Abstract
Bdelloid rotifers are important contributors to biogeochemical cycling and trophic dynamics of both aquatic and terrestrial ecosystems, but little is known about their biogeographic distribution and community structure in terrestrial environments. This lack of knowledge stems from a lack of phylogenetic information and assumptions that microbial eukaryotes are globally distributed and have very limited diversity across vast geographic distances. However, these assumptions have been based more on assessments of their morphology than any measure of their true genetic diversity and biogeographic distribution. We developed specific primers for the cytochrome c oxidase subunit 1 (cox1) gene of bdelloid rotifers and amplified and cloned sequences using a nested sampling scheme that represented local (0–10 m) to global (up to 10,000 km) scales. Using phylogenetic community analyses (UniFrac) and geospatial statistics (semivariograms, mantel tests), we were able to reject the hypothesis that communities of rotifers are the same across even fairly small geographic distances. Bdelloid communities showed highly significant spatial structuring with spatial autocorrelation ranges of 54–133 m, but beyond that distance communities were extremely dissimilar. Furthermore, we show that these spatial patterns are driven not only by changes in relative abundance of phylotypes but also by absolute changes in phylotype occurrence (richness). There is almost no overlap in phylotype [or operational taxonomic unit (OTU)] occurrence between communities at distances beyond the autocorrelation range (~133 m). Such small species ranges, combined with their ubiquity in soils, make it increasingly clear that the biodiversity of bdelloid rotifers (and other less easily dispersed microbes) is much higher than previously thought.
Keywords: phylogeography, microfauna, Bdelloidea
Microbiota have been described dogmatically as having similar global and local distributions, i.e., “everything is everywhere, and the environment selects,” as initially discussed by Beijerinck and Baas Becking (1, 2). This view is supported by the massive population sizes of microorganisms, by the general ease with which they can disperse (e.g., via wind), and by data based on the distribution of species defined by morphology. However, recent work has suggested significant dispersal limitation in some groups of microorganisms including bacteria and archaea (3, 4) and microbial eukaryotes (5, 6).
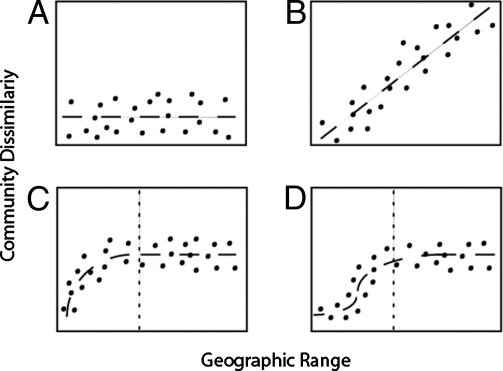
As reviewed elsewhere (3, 7, 8), the case of whether or not small organisms have any discernable biogeography can be split into two camps: those that think “everything is everywhere” (EisE) (Fig. 1A), and those that think all organisms have some level of biogeography, regardless of size (Fig. 1B). Basically, the issue is rooted in whether historical or contemporary effects are responsible for present patterns of distribution (7). For those on the side of EisE, the local environment plays a role larger than that of any historical biogeographical effects. The degree of similarity between any two communities should be independent of the geographical distance between them, given similar environments. Therefore, no matter the geographic scale sampled, all communities should maintain the same level of average, but low, dissimilarity (Fig. 1A). Those who maintain historical effects trump local environment effects would expect that communities increasingly further from each other should become increasingly dissimilar (Fig. 1B). Debates often revolve around which of these two community patterns is the rule.
Fig. 1.
Hypothetical community versus biogeographic relationships as they relate to microorganisms. (A) Assuming EisE, community relatedness is not dependent upon distance, so all communities appear similar to each other. (B) Assuming a continuous distance–decay relationship of community dissimilarity over geographic distance. (C) Spatial autocorrelation up to the autocorrelation range (vertical dashed line), after which the limit of spatial dependence is reached (sill), and communities are just as likely to be as similar to or as different from communities before the range. (D) Spatial autocorrelation as in C but with a lag before the start of autocorrelation indicating we are sampling from within the same community.
