YFP tagged with the E. coli DDD in P. falciparum. C-terminal and N-terminal fusions were driven by the constitutive heat shock protein 86 (hsp86) promoter (YFP-DDD and DDD-YFP, respectively). Parasites were grown in the presence of varying TMP concentrations for 24 h to test the ability of the ligand to stabilize the DDD in the parasite. Whole-cell protein extracts from these parasites then were analyzed by Western blots probed with anti-GFP antibody (Fig. 1B). Parasites transfected with DDD-YFP showed a nearly complete absence of YFP when grown with no TMP, whereas YFP-DDD parasites showed a minimal amount of protein with no TMP (Fig. 1B). In both cases, there was a dose-dependent increase in protein stabilization.
RFA Tagging of Rpn6 and Rpn6 Asparagine Repeat Deletion Mutant.
Several tags have been used to study protein function in P. falciparum. Individually these tags allow the study of one aspect of protein function. We wanted to construct an all-inclusive system that allowed protein regulation, live fluorescence microscopy, and affinity purification. For this purpose we generated a plasmid (termed “pGDB”) in which GFP (in place of YFP in the episomal constructs above) was cloned in frame with the C-terminal DDD and an HA tag at its 3′ end to make the RFA tag (Fig. 1A and Fig. S2). The plasmid also contained a blasticidin (BSD) resistance cassette that allows us to select for plasmid integration and protein stabilization using two orthogonal drugs (Fig. S2).
We used a single-crossover homologous recombination strategy to fuse the RFA tag to the endogenous Rpn6 gene, PF14_0025 (Fig. 1C). To generate Rpn6-RFA parasites, a 1.1-kb sequence at the 3′ end of the Rpn6 coding sequence was cloned in frame with the RFA tag in pGDB vector, creating the plasmid pRpn6GDB (Materials and Methods). To delete the asparagine repeat (amino acids 425–453), the complete coding sequence of Rpn6 was cloned into the pGDB vector in frame with the RFA tag, and the deletion mutant was generated via site-directed mutagenesis, creating the plasmid pRpn6ΔNGDB.
TMP, the drug that is used to stabilize the DDD, is toxic to parasites (Fig. S3). We therefore transfected pRpn6GDB and pRpn6ΔNGDB into parasites containing a human DHFR (hDHFR) marker integrated via double crossover into a nonessential gene, Plasmepsin I (PM1KO) (16). The presence of the hDHFR marker renders the parasites resistant to TMP (Fig. S3). After two cycles on and off BSD, successful integration of the RFA tag was obtained for both Rpn6 and Rpn6ΔN (Fig. 1D), creating Rpn6-RFA and Rpn6ΔN-RFA parasite lines. During drug cycling, parasites always were grown in the presence of 5 μM TMP to stabilize Rpn6 protein in parasites in which the RFA tag had integrated into the gene. The deletion of the asparagine repeat at the Rpn6 gene locus in Rpn6ΔN-RFA was confirmed by sequencing as well as by AflII digest of the PCR product obtained using a primer at the 5′ end of the Rpn6 gene and a reverse primer corresponding to the 5′ end of GFP (Fig. 1E).
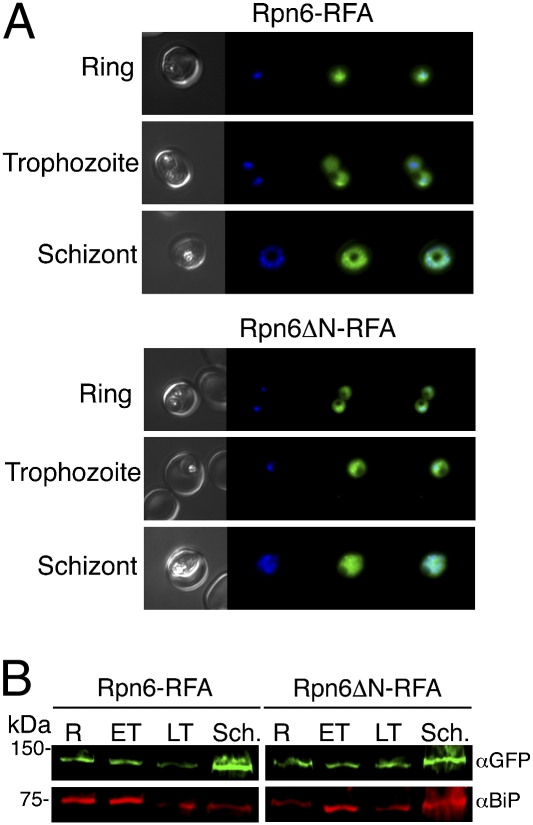
In Rpn6-RFA and Rpn6ΔN-RFA, the presence of the RFA-tagged proteins was detected via live fluorescence microscopy (Fig. 2A) and via Western blot of parasite extracts probed with anti-GFP (Fig. 2B) and anti-HA (Fig. S4). Rpn6 and Rpn6ΔN were expressed in all intraerythrocytic life cycle stages and were located in the cytoplasm (Fig. 2).
