Abstract
Aberrant promoter hypermethylation is frequently observed in cancer. The potential for this mechanism to contribute to tumor development depends on whether the genes affected are repressed because of their methylation. Many aberrantly methylated genes play important roles in development and are bivalently marked in ES cells, suggesting that their aberrant methylation may reflect developmental processes. We investigated this possibility by analyzing promoter methylation in 19 breast cancer cell lines and 47 primary breast tumors. In cell lines, we defined 120 genes that were significantly repressed in association with methylation (SRAM). These genes allowed the unsupervised segregation of cell lines into epithelial (EPCAM+ve) and mesenchymal (EPCAM−ve) lineages. However, the methylated genes were already repressed in normal cells of the same lineage, and >90% could not be derepressed by treatment with 5-aza-2′-deoxycytidine. The tumor suppressor genes APC and CDH1 were among those methylated in a lineage-specific fashion. As predicted by the epithelial nature of most breast tumors, SRAM genes that were methylated in epithelial cell lines were frequently aberrantly methylated in primary tumors, as were genes specifically repressed in normal epithelial cells. An SRAM gene expression signature also correctly identified the rare claudin-low and metaplastic tumors as having mesenchymal characteristics. Our findings implicate aberrant DNA methylation as a marker of cell lineage rather than tumor progression and suggest that, in most cases, it does not cause the repression with which it is associated.
Aberrant CpG island methylation occurs in cancer and is implicated in tumor progression (1), particularly when methylation of a tumor suppressor gene appears to phenocopy the equivalent genetic mutation. Examples include MLH1 methylation in sporadic microsatellite unstable colon cancer (2) and Rb in retinoblastoma (3).
Several tumor suppressor genes and putative tumor suppressor genes have been reported to be methylated in breast cancer (4), but in most cases, evidence for a functional role in tumorigenesis is lacking. BRCA1, which is mutated in familial breast cancer, is reported to be methylated in ~10% of sporadic tumors. In BRCA1-associated familial tumors, the wild-type BRCA1 allele is frequently lost. One report suggested that the loss of function could occur through methylation of the remaining wild-type allele (5), but this finding has not been supported by subsequent, larger studies (6, 7).
Breast development begins in embryonic life when epidermal cells of ectodermal origin project into the mesenchyme underlying the mammary ridge and form lactiferous ducts. Mammary stem cells give rise to both the inner luminal-epithelial and the outer “basal” myoepithelial cells of the lobulo-ductal system (8). Primary breast tumors can been subdivided into many different types by histology and by molecular profiling, but most tumors are thought to be epithelial in origin, deriving either from luminal-epithelial cells or from their progenitors (9).
It is known that many genes de novo methylated in cancer have “bivalent” histone marks (combined histone H3 lysine-27 and lysine-4 trimethylation) in embryonic stem (ES) cells (10). Because bivalently marked genes frequently have a role in development, we asked whether cancer-associated aberrant methylation might reflect the particular cell lineage from which a breast tumor was derived (its ontogeny). We show that aberrant DNA methylation occurs in genes down-regulated through normal lineage commitment and that the genes affected can be used to distinguish breast tumors of epithelial and mesenchymal lineage. We propose that most aberrant methylation reflects lineage commitment rather than tumor progression.
Results
DNA Methylation Occurs Variably Across Breast Cancer Cell Lines and Is Associated with Gene Repression.
We correlated promoter methylation with gene expression by analyzing 19 breast cancer cell lines on Infinium arrays and combining these results with published transcriptome data (11). Infinium arrays assay the proportion of 5-methylcytosine to total cytosine at 27,578 different CpG dinucleotides in >14,000 genes after bisulfite conversion (12). We validated the capacity of the Infinium arrays to detect changes in DNA methylation by using DNA from wild-type and DNA methyltransferase deficient HCT116 colon cancer cell lines (Fig. S1A). The methylation levels reported by the arrays also corresponded well to those assayed by bisulfite sequencing, both for the individual CpGs interrogated and for neighboring CpGs (Fig. S1 B and C). We restricted our analysis to probes within 200 bp of transcription start sites because we were interested in the effects of methylation on expression. As expected, genes associated with methylated promoters were less expressed than genes with unmethylated promoters (Fig. S1D).