However, it also is possible that both historical and environmental effects determine the biogeographic distribution of microbes (7). If so, then one would expect local communities to show more similarity than that seen with geographically distant communities. In other words, local communities should show a high degree of autocorrelation, and distant communities should present a random array of pair-wise community relatedness. This latter case is depicted graphically in Fig. 1C. However, to detect a pattern as shown in Fig. 1C, a nested sampling strategy is essential to reveal the spatial structure of communities at scales ranging from local (centimeter to meter scales) to global. A fourth and final possibility is one that resembles the trend in Fig. 1C with the addition of a flat lag at very small geographic distances (Fig. 1D) before spatial autocorrelation is observed. This pattern reflects the case in which, at small spatial distances, communities would be identical. That is, what we think are different communities are really independent spatial replicates from the same community at a local site (9); this case is represented in Fig. 1D.
One method to describe biogeographic patterns quantitatively at all scales is through the use of spatial autocorrelation statistics (10). Spatial autocorrelation is defined as the level of dissimilarity in a variable as the distance of separation between sample locations increases. Thus, all the patterns depicted in Fig. 1 can be described using spatial autocorrelation statistics. For example, Fig. 1C, shows a hypothetical dataset that is spatially correlated only up to a certain autocorrelation range (denoted by the vertical dashed line). After this range any two communities are equally likely to be as similar to or as different from other pair-wise comparisons because the limit of spatial dependence has been surpassed.
Here we use spatial autocorrelation statistics to describe the biogeographic distributions of bdelloid rotifers, a ubiquitously distributed group of microbial eukaryotes that is a prey item for larger organisms and is an important predator of smaller microbial species in aquatic and terrestrial systems (11–13). Fortunately, despite the debates on issues of cryptic speciation within this group, exact species identification is not necessary to describe the spatial patterning and community diversity based on phylogenetic relatedness (6, 14, 15). We describe a culture-independent sequencing strategy that resulted in the generation of many long-read sequences directly from environmental samples and allowed us to elucidate the spatial scale at which microbial eukaryote communities are structured.
Results
Amplification and Utility of the cox1.
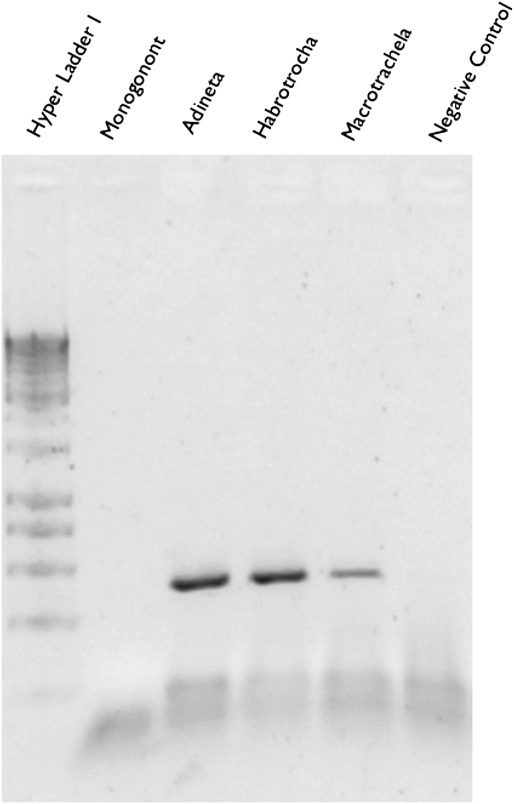
We developed primers specific to the cytochrome oxidase subunit 1 (cox1) gene of bdelloid rotifers. PCR, BLAST (16), and phylogenetic analyses confirmed that the cox1 primers amplify bdelloid rotifer DNA with specificity and fidelity (Fig. 2 and Dataset S1). Our local-to-global scale sampling resulted in 1,024 sequences comprising 790 unique sequences (GenBank accession numbers HQ174968–HQ175991) (Table S1).