Fig. 2.
Intraerythrocytic expression of Rpn6 and Rpn6ΔN. (A) Stages of the erythrocytic life cycle of Rpn6-RFA (Upper) and Rpn6ΔN-RFA (Lower) parasites observed by live fluorescence microscopy. Images left to right are phase, DAPI, GFP, and fluorescence merge. (B) Western blot of lysates from different erythrocytic life cycle stages of Rpn6-RFA (Left) and Rpn6ΔN-RFA (Right) parasites, probed with anti-GFP antibody. BiP is the loading control. ET, early trophozoites; LT, late trophozoites; R, rings; Sch., schizonts.
Stabilization of Rpn6-RFA and Rpn6ΔN-RFA with TMP.
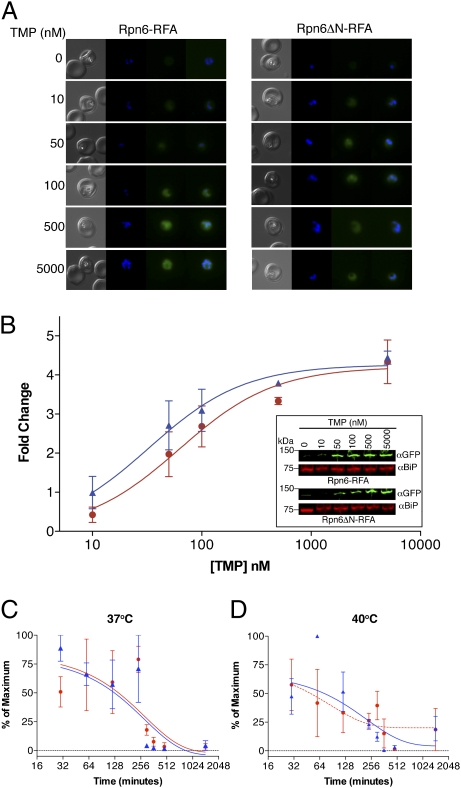
To assess the efficacy of the DDD within the RFA tag, synchronized early-trophozoite-stage Rpn6-RFA and Rpn6ΔN-RFA parasites were incubated with varying amounts of TMP for 24 h. We monitored protein levels by live fluorescence microscopy (Fig. 3A) and by Western blots (Fig. 3B). There was a dose-dependent response to TMP (Fig. 3 A and B). Synchronized early-trophozoite-stage Rpn6-RFA and Rpn6ΔN-RFA parasites were washed and incubated in medium without TMP to test the time course of protein degradation (Fig. 3 C and D). The half-life of tagged Rpn6 was about 6 h. Thus, the RFA tag can promote rapid degradation of fused proteins upon removal of the stabilizing ligand TMP.
Fig. 3.
Regulation of RFA-tagged proteins. (A) Live fluorescence images of Rpn6-RFA (Left) and Rpn6ΔN-RFA (Right) parasites incubated for 24 h in different TMP doses (as indicated). Images left to right are phase, DAPI, GFP, and fluorescence merge. (B) Stabilization of Rpn6-RFA (red filled circle) and Rpn6ΔN-RFA (dark blue filled triangle) by TMP. Data are shown as fold change over the no-TMP sample. GFP signal from Western blots was normalized against BiP. (Inset) One of four independent TMP dose–response Western blots. BiP served as the loading control. (C and D) Change in the amount of protein over time when Rpn6-RFA (red filled circle) and Rpn6ΔN-RFA (dark blue filled triangle) parasites were incubated without TMP and quantified in three independent experiments. (Sample Western blots are shown in Fig. S5.) BiP served as the loading control. The experiment was done at two temperatures, 37 °C (C) and 40 °C (D). Error bars represent SE from three independent experiments.
Asparagine repeats are known to affect protein stability and induce amyloid fibril formation (3). We compared Rpn6-RFA and Rpn6ΔN-RFA protein levels at different TMP concentrations (Fig. 3 A and B) and at different times after removal of TMP (Fig. 3 C and D). Protein stability also can be affected by stress, especially heat shock, and P. falciparum is known to be very sensitive to heat shock stress (17). Therefore Rpn6-RFA and Rpn6ΔN-RFA protein half-lives also were measured at 40 °C (Fig. 3D). Stability at an intermediate TMP concentration, 100 nM, was tested also (Fig. S5). In all cases, levels of tagged protein with or without the asparagine repeat were similar.