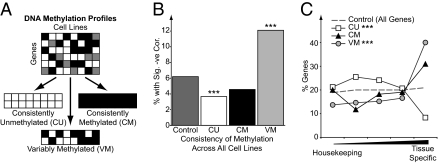
To understand the factors that might be influencing methylation in the cell lines, we categorized the CpG probes into three groups depending on their consistency of methylation across the cell lines (Fig. 1A and Materials and Methods) and determined the proportion of CpG island genes with each group. Most consistently unmethylated (CU) probes (3,901 genes) were located within CpG islands, whereas consistently methylated (CM) probes (259 genes) were mostly located at non-CpG island promoters (Fig. S1E). Variably methylated (VM) probes (1,023 genes) were significantly more likely to be in CpG islands than CM probes (51% vs. 24%). VM genes were frequently not expressed even when unmethylated, with 45% being unexpressed in all 19 cell lines (Fig. S1F). However, a significant proportion (12%) of the VM genes did show the expected inverse relationship between DNA methylation and expression (Fig. 1B).
Fig. 1.
VM genes have tissue-specific expression patterns. (A) An illustration of the general strategy used to segregate genes into sets with different methylation patterns. (B) The proportion of CU, CM, and VM genes, which show a significantly negative correlation between expression and methylation compared with the percentage found on the whole array (Fisher's exact tests). (C) The expression patterns of genes in different methyl gene sets in normal tissues as defined using a specificity score (SI Materials and Methods). The distributions of CU and VM genes were significantly different from the profiles of all genes (***P < 0.001, χ2 test).
Methylated and Variably Methylated Genes Have Tissue-Specific Expression Patterns.
We functionally characterized the gene groups using Gene Ontology (GO) terms (Fig. S1G). CU genes were associated with metabolic or housekeeping processes, whereas CM genes were associated with more specialized, lineage-restricted terms, such as meiosis, and mast cell activation. In contrast, VM genes were significantly associated with general developmental processes.
Given that genes with different methylation patterns were associated with different functions, we examined whether they also had different patterns of expression in normal tissues by scoring them according to their degree of tissue specificity (SI Materials and Methods). CU genes were significantly enriched in genes showing a housekeeping expression pattern (Fig. 1C). In contrast, VM genes were significantly enriched for tissue-specific expression. CM genes displayed a similar pattern to VM genes but did not quite reach significance (P = 0.065). The tissue specificity of VM genes was also apparent when VM genes with CpG island and non-CpG island promoters were analyzed separately (Fig. S1H).
CpGs That Are Variably Methylated in Cell Lines Are Frequently Unmethylated in Normal Breast Tissue and Normal Mammary Epithelial Cells.
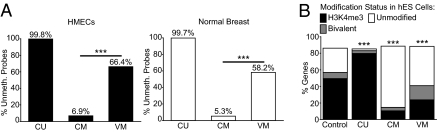
We next asked whether it was the CM or VM probes that could be regarded as aberrantly methylated in cancer because they were unmethylated in normal human mammary epithelial cells (HMEC) and normal breast tissue. CU probes were nearly always unmethylated in the normal DNA samples. A high proportion of VM probes (58–66%) were also unmethylated in these normal samples, and this was a significantly greater proportion than was found for the CM probes (5–7%, P < 2.2 × 10−16, Fisher's exact tests; Fig. 2A). As there were also ~4 times more VM genes (n = 1,023) than CM genes (n = 259), aberrant methylation was significantly more likely to occur at VM genes. VM probes were also more likely to be unmethylated than CM probes in a panel of nine normal tissues and in human ES (hES) cells (Fig. S2 A and B).
Fig. 2.
VM genes are usually unmethylated in normal breast tissues and are enriched for bivalent histone marks in hES cells. (A) The proportions of CU, CM, and VM probes that are unmethylated in either HMECs or the normal breast are shown. Significantly more VM than CM probes are unmethylated in the normal samples (Fisher's exact tests). (B) The proportions of CU, CM, and VM genes that have different histone modification patterns in hES cells are shown. All three groups show a distribution that is significantly different from the control (all genes on the array, χ2 tests). ***P < 0.001.