Fig. 2.
Gel image of amplification products obtained with bdelloid-specific cox1 primers. Adineta, Habroctrocha, and Macrotrachela are from morphologically identified bdelloids. The negative control and the outgroup Monogonont rotifer Brachionus plicatilis produced no bands.
Spatial and Community Analysis.
Semivariogram plots (10) of the decay in community similarity with increasing geographic distance (Figs. 3 and 4) were best fit (best r2 values) by the exponential autocorrelation model for both the weighted and unweighted UniFrac (17) metrics (using the phylogeny from Dataset S2). There was strong linear relationship [Mantel tests (18) (P < 0.002)] in the data across geographic distances up to autocorrelation range (see below). This result was supported by more traditional operational taxonomic unit (OTU)-based methods, which, despite overestimation of differences between communities, showed the same spatial patterns over similar distances (Figs. S1 and S2).
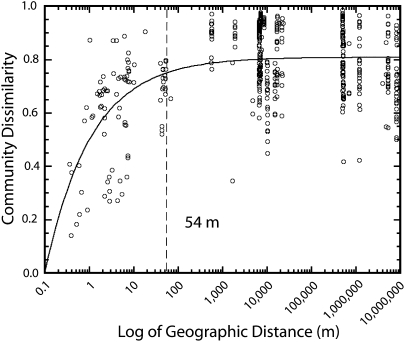
Fig. 3.
Weighted UniFrac (17) variogram (bdelloid abundance) plotted as the UniFrac metric (Community Dissimilarity) versus the log of geographic distance. Values close to 1 indicate completely different communities, and values close to 0 indicate identical communities. The dashed line indicates the autocorrelation range.
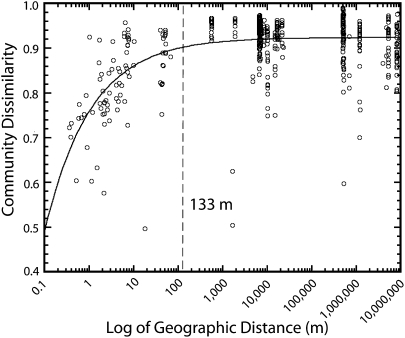
Fig. 4.
Unweighted UniFrac (17) variogram (bdelloid occurrence) plotted as the Unifrac metric (Community Dissimilarity) versus the log of geographic distance. Values close to 1 indicate completely different communities, and values close to 0 indicate identical communities. The dashed line indicates the autocorrelation range.
Overall, our sampling of bdelloid communities from local to global scales demonstrates that local communities show strong spatial structuring, whereas more distant communities are very different from one another, even in similar environments, thus best matching the theoretical model depicted in Fig. 1C. This local effect was evident at distances up to 54 m (autocorrelation range) when relative abundance of sequences was taken into account (weighted UniFrac; Fig. 3) and at distances up to 133 m when occurrence (richness) data were analyzed (unweighted UniFrac; Fig. 4). Beyond the autocorrelation range, all communities averaged community dissimilarity of about 0.8 and 0.9 (Figs. 3 and 4). This high level of dissimilarity suggests that unique clades of bdelloids exist at each location beyond the autocorrelation range, a conclusion independently supported by the decreasing probability of sampling specific OTUs at increasing distances (Fig. S3). We further tested this idea by using the Net Relatedness Index (NRI) (19, 20); the large positive NRI values obtained for geographically distant communities (Table S2) indicate that communities are “phylogenetically constrained,” i.e., each community is composed of unique clades compared with all others.
We failed to observe a noticeable lag at small spatial scales (Fig. 1D), indicating that we may not have sampled at small enough spatial distances to resample the same community. This result is informative, because our closest samples were 0.16 m apart. To observe such disparate bdelloid clones and communities at such close distances reveal that these limnoterrestrial bdelloid communities are heterogeneous at small scales (21), being composed of similar but not identical bdelloid clones.