Rpn6 Is an Essential Protein.
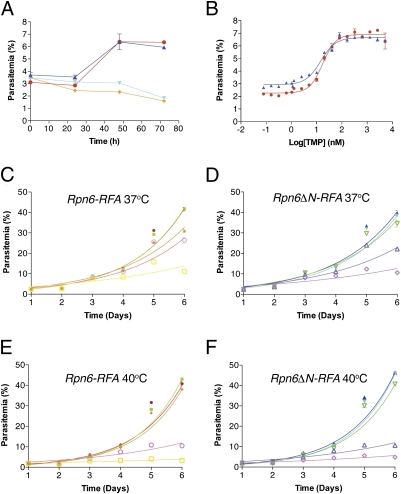
Rpn6 is known to be essential for growth in some organisms (8, 9), but the importance of this protein in P. falciparum is not known. We grew synchronized ring-stage Rpn6-RFA and Rpn6ΔN-RFA in the presence or absence of 5 μM TMP and monitored parasite growth by flow cytometry over several days (Fig. 4A). In the absence of TMP, parasites did not grow, and no live parasites were seen when Giemsa-stained blood smears were observed by microscopy. Parasite growth also was measured after 48 h in different TMP doses; growth was dependent on the presence of TMP with an EC50 of about 15 nM (Fig. 4B). Rpn6 is essential for the intraerythrocytic growth of P. falciparum.
Fig. 4.
Rpn6 is an essential gene. (A) Synchronous ring-stage parasites were grown with (Rpn6-RFA, red filled circle; Rpn6ΔN-RFA. dark blue filled triangle) or without (Rpn6-RFA, orange filled diamond; Rpn6ΔN-RFA, light blue inverted filled triangle) 5 μM TMP, and their growth was monitored over 3 d by flow cytometry. (B) Asynchronous Rpn6-RFA (red filled circle) and Rpn6ΔN-RFA (dark blue filled triangle) parasites were incubated with different TMP concentrations, and their growth was measured after 48 h by flow cytometry. (C) Asynchronous Rpn6-RFA parasites were incubated with different TMP concentrations [5 μM (red filled circle); 0.312 μM (green filled square); 0.156 μM (orange filled diamond); 0.078 μM (open purple circle); or 0.039 μM (open yellow square)] at 37 °C, and their growth was monitored over 6 d by flow cytometry. (D) Asynchronous Rpn6ΔN-RFA parasites were incubated with different TMP concentrations [5 μM (dark blue filled triangle); 0.312 μM (light blue inverted filled triangle); 0.156 μM (inverted open green triangle); 0.078 μM (open purple triangle); and 0.039 μM (open purple diamond)] at 37 °C, and their growth was monitored over 6 d by flow cytometry. (E) Asynchronous Rpn6-RFA parasites were incubated with different TMP concentrations (symbols as in C) and were heat shocked for 6 h at 40 °C. After 6 h the cultures were transferred to 37 °C, and their growth was monitored over 6 d by flow cytometry. (F) Asynchronous Rpn6ΔN-RFA parasites were incubated with different TMP concentrations (symbols as in D) and were heat shocked for 6 h at 40 °C. After 6 h the cultures were transferred to 37 °C, and their growth was monitored over 6 d by flow cytometry. Error bars represent SE from one of three independent experiments done in triplicate.
To test if the presence of the asparagine repeat in Rpn6 has an effect on the ability of parasites to deal with a global protein unfolding stress, Rpn6-RFA and Rpn6ΔN-RFA were incubated at 37 °C or 40 °C for 6 h, then were maintained at 37 °C, and their growth was monitored over several days (Fig. 4 C–F). There was no difference in the growth characteristics of the two parasite lines either with or without heat shock. There was a threshold of TMP concentration, about 150 nM, that was absolutely required to deal with the heat shock stress, and this threshold was the same for both Rpn6-RFA and Rpn6ΔN-RFA. The parasite requires Rpn6 for growth within the red blood cell.
Asparagine Repeat of Rpn6 Is Dispensable for Proteasome Function.