VM Genes Are Enriched for “Bivalent” Histone Marks in hES Cells.
Cancer-associated aberrant methylation frequently occurs at genes with bivalent histone marks in hES cells (histone H3K4me3 and H3K27me3; ref. 10). We noticed a striking similarity between functional terms associated with VM genes and those previously associated with bivalently marked genes in hES cells (Fig. S2C; ref. 13). We used data from this study to determine the histone marks associated with CU, CM, and VM genes in hES cells. A significant proportion of CU genes were marked by H3K4me3 alone (P < 2 × 10−16, Fisher's exact test), whereas most CM and VM genes lacked H3K4me3 and H3K27me3 (Fig. 2B). However, the VM group was significantly enriched for bivalent marks (16.9% of the total; P = 7 × 10−22, Fisher's exact test) compared with the control. This enrichment was not seen in the CM group.
Genes That Are Significantly Repressed in Association with Methylation (SRAM) Segregate Breast Cancer Cell Lines into Epithelial and Mesenchymal Lineages.
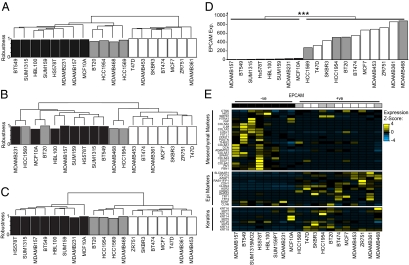
As VM genes were lineage-specific, we asked whether they could be used to categorize the cell lines according to lineage. The expression levels of the 1,000 most variably expressed genes segregated the 19 breast cancer cell lines into the previously described luminal, basal A, and basal B subtypes (Fig. 3A; ref. 11). However, hierarchical clustering using methylation levels of the 1,023 VM genes derived different groupings (Fig. 3B): Two of the basal A cell lines (MDAMB468 and HCC1954) now clustered with the luminal cell lines. As not all VM genes showed a good correlation with repression (Fig. 1B), we repeated the analysis using the expression levels of those VM genes that were significantly repressed in association with methylation (SRAM; 120 genes; Fig. 3C and Dataset S1). In this analysis, all of the basal A cell lines clustered with the luminal cell lines. Similar results were observed when we used only those SRAM genes with CpG island promoters (67 genes; Fig. 3A).
Fig. 3.
SRAM gene expression segregates breast cancer cell lines into cells of epithelial and mesenchymal lineage. (A–C) Dendrograms derived from unsupervised hierarchical clustering of the cell lines based on expression of the 1,000 most variably expressed genes (A), percentage methylation of the 1,023 VM genes (B), and expression values from a subset of genes that are SRAM (120 genes; C). The robustness of each sample's cluster membership is shown below the dendrogram, expressed as the percentage of permutations in which that sample grouped in its cluster (consensus clustering, see supplementary methods). White, luminal A; gray, basal A; black, basal B (according to ref. 11). (D) The expression of EPCAM correlates with the two main clusters derived in C (P < 2.2 × 10−16, Wilcoxon test). The cell lines are ordered based upon their expression of EPCAM. Color coding as for A–C. (E) Markers of epithelial and mesenchymal lineages (SI Materials and Methods) are differentially expressed between the cell lines. The cell lines are ordered and color-coded as in D. Genes that were silent in all 19 cell lines were excluded from the analysis.
The classification based on SRAM genes correlated well with cell morphology; the luminal group cells generally grew as tight clusters typical of epithelial cells, whereas the other group showed less cell–cell contact and were spindle-shaped (Fig. S3B). The epithelium-like cells were all exclusively positive for the epithelial marker EPCAM (also known as TACSTD1 and recognized by the BerEP4 antibody; Fig. 3D) and, with the exception of HCC1569, all expressed cytokeratin 19 and other markers expressed by normal epithelial cells (Fig. 3E; ref.14). In contrast, the other group was negative for EPCAM expression and, with the exception of MCF10A cells, did not consistently express keratins. However, they did express genes associated with mesenchyme (Fig. 3E; ref. 15). These data indicated that EPCAM−ve cells were likely to be of mesenchymal lineage. Thus, the differential expression of SRAM genes classified breast cancer cell lines into those of epithelial and mesenchymal lineage.