To verify that we sampled communities to a level sufficiently deep to characterize phylogenetic differences adequately, we performed both rarefaction analyses (Fig. S4) and jackknife analyses (randomly resampling sequences without replacement) (Figs. S5 and S6). These analyses demonstrated that we sampled a majority of the phylotypes (48–100%) at most of our sites (Fig. S4) and that, even if we jackknifed all our sites using the minimum sampling intensity for any site, we still recovered the same community patterns (compare Figs. 3 and 4 and Figs. S5 and S6).
Discussion
Based on most previous studies of the biogeography of small eukaryotes (3, 6, 7, 22–27), our expectation was that bdelloid rotifers either would show no spatial structure (Fig. 1A) or would show increasingly different communities as the distance between pair-wise samples became greater (Fig. 1B). Instead, we found that these communities showed the pattern depicted in Fig. 1C: Local communities show a high degree of spatial autocorrelation, whereas geographically more distant communities show a high level of community dissimilarity. Our analyses also produced estimates of spatial autocorrelation ranges for microbial eukaryotes; quantitative (weighted UniFrac) and qualitative (unweighted UniFrac) metrics indicate that this range is around 54–133 m. This unexpectedly small autocorrelation range highlights the importance of sampling at scales that pick up local, regional, and global biogeographic patterns. It is apparent that if we had not sampled intensively at local scales (0.2–100 m), we would have concluded that all communities at all scales are equally dissimilar.
Even more surprising than the small spatial autocorrelation ranges for bdelloid communities was the lack of similarity between communities at geographic distances beyond the autocorrelation range (Figs. 3 and 4). This pattern indicates not that “everything is everywhere” but just the opposite: There is little overlap in community composition at distances greater than about 54–133 m. This unexpected pattern is driven not just by changes in abundance (weighted UniFrac; Fig. 3) of the dominant clades, as has been documented for the geographic distribution of soil bacterial communities (28), but also by the presence of many novel phylotypes at each site so that there is almost no overlap in OTUs (defined at the 97% level of similarity; Figs. S1 and S2) at sites further apart than the autocorrelation range. This finding directly contradicts the idea that rotifers (and other less easily dispersed microbial eukaryotes) are cosmopolitan and supports the idea that there are vastly more “cryptic species” (8, 29) of bdelloid rotifers than previously was thought.
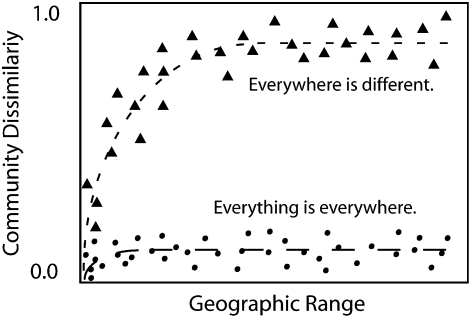
Of the models depicted in Fig. 1, our data are most similar to Fig. 1C, but, unexpectedly, the level of sequence dissimilarity between communities (at distances beyond the autocorrelation range) was much greater than predicted or observed in previous studies (7, 28). Therefore in Fig. 5 we expand upon the model shown in Fig. 1C to contrast the extreme cases of EisE and the very different findings of the current study. EisE would result in very low community dissimilarity values (circles in Fig. 5). In contrast, the pattern we observed is that every community is mostly unique (triangles in Fig. 5) at distances beyond the autocorrelation range. Our phylogenetic (Figs. 3 and 4) and OTU-based (Figs. S1 and S2) analyses strongly support a global view of microbial distribution in which communities are dominated by endemic species and share very few common clades between sites (circles in Fig. 5). Local endemicity is supported further by the largely positive NRI (19, 20) values (Table S2), which show that local communities tend to be comprised of closely related lineages relative to the phylogenetic diversity of rotifers across the planet. This world view directly contradicts the idea of EisE and suggests that the diversity of microbial eukaryotes such as rotifers may be vast beyond our imagining, especially given that endemic microbes may have species ranges of about 100 m (54–133 m).