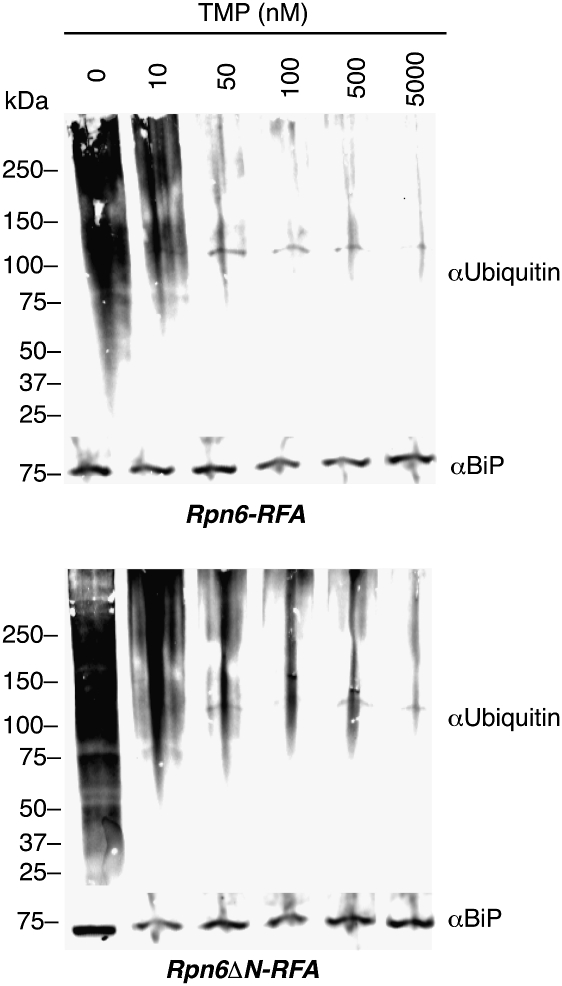
Rpn6 is part of the proteasome lid. To test if its essential nature results from a role in proteasome function, we incubated synchronized early-trophozoite-stage Rpn6-RFA and Rpn6ΔN-RFA parasites with varying TMP concentrations. After 24 h, whole-cell protein lysates were analyzed for ubiquitinated proteins via Western blot (Fig. 5). Ubiquitinated proteins started to accumulate as the dose of TMP decreased and the levels of Rpn6 decreased (Fig. 5). Rpn6, but not the asparagine repeat, was essential for proper disposal of ubiquitinated proteins.
Fig. 5.
Ubiquitinated protein degradation depends on Rpn6. Synchronous early-trophozoite-stage Rpn6-RFA (Upper) and Rpn6ΔN-RFA (Lower) parasites were incubated for 24 h in different TMP concentrations (as indicated). Western blots of whole-cell protein extracts were probed with anti-ubiquitin antibody to assess the accumulation of ubiquitinated proteins in parasites. BiP was the loading control.
To test further if Rpn6 is a proteasome subunit in P. falciparum, Rpn6-associated proteins were isolated by pull down using an anti-GFP antibody and were identified by MS (Table 1). Profiles from Rpn6-RFA and Rpn6ΔN-RFA parasites were nearly identical. We identified nearly 30 proteasome subunits in pull downs with whole-cell protein extracts from both Rpn6-RFA and Rpn6ΔN-RFA parasites. This result strongly suggests that Rpn6 is indeed a component of the proteasome and that the asparagine repeat does not affect its associations.
Table 1.
Proteins associated with Rpn6-RFA and Rpn6ΔN-RFA
| PlasmoDB I.D. | Protein function | Unique peptides with Rpn6-RFA | Unique peptides with Rpn6ΔN-RFA |
| PF14_0632 | 26S proteasome subunit | 29 | 62 |
| PFB0260w | Proteasome 26S regulatory subunit | 28 | 42 |
| PFL2345c | Tat-binding protein homolog (proteasome subunit) | 28 | 29 |
| MAL13P1.190 | Proteasome regulatory component | 22 | 23 |
| PFD0665c | 26S proteasome AAA-ATPase subunit RPT3 | 23 | 22 |
| PF11_0314 | 26S protease subunit regulatory subunit 6a | 21 | 26 |
| PF13_0063 | 26S proteasome regulatory subunit 7 | 17 | 17 |
| PF14_0025 | Proteasome subunit, Rpn6 | 25 | 32 |
| PF11_0303 | 26S proteasome regulatory complex subunit | 19 | 23 |
| PF10_0081 | 26S proteasome regulatory subunit 4 | 17 | 22 |
| PF10_0298 | 26S proteasome subunit | 19 | 21 |
| PF08_0109 | Proteasome subunit α type 5 | 12 | 18 |
| PF13_0033 | 26S proteasome regulatory subunit | 18 | 19 |
| MAL8P1.128 | Proteasome subunit α | 13 | 11 |
| MAL13P1.343 | Proteasome regulatory subunit | 15 | 14 |
| PF10_0174 | 26s proteasome subunit p55 | 12 | 21 |
| PF07_0112 | Proteasome subunit α type 5 | 10 | 10 |
| PF08_0054 | heat shock 70-kDa protein | 13 | 11 |
| PF14_0716 | Proteasome subunit α type 1 | 8 | 7 |
| PFI0630w | 26S proteasome regulatory subunit | 8 | 10 |
| MAL13P1.270 | Proteasome subunit | 8 | 7 |
| PFC0520w | 26S proteasome regulatory subunit S14 | 7 | 10 |
| PF11_0177 | Deubiquinating/deneddylating enzyme | 7 | 11 |
| PF13_0304 | Elongation factor-1 α | 9 | 6 |
| PFC0745c | Proteasome component C8 | 6 | 6 |
| PFL2215w | Actin I | 5 | 3 |
| PFI1545c | Proteasome precursor | 6 | 5 |
| PFF0420c | Proteasome subunit α type 2 | 7 | 9 |
| PF14_0676 | 20S proteasome β 4 subunit | 6 | 8 |
| MAL8P1.142 | Proteasome β-subunit | 6 | 3 |
| PFE0915c | Proteasome subunit β type 1 | 6 | 10 |
| PF13_0282 | Proteasome subunit | 4 | 5 |