SRAM Genes Undergo Lineage-Specific Repression.
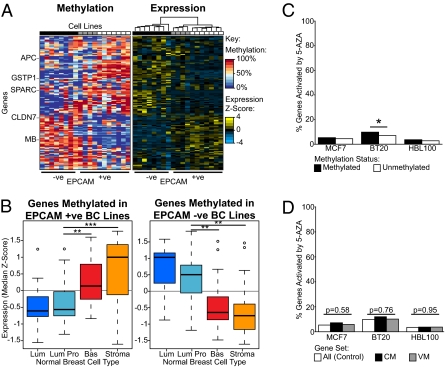
Heat maps of SRAM gene expression and methylation illustrate the striking patterns that differentiate epithelial and mesenchymal cell lines (Fig. 4A; larger heat maps are presented in Fig. S4A). The SRAM gene list contains APC, GSTP1, and PYCARD (16, 17), which have been reported to be methylated in breast cancer, and CLDN7, a tight junction protein expressed in epithelial cells that is methylated in some breast cancer cell lines (18). It also contains genes that have been shown to be differentially expressed in different subcompartments of the normal breast (for example, SPARC and MB; refs. 19 and 20). Indeed, 71 SRAM genes are included in published signatures of different cell populations purified from normal breast tissue (21), a highly significant enrichment (P = 7.1 × 10−16, Fisher's exact test).
Fig. 4.
In cell lines SRAM genes are repressed and methylated in a lineage-dependent manner, and most are not controlled by DNA methylation. (A) Heat maps showing the expression and methylation levels of SRAM genes in breast cancer cell lines (color coded as in Fig. 3) together with their EPCAM status. The cell lines and genes are clustered using hierarchical clustering. See Fig. S4A for larger heat maps. (B) Expression levels of differentially methylated SRAM genes in different cell types in the normal breast. Lum, luminal epithelial cells; Lum Pro, luminal epithelial progenitors (both EPCAM+ve); Bas, basal myoepithelial cells; Stroma, mesenchymal stromal cells (both EPCAM−ve). Expression values are median z scores, and differences between groups were tested using Wilcoxon tests. (C) The percentages of methylated and unmethylated genes that were reactivated by 5-aza-dC treatment in three breast cancer cell lines (Fisher's exact tests). (D) The percentage of CM and VM genes reactivated by 5-aza-dC compared with the percentage of all genes reactivated by 5-aza-dC. No significant differences were detected (χ2 tests). *P < 0.05; **P < 0.01; ***P < 0.001.
To determine whether the SRAM genes were coordinately repressed in association with lineage in the normal breast, we interrogated the same dataset of normal cell populations (21). SRAM genes preferentially methylated in EPCAM+ve breast cancer cell lines had significantly lower levels of expression in cellular fractions corresponding to differentiated luminal and luminal progenitor cells (both EPCAM+ve, Wilcoxon test; Fig. 4B). In contrast, genes methylated in EPCAM−ve breast cancer cell lines had significantly lower levels of expression in the basal/myoepithelial cell fraction and even lower levels of expression in the mesenchymal stromal fraction (both EPCAM−ve, Wilcoxon test; Fig. 4B). A similar pattern was observed when we considered SRAM genes with CpG island and non-CpG island promoters separately (Fig. S4B). Thus, genes prone to methylation in cell lines of different lineages are generally already repressed in normal cells of the corresponding lineage.
Majority of Genes Methylated in Breast Cancer Cell Lines Are Not Derepressed by Demethylation.