Fig. 5.
Theoretical expectations for “everything is everywhere” (circular points) and “everywhere is different” (triangular points). One would expect very low community dissimilarity values if most bdelloid communities are composed predominately of ubiquitous bdelloid species that are not impeded in their dispersal (circles). An equal but opposite extreme is that all communities are highly unique and are composed predominately of unique endemic bdelloid rotifers (triangles), revealing differences in dispersal and or habitat.
Bdelloids are known to produce small, resistant resting stages (30, 31) that should disperse easily by wind. The fact that our results show that bdelloids are not widely distributed implies that other microbial eukaryotes with less resistant stages (23, 32) should have even more geographically restricted distributions; unfortunately, few phylogenetically based studies have been done at the range of spatial scales needed to see the patterns observed in the present study (6, 7). Importantly, the ability to form resistant stages probably evolved not for dispersal but to survive periods of unfavorable environmental conditions (i.e., dry and cold conditions) (30), thereby maintaining unique local communities (21). Resistant survival structures also result in large banks of propagules (30) which can undergo rearrangement of nuclear genes during rehydration (33), allowing long-term monopolization (21) by local cox1 phylotypes. Monopolization could be especially important in intermittently wet soils (such as those studied here), because indigenous propagules would be numerically dominant in the soil on the rare occasions when abundant water becomes available. Thus, our observation of strong geographic differentiation of bdelloid communities presumably reflects local numerical abundance of locally adapted clones that persist over relatively long periods of time.
In community spatial studies it is important to distinguish between conclusions based on relative abundance (diversity) data and those based on occurrence (richness) data. In our analyses, relative abundance data (weighted UniFrac) gave a stronger spatial signal (Fig. 3 and Fig. S1) than our analyses based on richness (unweighted UniFrac; Fig. 4 and Fig. S2). This finding could indicate that large-scale patterns of microbial diversity are driven mostly by changes in relative abundance, so that the same organisms occur everywhere, but the dominant organisms are different in different sites. However, our data do not support this model because, even though relative abundance does have a strong effect, we still see spatial patterning when only species richness is taken into account (Fig. 4 and Fig. S2). As discussed above, there are novel clades or OTUs at every site we sampled. In fact, the probability of encountering the same OTU (defined at the 97% similarity level) drops from near 100% to zero as geographic distance increases beyond the autocorrelation range (Fig. S3), and this finding is not attributable to undersampling. Undersampling would account for the observed patterns only if dominant phylotypes at one site were rare at other sites (and vice versa) and therefore were missed because of undersampling. However, rarefaction curves show that we sampled many of our sites to near saturation, encountering 48–100% of the estimated number of phylotypes (Fig. S4) (34), making it highly unlikely that changes in abundance alone explain the spatial patterns we observed.
Finally, it is important to note that the phylogenetic approach used in the present study (UniFrac) actually underestimates spatial structuring of communities compared with traditional OTU-based metrics. When we analyzed all our data using traditional OTU-based metrics, the spatial patterns were exaggerated compared with UniFrac (which preserves actual phylogenetic relatedness in the analyses). This difference is easily seen by comparing Figs. 3 and 4 with the OTU-derived views of spatial structure (Figs. S1 and S2). Collapsing the data into OTUs gives the false impression that almost all communities are 100% different (dissimilarity values of 1) at distances beyond the autocorrelation range (Figs. S1 and S2). This false impression occurs because collapsing data into OTUs makes all OTUs equally dissimilar, obscuring deeper levels of genetic relatedness (35, 36). Thus, our phylogenetic analyses reveal that, although bdelloid communities are very different at large spatial scales, they still show deeper levels of phylogenetic relatedness than one would predict from OTU-based metrics. Nonetheless both OTU and phylogenetic metrics support our conclusion that bdelloid rotifer communities are highly spatially autocorrelated at local scales and are very different at larger spatial scales.