| PF13_0346 | 60S ribosomal protein L40/UBI | 3 | 6 |
| PF14_0138 | Conserved protein | 5 | 6 |
| MAL13P1.351 | Conserved Plasmodium protein | 13 | |
| PFL0340w | Conserved Plasmodium protein | 5 | |
| PF14_0598 | Glyceraldehyde-3-phosphate dehydrogenase | 6 |
All proteins for which more than four unique peptides were identified in Rpn6-RFA or Rpn6ΔN-RFA pulldowns are shown.
Several proteasome inhibitors have been well characterized, and some, such as MG132, have been shown to be toxic to P. falciparum (18). The toxicity of MG132 is assumed to result from its inhibition of the proteasome, which is required for parasite growth. We tested MG132 for its effect on parasite growth after 24 h in different TMP doses (Fig. S6). Different TMP doses correspond to different Rpn6 levels, and Rpn6 levels have a direct effect on proteasome function. We expected parasites in higher TMP doses to be more resistant to MG132, and vice versa. However, the IC50 values for MG132 were unaffected by the TMP dose in both Rpn6-RFA and Rpn6ΔN-RFA parasites (Fig. S6C). This result suggests that MG132 kills parasites not by its inhibition of proteasome function but via some other mechanism.
Discussion
The study of essential genes in P. falciparum has met with limited success. The only conditional expression technology that is widely applicable in the parasite is the ddFKBP-based system (12–14), whose ligand is prohibitively expensive and toxic to the parasite (13). In this study, we tested the feasibility of using the DDD in P. falciparum. The DDD is stabilized by a commercially available and inexpensive ligand, TMP. We found that this domain can modulate protein levels, whether episomally expressed or integrated into the genome.
Because of the difficulties in tagging genes in P. falciparum, we wanted to use the DDD in combination with other tags to provide a one-stop approach for functional characterization of genes. In this work, we used GFP and an HA sequence along with the C-terminal DDD to generate the RFA tag (Fig. 1A). We used the RFA tag to tackle an important biological question in the malaria parasite. About one fourth of the entire proteome of P. falciparum contains asparagine repeats (2). Although most of the work on these repeats has relied on bioinformatic and statistical analysis (2, 4–6), almost nothing is known about the effect of these repeats on the cellular function of proteins. In the present work we carried out detailed characterization of the asparagine repeat in P. falciparum Rpn6 protein. We chose to study the Rpn6 protein because it has a single, large, well-defined asparagine repeat (28 asparagines) and a predicted function. However, despite a predicted function, the homology of P. falciparum Rpn6 to that from other organisms is low, and the parasite Rpn6 contains a large insertion that could be involved in a parasite-specific function (Fig. S1).
Our results demonstrated that we could generate parasite lines in which Rpn6 was fused to the RFA tag (Fig. 1 C–E). The presence of GFP in the RFA tag allowed us to follow Rpn6 fused to it by live fluorescence microscopy (Fig. 2A), and HA allowed us to probe for the protein in Western blots (Fig. S4). Rpn6 is a highly expressed protein (Fig. 2B and Fig. S4) and is located within the cytoplasm of the parasite (Fig. 2A). The degradation of Rpn6-RFA and Rpn6ΔN-RFA was rapid upon TMP removal, disappearing within about 8 h, even under heat shock (Fig. 3 and Fig. S5).