As our results suggested that genes prone to methylation might already be repressed by lineage-specific factors, we investigated the extent to which DNA methylation might be important for their repression using the demethylating agent 5-aza-2′-deoxycytidine (5-aza-dC). Treatment of three breast cancer cell lines with 5-aza-dC led to the demethylation and reexpression of DAZL, a gene whose expression is known to be directly controlled by DNA methylation in normal development (Fig. S4 C and D; ref. 22). The cancer testis antigen GAGE4 was also derepressed as expected (23). We profiled gene expression levels after 5-aza-dC or mock treatment using microarrays, combining this with our methylation data to ascertain, in an unbiased manner, the proportion of methylated genes that were reactivated. Less than 10% of the silenced methylated genes were derepressed by 5-aza-dC in the three breast cancer lines, and derepression did not show a greater specificity for VM genes (Fig. 4 C and D). A similar proportion of genes with unmethylated promoters were derepressed by 5-aza-dC exposure (Fig. 4C). Our arrays indicated that methylated CDH1 gene was not reexpressed by 5-aza-dC in HBL100 cells; this result was verified using quantitative RT-PCR (Fig. S4D).
As would be expected, 5-aza-dC treatments lead to significant but incomplete demethylation (Fig. S4C). To be sure that we were not missing transcription effects because of inadequate demethylation, we took advantage of the DNA methyltransferase-deficient HCT116 DKO cells where DNA methylation is reduced to 3–4% of that seen in wild-type (24, 25). We compared genes reactivated in DKO cells with those reactivated by treating wild-type HCT116 cells with 1 μM 5-aza-dC for 3 d, a dose shown to reduce global methylation to 35% of control (26). As expected, there was a significant overlap in the methylated genes derepressed by these two methods (Fig. S4E). However, despite the fact that more methylated genes were derepressed in the DKO cells than by 5-aza-dC treatment, this result still only represented 16.5% of methylated genes (Fig. S4F). These data suggest that DNA methylation at promoters is not the primary mechanism responsible for the repression of most methylated genes in cancer.
Lineage-Specific Aberrant Methylation Occurs in Primary Tumors.
To examine whether lineage-specific methylation also occurred in primary tumors, we generated methylation profiles from 47 primary breast tumors. Firstly, we analyzed SRAM gene methylation in the samples. After excluding probes that were methylated in normal breast, SRAM probes that were methylated in EPCAM+ve cell lines were significantly more frequently methylated in primary tumors than those specific for EPCAM−ve cell lines (Fig. 5 A and B and Fig. S5A). A further analysis using all genes that showed a significant preference for methylation in EPCAM+ve or −ve cell lines produced a similar result (Fig. S5B). Furthermore, genes that were specifically repressed in normal luminal epithelial cells (compared with stroma) were also significantly more frequently methylated in primary tumors than those genes that were active (Fig. S5C). Within this list of genes we also found significant enrichments in genes previously reported to be frequently methylated in breast tumors (Table S1).
Fig. 5.
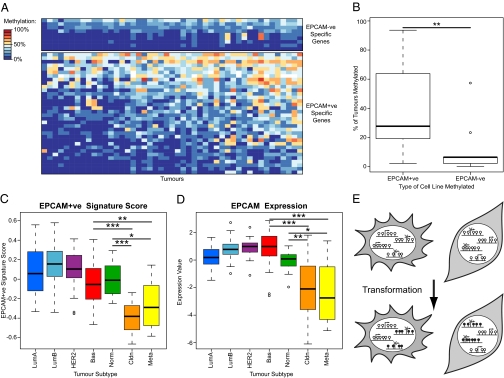
Lineage-specific aberrant methylation occurs in primary breast tumors. (A) Heat map indicating methylation frequency of differentially methylated SRAM genes in 47 primary breast tumors. Only genes that are unmethylated in the normal breast are shown. Genes and samples are ordered by their frequency of methylation. A larger version of the heat map is in Fig. S5A. (B) Genes methylated in EPCAM+ve cell lines are more frequently methylated in primary tumors. The frequency of methylation in tumors of the groups of genes shown in A was compared. Significance was assessed using a Wilcoxon test. (C) Boxplot of EPCAM+ve SRAM expression signature scores by tumor type for a series of breast tumors. Claudin-low (Cldn) and metaplastic tumors (Meta) have scores that are significantly lower than all other subtypes (Wilcoxon tests). A plot using an EPCAM−ve signature is in Fig. S5E. (D) Boxplot of EPCAM expression by tumor subtype. Claudin-low and metaplastic tumors have significantly lower EPCAM expression than the other subtypes (Wilcoxon tests). (E) Model showing that normal lineage commitment leads to the repression of genes in a lineage-specific manner. Lineage-repressed genes are prone to hypermethylation upon transformation. *P < 0.05; **P < 0.01; ***P < 0.001, Wilcoxon tests.