Methods
DNA Extraction and PCR.
Soil samples were collected from the sites listed in Table S1 and represented seasonally dry, high-elevation ecosystems across the western United States and similar sites at greater geographic distances (e.g., the high Andes). Samples consisted of 100-g soil cores of the top 4 cm of soil, which subsequently were homogenized; 10-g subsamples were used for DNA extraction. Total cellular DNA was extracted from soil using the UltraClean Mega Soil DNA Isolation Kit (#12900; Mo Bio Laboratories, Inc.). Bdelloid rotifer DNA was amplified from these soil DNA extractions by using a two-step PCR protocol to amplify cox1.
We chose the cox1 gene because it provides some analytical advantages with regard to bdelloid rotifers (37), not only in what one would expect to find regarding branch length and tree topology (38, 39) but also because (i) bar-coding initiatives have been accruing cox1 data (40) to which our sequence data can be compared; (ii) robust primers (41) make it possible to amplify the cox1 gene from almost any invertebrate, and these sequences then can be used to design taxon-specific primers, as in the present study; and (iii) cox1 is an effectively haploid gene which eliminates the “Meselson effect” that can make it difficult or impossible to recover the correct phylogenetic tree using nuclear gene sequences (37, 42) (also see ref. 43 for a discussion of problems with elucidating bdelloid diversity with the 18S rDNA gene).
The first PCR made use of primers from Folmer et al. (40). The second PCR made use of bdelloid-specific primers developed for this study: Bdell_CO1_FW: 5′-CGT ACW GAG TTA GGA ATR GTA-3′, and Bdell_CO1_Rev: 5′-CCA AAA TTW CGA TCT AAY A-3′. Touch-down PCR was used for both reactions and was set up as follows: 94 °C for 5 min, followed byeight cycles of (−1 °C annealing per cycle) 94 °C for 30 s, 55 °C for 30 s, and 62 °C for 1 min, followed by 30 cycles of 94 °C for 30 s, 48 °C for 30 s, and 62 °C for 1 min. The amplified template from the first primers (41) was diluted 20-fold and used as a template for the second PCR using our bdelloid-specific primers.
A 50-μL PCR contained the following: 10× PCR buffer, 0.5 units Taq polymerase, 1.5 mM MgCl2 (catalog nos. M0267L and B9021S; New England Biolabs), 0.2 μM dNTPs (dNTP stock was made as follows from individual 100-mM stocks: 12 μL A, 12 μL T, 8 μL G, 8 μL C, and 360 μL water; catalog no. 10297–018; Invitrogen), 0.4 μM of each primer (1–2 μL template DNA). We want to emphasize that the thermocycling protocol listed above in combination with the dNTP mix ratios produced the best-quality sequence for this very AT-rich region (44).
DNA Purification, Cloning, and Sequencing.
The final PCR product was purified using the UltraClean GelSpin DNA Extraction Kit (#12400; Mo Bio Laboratories, Inc.). Purified PCR product then was cloned using the Invitrogen TOPO TA Kit (with pCR4-TOPO vector) with One Shot TOP10 chemically competent Escherichia coli (K4575-01). Pelleted cells were sent to Functional Biosciences, Inc. for sequencing. Sequence data were assembled, the vector sequence was removed, and data were edited by hand using Sequencher 4.7 (Gene Codes Corporation). Data then were exported for use in various phylogenetic and community-based analysis programs. Table S2 lists the 1,024 sequences generated from this study.
Phylogenetic and OTU-Based Analyses.