The presence of the asparagine repeat did not affect expression levels, localization, or half-life of P. falciparum Rpn6. The ability to delete the asparagine repeat in the endogenous locus suggested either that Rpn6 is not essential or that the repeat is dispensable during intraerythrocytic growth. The latter possibility is supported by the fact that the removal of TMP results in parasite death (Fig. 4A), because TMP modulates protein levels via RFA-tag stabilization, and removal of TMP results in degradation of RFA-tagged Rpn6 protein within the cell (Fig. 3 A and B). Indeed, Rpn6-RFA and Rpn6ΔN-RFA parasite lines are completely dependent on TMP for their survival (Fig. 4 B–D) and for surviving a heat shock stress (Fig. 4 E and F).
Because Rpn6 is a subunit of the proteasome lid, we investigated its role in proteasome function. We found that it is required for disposal of ubiquitinated proteins by the proteasome (Fig. 5), suggesting that it is required for proper proteasome function. Indeed, we find that P. falciparum Rpn6 immunoprecipitation selectively brings down the entire proteasome (Table 1). This result suggests that the essentiality of the Rpn6 gene is tied to its requirement for proteasome function. The asparagine repeat in Rpn6 was not required for its interaction with other subunits of the proteasome, its function in the proteasome, or the survival of the parasite.
One protein, PF14_0598, was found only in the pulldown of the asparagine repeat-containing Rpn6. It is an abundant glycolytic protein that frequently comes down in pulldowns with unrelated proteins (19). The other two proteins come down only with the asparagine-deletion mutant. It is unlikely that the mutant has a gain of function (i.e., the asparagine repeat protects against association). Therefore, we do not believe that these proteins are significant differential interactors, although the current evidence does not rule out this possibility.
Our data suggest that the asparagine repeat in one highly expressed protein, Rpn6, does not have a discernable effect on protein function or cellular biology. It is, of course, possible that asparagine repeats may not have the same role in every protein in P. falciparum, but the results do support the notion that the evolution of these repeats is happening via protein-neutral mechanisms (5). Whether there is selection at the genome level is not addressed by our experiments (6). We have demonstrated the utility of the RFA tag in interrogating the role of the Rpn6 protein in P. falciparum biology. The data presented here establish the feasibility of using the RFA tag to study essential genes in P. falciparum. The RFA tag facilitates comprehensive analysis of gene product function in this important but difficult-to-study organism.
Materials and Methods
Plasmid Construction.
The episomal vector was constructed from the plasmid containing Plasmepsin II in frame with GFP (pPM2GT) (20) by inserting the Hsp86 promoter between the AatII and XhoI (New England Biolabs) sites. YFP-DDD and DDD-YFP were introduced into this vector using XhoI and EagI (New England Biolabs) sites.
The DDD (mutations: N18T and A19V) was amplified using the primer 5′-TACAAATAAGCGGCCGATCAGTCTGATTGCGGCGTTAGCGGTAGATCACG-3′ and another primer encoding the HA-tag, 5′-TAACTCGACGCGGCCGTCAAGCGTAATCTGGAACATCGTATGGGTATCGCCGCTCCAGAATCTCAAAGCAATAGCTGTGAGAG-3′. The PCR product was introduced into the PM2GT vector (20) using the EagI restriction site and the In-Fusion cloning system (Clontech) that ensured insertion in the proper orientation. The pGDB plasmid was generated by replacing the hDHFR cassette from pPM2GT (20) plasmid with a BSD cassette from pRZ-TK-BSD2 (21) using SalI and BglII (New England Biolabs) restriction sites. The integration plasmid, pRpn6GDB, was created by introducing a 1.1-kb PCR product from the 3′ end of the Rpn6 ORF into pGDB. The PCR was done using the forward primer 5′-CACTATAGAACTCGAGCTTCTCAAAATAAAACAAAAAGTATAAGTCAAATAAGTCCC-3′ and the reverse primer 5′- CTGCACCTGGCCTAGGTATCGTCAATTGAGCTTTTTCATATAATATGTCTACAGACTCC-3′. Inserting the entire 2-kb Rpn6 ORF (forward primer, 5′- CACTATAGAACTCGAGATGGATAACTTTGAAGAAATCGAAAGAGGTTACAAAGAAATAG-3′ and the same reverse primer as before) into pGDB created the integration plasmid, pRpn6ΔNGDB. The PCR products in both cases were introduced into pGDB between the XhoI/AvrII (New England Biolabs) restriction sites using the In-Fusion PCR cloning kit (Clontech). The Rpn6 deletion mutant Rpn6ΔN was generated using the QuikChange site directed mutagenesis kit (Stratagene) with the forward primer 5′-ACGTTAAATAGAGAAAATGAAAACATTGATCTAAAAAATGATAGTAGTAGTAGTAGTTTTTGTAGT-3′ and the reverse primer 5′-ACTACAAAAACTACTACTACTACTATCATTTTTTAGATCAATGTTTTCATTTTCTCTATTTAACGT-3′.