We then looked specifically at the methylation of important tumor suppressor genes in breast cancer (BRCA1 and CDH1), as well as other genes that have been frequently reported to be methylated and that might also be important in breast cancer biology (APC, GSTP1, and ESR1). GSTP1 and APC are both SRAM genes methylated predominantly in EPCAM+ve cell lines (Fig. 4A) and were frequently methylated in primary tumors (Fig. S5D). Both are expressed in luminal progenitor cells but are down-regulated in differentiated luminal cells, suggesting that their methylation could be linked to terminal differentiation. BRCA1 displayed a similar expression pattern and was methylated to a level of >30% in 4 of the 47 (8.5%) primary tumors, a frequency consistent with previous reports (27). In contrast, CDH1 and ESR1, which are both expressed in epithelial cells, were infrequently methylated (2/47 and 0/47, respectively; Fig. S5D). The level of CDH1 methylation in the two tumors was also comparatively low (31% and 34%). This result is consistent with methylation rarely affecting genes that are ordinarily expressed in that lineage. In cell lines, CDH1 methylation was specific to those with low EPCAM expression (Fig. S5D).
Our results demonstrate that primary tumors have epithelial-specific methylation patterns. However, recent reports have suggested that certain rare tumor types, claudin-low and metaplastic tumors, might have mesenchymal characteristics (28). We tested whether an expression signature composed of SRAM genes could distinguish these tumors in that dataset. As predicted, most tumor subtypes had a high EPCAM+ve score, but claudin-low and metaplastic tumors more closely resembled the SRAM expression profile of EPCAM−ve cell lines (Fig. 5C and Fig. S5E). Our signature was also predictive of EPCAM expression in tumors, as had been the case for the cell lines (Fig. 5D). The expression levels of a larger panel of marker genes further supported a mesenchymal origin for claudin-low tumors and metaplastic tumors, although the latter also expressed some epithelial markers (Fig. S5F).
Discussion
The methylation of CpG island promoters is a normal developmental process that is essential for repression of some genes, such as those on the inactive X chromosome, imprinted genes, and some tissue-specific genes (29). In cancer, many additional promoters are both repressed and methylated. It is often argued that methylation could also be instrumental in their repression. However, our data suggest a model whereby in breast cancer aberrant methylation occurs at genes that are already repressed through normal lineage commitment and methylation is generally not required for their repression (Fig. 5E). Lineage-specific aberrant methylation has not been previously reported but can be found in datasets of breast cancer methylation patterns from a number of other studies (Table S1).
The finding that most cancer-associated aberrant methylation occurs in genes that are already down-regulated has been alluded to previously (30), and this phenomenon also occurs in normal cultured neural cells (31). However, the literature contains many examples of methylated genes being derepressed by 5-aza-dC in cell lines, which has been central to the argument that aberrant methylation causes tumor progression by silencing genes. Indeed, one study assumed that in HCT116 cells, all methylated genes are repressed because of methylation and used the amount of deregulation induced by 5-aza-dC to estimate the size of the methylome (5% of all genes; ref. 32). However, by using an unbiased approach and directly measuring the proportions of methylated and unmethylated genes that are actually derepressed by 5-aza-dC, our results challenge this view. We find that 5-aza-dC derepresses <10% of all methylated genes and that 25.5% of all genes are methylated in HCT116 cells. Repressive histone marks may remain after treatment with 5-aza-dC (33), indicating that DNA methylation may be one of many epigenetic mechanisms involved in repression.