Parasite Culturing and Transfection.
Parasites were cultured and synchronized as described earlier (22). Transfections were carried out as described (23). Parasites transfected with episomally maintained plasmids (YFP-DDD and DDD-YFP) were selected and maintained as described earlier (23). Parasites transfected with the pGDB vectors for integration underwent positive selection 48 h after transfection with 2.5 μg/mL BSD (Calbiochem) and 5 μM TMP (Sigma). Integration was detected after two rounds of BSD cycling. TMP always was present in the medium after its initial introduction. Integrant clones were isolated by limiting dilution.
Southern Blot.
Genomic DNA for Southern blots was isolated using the Qiagen Blood and Cell Culture kit. Southern blots were performed as described earlier (20) using 1 μg of DNA digested overnight with the restriction enzymes AatII and BstBI (New England Biolabs). Integrations were screened using a probe for the 3′ end of the Rpn6 ORF.
Microscopy.
Live parasites were stained with 2 μM Hoescht 33342 (Molecular Probes) and were observed using an Axioscope Microscope (Carl Zeiss Microimaging) as described previously (20). The images were collected using a fixed exposure time in both GFP and DAPI channels for all samples. Images were analyzed and processed using ImageJ (National Institutes of Health) and Adobe Photoshop.
Western Blot, Protein Quantification, and Immunoprecipitation.
Parasites were collected, and the host erythrocytes were permeabilized selectively by treatment with cold 0.04% Saponin in PBS for 10 min, followed by a wash in PBS. Western blots were performed as described previously (20). Lysates from 1 × 107 parasites were loaded per lane. The antibodies used in this study were mouse monoclonal anti-GFP (1:4,000), JL8 (Clontech); mouse monoclonal anti-HA (1:3,000), 3F10 (Roche); mouse monoclonal anti-ubiquitin (1:1,000), P4D1 (Santa Cruz); rabbit polyclonal anti-binding immunoglobulin protein (anti-BiP) (1:20,000), MRA-20 (MR4; ATCC). The signal was detected using IRDye 680CW (1:15,000) conjugated donkey anti-rabbit IgG (LI-COR Biosciences) and IRDye 800CW (1:20,000) conjugated goat anti-mouse IgG (LI-COR Biosciences) on the Odyssey infrared imager (LI-COR Biosciences). The Western blot images were processed and analyzed and GFP and BiP signals were quantified using the Odyssey infrared imaging system software. The protein dose–response and decay data were fit using standard dose–response and single exponential decay equations in GraphPad Prism, v. 5.0 (nonlinear least-squares analysis).
For immunoprecipitation, 4 × 108 parasites were treated with cold 0.04% saponin in PBS for 10 min and washed once with PBS. The purified parasites were lysed with cold 0.5% Triton X-100 in PBS with protease inhibitor mixture (Roche), and the insoluble fraction was separated by centrifuging the lysates at 4,000 × g for 5 min at 4 °C. The soluble fraction was incubated with 150 μL Protein A Dynabeads (Invitrogen) and 0.5 μg mouse monoclonal anti-GFP, 3E6 (Invitrogen), for 1 h at 4 °C. The beads then were washed four times in PBS containing protease inhibitor mixture (Roche). The immunoprecipitated complex was solubilized with SDS/PAGE sample buffer, and the proteins were fractionated by 10% SDS/PAGE. The proteins resolved on 10% SDS/PAGE were excised, trypsinized, and identified by MS-MS (Fingerprints Proteomics Facility, College of Life Sciences, University of Dundee, Dundee, United Kingdom) (24). A pull down using anti-GFP from PM1KO (parent) lysates acted as the control.
Flow Cytometry.
Parasite culture aliquots (5 μL) were stained with 1.5 μg/mL Acridine Orange (Molecular Probes) in PBS, and the fluorescence profiles of infected erythrocytes were analyzed on a BD FACSCanto flow cytometer (BD Biosystems). The parasitemia data were fit to standard growth curve or dose–response equations in GraphPad Prism, v. 5.0 (nonlinear least-squares analysis).
Supplementary Material
Acknowledgments
We thank B. Vaupel for technical assistance, P. A. Sigala and J. P. Mallari for comments on the manuscript and D. Lamont and K. Beattie for assistance with mass spectrometry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018449108/-/DCSupplemental.