We found CDH1 and ESR1 to be rarely methylated in primary tumors, and as these genes are expressed in epithelial cells, this finding would be predicted by our model. Some studies using the nonquantitative methylation-specific PCR (MSP) technique have reported higher methylation frequencies of CDH1 and ESR1 (16, 34), including 72% in the case of CDH1 (35). However, MSP may be prone to detecting low-level methylation at some genes. A study using a quantitative version of MSP agrees with our finding of infrequent methylation at CDH1 and ESR1 (36). We found genuine BRCA1 methylation in 8.5% of tumors, consistent with a previous report (27). BRCA1 is down-regulated during terminal epithelial differentiation, which could make it susceptible to methylation. We also note that disproportionately frequent BRCA1 methylation is observed in metaplastic carcinomas (63% of a series of 27 tumors; ref. 27). As these tumors appear to be mesenchymal and BRCA1 is repressed in normal mesenchymal cells, this finding is consistent with our model that methylation affects lineage-repressed genes.
The detection of CDH1 methylation by sensitive nonquantitative techniques (MSP) could be due to the presence of stromal cells, contaminating blood cells (37), or tumor cells that have undergone epithelial-to-mesenchymal transition (EMT). The existence of EMT is breast cancer is contentious, but it would be predicted to down-regulate CDH1 and induce metastasis. EMT might also lead to CDH1 methylation under our model. However, as we did not detect significant CDH1 methylation, our data do not support extensive EMT in most breast tumors. A previous study also found no differences in CDH1 expression between primary tumors and their metastases (38). Whether EMT is responsible for the mixed epithelial and mesenchymal characteristics in rare metaplastic tumors (Fig. S5E) remains to be determined.
Although we observed that aberrantly methylated genes were significantly enriched for those that are bivalently marked in ES cells (10), most of the affected genes lacked these marks. It is possible, therefore, that in cancer, bivalent genes are prone to methylation because they are lineage-specific and repressed, rather than because of a direct interaction of the polycomb and DNA methylation machineries.
In summary, our data indicate that aberrant methylation is a marker of lineage restriction in cancer. Although we cannot claim that this finding applies to every aberrantly methylated gene, our unbiased approach clearly demonstrates that normal developmental repression influences whether genes become aberrantly methylated in cancer. Our findings force a reappraisal of the likely efficacy of DNA demethylating agents in cancer therapy.
Materials and Methods
A brief summary of methods used is given below. For full details, see SI Materials and Methods and Tables S2–S4.
Breast Cancer Cell Lines and Samples.
Breast cancer cell lines were obtained from Cancer Research UK or ATCC. Wild-type and DKO (Dnmt1−/−, Dnmt3b−/−) HCT116 cells were kind gifts from B. Vogelstein (24). HMECs were a gift from E. Katz at the Edinburgh Breakthrough Breast Cancer Research Unit. SHEF-6 hES cell DNA was a gift from D. Hay (MRC Centre for Regenerative Medicine). DNA from normal breast, fetal and adult brain, testis, liver, placenta, spleen, blood, and colon were from Biochain, After approval by our ethical board, 47 fresh frozen unselected tumor samples were obtained through the Experimental Cancer Medicine Centre in Edinburgh.
5-aza-dC Treatment.
Cell lines were exposed to 1 μM 5-aza-dC, refreshed every 24 h, for a total of 72 h.
Microarrays.
We used Illumina Infinium Human Methylation27 beadarrays and Illumina human HT12 Expression beadarrays. All data have been submitted to the Gene Expression Omnibus (GEO) database (accession no. GSE26990).
Bioinformatic Analyses.
Bioinformatic analyses were performed using R (version 2.9.2). Ensembl 54 (NCBI36) gene annotations were used throughout. In cell lines unmethylated genes were defined as those with ≤30% methylation and methylated ones as ≥70%. In tumor samples methylated genes were defined as those with >30% methylation due to the heterogeneity of the samples. We defined groups of genes with different methylation patterns as follows: CU, unmethylated in all cell lines; CM, methylated in all cell lines or all but one cell line; VM, methylated in at least four and unmethylated in at least four cell lines. Only CpGs within 200 bp of transcription start sites were considered in our analyses. SRAM genes were VM genes that had significantly lower expression when methylated (one-sided Wilcoxon test). The specificity of a gene expression pattern was measured using a method based on information theory (SI Materials and Methods). Datasets were downloaded from data repositories or individual papers as appropriate.