References
- 1.Breman JG. The ears of the hippopotamus: Manifestations, determinants, and estimates of the malaria burden. Am J Trop Med Hyg. 2001;64(Suppl1–2):1–11. doi: 10.4269/ajtmh.2001.64.1. [DOI] [PubMed] [Google Scholar]
- 2.Singh GP, et al. Hyper-expansion of asparagines correlates with an abundance of proteins with prion-like domains in Plasmodium falciparum. Mol Biochem Parasitol. 2004;137:307–319. doi: 10.1016/j.molbiopara.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Perutz MF, Pope BJ, Owen D, Wanker EE, Scherzinger E. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid beta-peptide of amyloid plaques. Proc Natl Acad Sci USA. 2002;99:5596–5600. doi: 10.1073/pnas.042681599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DePristo MA, Zilversmit MM, Hartl DL. On the abundance, amino acid composition, and evolutionary dynamics of low-complexity regions in proteins. Gene. 2006;378:19–30. doi: 10.1016/j.gene.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 5.Zilversmit MM, et al. Low-complexity regions in Plasmodium falciparum: Missing links in the evolution of an extreme genome. Mol Biol Evol. 2010;27:2198–2209. doi: 10.1093/molbev/msq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalby AR. A comparative proteomic analysis of the simple amino acid repeat distributions in Plasmodia reveals lineage specific amino acid selection. PLoS ONE. 2009;4:e6231. doi: 10.1371/journal.pone.0006231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isono E, Saito N, Kamata N, Saeki Y, Toh-E A. Functional analysis of Rpn6p, a lid component of the 26 S proteasome, using temperature-sensitive rpn6 mutants of the yeast Saccharomyces cerevisiae. J Biol Chem. 2005;280:6537–6547. doi: 10.1074/jbc.M409364200. [DOI] [PubMed] [Google Scholar]
- 8.Lier S, Paululat A. The proteasome regulatory particle subunit Rpn6 is required for Drosophila development and interacts physically with signalosome subunit Alien/CSN2. Gene. 2002;298:109–119. doi: 10.1016/s0378-1119(02)00930-7. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Wang CC. Functional characterization of the 11 non-ATPase subunit proteins in the trypanosome 19 S proteasomal regulatory complex. J Biol Chem. 2002;277:42686–42693. doi: 10.1074/jbc.M207183200. [DOI] [PubMed] [Google Scholar]
- 10.Crabb BS, et al. Transfection of the human malaria parasite Plasmodium falciparum. Methods Mol Biol. 2004;270:263–276. doi: 10.1385/1-59259-793-9:263. [DOI] [PubMed] [Google Scholar]
- 11.Meissner M, et al. Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators. Proc Natl Acad Sci USA. 2005;102:2980–2985. doi: 10.1073/pnas.0500112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat Methods. 2007;4:1007–1009. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- 13.Russo I, Oksman A, Vaupel B, Goldberg DE. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc Natl Acad Sci USA. 2009;106:1554–1559. doi: 10.1073/pnas.0806926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dvorin JD, et al. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010;328:910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwamoto M, Björklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol. 2010;17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Gluzman IY, Drew ME, Goldberg DE. The role of Plasmodium falciparum food vacuole plasmepsins. J Biol Chem. 2005;280:1432–1437. doi: 10.1074/jbc.M409740200. [DOI] [PubMed] [Google Scholar]
- 17.Kwiatkowski D. Febrile temperatures can synchronize the growth of Plasmodium falciparum in vitro. J Exp Med. 1989;169:357–361. doi: 10.1084/jem.169.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreidenweiss A, Kremsner PG, Mordmüller B. Comprehensive study of proteasome inhibitors against Plasmodium falciparum laboratory strains and field isolates from Gabon. Malar J. 2008;7:187. doi: 10.1186/1475-2875-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russo I, et al. Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature. 2010;463:632–636. doi: 10.1038/nature08726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klemba M, Beatty W, Gluzman I, Goldberg DE. Trafficking of plasmepsin II to the food vacuole of the malaria parasite Plasmodium falciparum. J Cell Biol. 2004;164:47–56. doi: 10.1083/jcb200307147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Bissati K, et al. The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite Plasmodium falciparum. Proc Natl Acad Sci USA. 2006;103:9286–9291. doi: 10.1073/pnas.0602590103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drew ME, et al. Plasmodium food vacuole plasmepsins are activated by falcipains. J Biol Chem. 2008;283:12870–12876. doi: 10.1074/jbc.M708949200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russo I, Oksman A, Goldberg DE. Fatty acid acylation regulates trafficking of the unusual Plasmodium falciparum calpain to the nucleolus. Mol Microbiol. 2009;72:229–245. doi: 10.1111/j.1365-2958.2009.06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.