Supplementary Material
Acknowledgments
We thank Dr. T. I. Simpson for the use of his consensus clustering algorithm, L. Renshaw for assistance in the collection of clinical samples, and Dr. J.S. Thomas for assistance with breast cancer pathology. This work was supported by Breakthrough Breast Cancer and the Medical Research Council. Central services utilized in the study are funded by Cancer Research UK and the Wellcome Trust.
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE26990).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013224108/-/DCSupplemental.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohtani-Fujita N, et al. Hypermethylation in the retinoblastoma gene is associated with unilateral, sporadic retinoblastoma. Cancer Genet Cytogenet. 1997;98:43–49. doi: 10.1016/s0165-4608(96)00395-0. [DOI] [PubMed] [Google Scholar]
- 4.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 6.Dworkin AM, Spearman AD, Tseng SY, Sweet K, Toland AE. Methylation not a frequent “second hit” in tumors with germline BRCA mutations. Fam Cancer. 2009;8:339–346. doi: 10.1007/s10689-009-9240-1. [DOI] [PubMed] [Google Scholar]
- 7.Tung N, et al. Prevalence and predictors of loss of wild type BRCA1 in estrogen receptor positive and negative BRCA1-associated breast cancers. Breast Cancer Res. 2010;12:R95. doi: 10.1186/bcr2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 9.Gusterson B. Do 'basal-like' breast cancers really exist? Nat Rev Cancer. 2009;9:128–134. doi: 10.1038/nrc2571. [DOI] [PubMed] [Google Scholar]
- 10.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 13.Zhao XD, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Allinen M, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 17.Conway KE, et al. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 2000;60:6236–6242. [PubMed] [Google Scholar]
- 18.Kominsky SL, et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–2033. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 19.Jones C, et al. Expression profiling of purified normal human luminal and myoepithelial breast cells: Identification of novel prognostic markers for breast cancer. Cancer Res. 2004;64:3037–3045. doi: 10.1158/0008-5472.can-03-2028. [DOI] [PubMed] [Google Scholar]
- 20.Kristiansen G, et al. Endogenous myoglobin in human breast cancer is a hallmark of luminal cancer phenotype. Br J Cancer. 2010;102:1736–1745. doi: 10.1038/sj.bjc.6605702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 22.Maatouk DM, et al. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development. 2006;133:3411–3418. doi: 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- 23.Kumagai T, et al. Epigenetic regulation and molecular characterization of C/EBPalpha in pancreatic cancer cells. Int J Cancer. 2009;124:827–833. doi: 10.1002/ijc.23994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhee I, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 25.Egger G, et al. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci USA. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel K, et al. Targeting of 5-aza-2'-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner NC, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 28.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 30.Keshet I, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 31.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuebel KE, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGarvey KM, et al. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 34.Parrella P, et al. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clin Cancer Res. 2004;10:5349–5354. doi: 10.1158/1078-0432.CCR-04-0555. [DOI] [PubMed] [Google Scholar]
- 35.Caldeira JR, et al. CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC Cancer. 2006;6:48. doi: 10.1186/1471-2407-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suijkerbuijk KP, et al. Methylation is less abundant in BRCA1-associated compared with sporadic breast cancer. Ann Oncol. 2008;19:1870–1874. doi: 10.1093/annonc/mdn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombaerts M, et al. Infiltrating leukocytes confound the detection of E-cadherin promoter methylation in tumors. Biochem Biophys Res Commun. 2004;319:697–704. doi: 10.1016/j.bbrc.2004.05.041. [DOI] [PubMed] [Google Scholar]
- 38.Kowalski PJ, Rubin MA, Kleer CG. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res. 2003;5:R217–R222. doi: 10.1186/bcr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.