MUSCLE (45) was used to generate two cox1 alignments (with and without outgroup taxa; Table S2 lists sequences used), which then were edited by hand to ensure sequences were aligned by codon. These alignment data then were analyzed in RAxML v.7.2.6 (46). The best-scoring likelihood tree, from 100 full-alignment inferences using the GTR + I + Γ model as chosen by MultiPhyl Online (47) and partitioned by codon position, was retained. Once placement of sequence data was confirmed with outgroup taxa (Dataset S1) using Dendroscope (48), the phylogeny containing only bdelloids (Dataset S2) was submitted as a midpoint-rooted tree to several phylogenetic community-comparison programs such as UniFrac (17), Phylocom (19, 20), and mothur (49). UniFrac and OTU-based metrics like Sorensen and Bray-Curtis were implemented via QIIME (50) to determine the relatedness of bdelloid communities. For OTU-based metrics, a 97% sequence similarity cutoff was implemented, based on the results of Birky et al. (51). Phylocom was used to generate Mean Phylogenetic Diversity (MPD) and NRI values. Mothur (49) was used to generate a distance matrix for Mantel tests (18) along with rarefaction and Chao1 (34) analysis.
Spatial Analyses.
To determine which model of biogeographical distribution applies to bdelloid rotifers in soil, we determined rotifer sequence diversity at scales ranging from 0.16 m to 9,100 km. Each 10-g soil sample was assumed a priori to represent a community. UniFrac (17) was used to generate community distance matrices taking into account sequence abundance (weighted; quantitative diversity measure taking into account relative abundance) or only occurrence (unweighted; qualitative diversity measure). Data were plotted against the log of geographic distance (Figs. 3 and 4). Spatial autocorrelation models were fit in Kaliedograph (Synergy Software) using the Levenberg–Marquardt algorithm (52, 53), and the best-fit model (out of nugget, spherical, Gaussian, and exponential) (54) was chosen based on the highest r2 value. Significance of spatial autocorrelation was tested using the Mantel test (18) to the maximum distance of spatial autocorrelation as given by the spatial model. General patterns of spatial autocorrelation were confirmed and validated via Jackknife analyses at varying sampling depths for all beta-diversity metrics (55).
Supplementary Material
Acknowledgments
We thank Ryan Lynch and Terry Campbell and two anonymous reviewers for comments on the manuscript, David Mark Welch for providing genomic controls for PCR and comments on the manuscript, and Catherine Lozupone and Greg Caporaso for assistance with the use and assessment of beta-diversity metrics and applications. This project was funded by the National Science Foundation Microbial Observatories Program (MCB-0455606) and by student grants from the Department of Ecology and Evolutionary Biology at the University of Colorado at Boulder.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HQ174968–HQ175991).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012678108/-/DCSupplemental.
References
- 1.O'Malley M. The nineteenth century roots of ‘everything is everywhere’. Nature. 2007;5:647–651. doi: 10.1038/nrmicro1711. [DOI] [PubMed] [Google Scholar]
- 2.de Wit R, Bouvier T. ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environ Microbiol. 2006;8:755–758. doi: 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- 3.Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 4.Cho JC, Tiedje JM. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol. 2000;66:5448–5456. doi: 10.1128/aem.66.12.5448-5456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontaneto D, Ricci C. Spatial gradients in species diversity of microscopic animals: The case of bdelloid rotifers at high altitude. J Biogeogr. 2006;33:1305–1313. [Google Scholar]
- 6.Green JL, et al. Spatial scaling of microbial eukaryote diversity. Nature. 2004;432:747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- 7.Martiny JBH, et al. Microbial biogeography: Putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 8.Fontaneto D, Barraclough TG, Chen K, Ricci C, Herniou EA. Molecular evidence for broad-scale distributions in bdelloid rotifers: Everything is not everywhere but most things are very widespread. Mol Ecol. 2008;17:3136–3146. doi: 10.1111/j.1365-294X.2008.03806.x. [DOI] [PubMed] [Google Scholar]
- 9.Mackas D. Spatial autocorrelation of plankton community composition in a continental shelf ecosystem. Limnol Oceanogr. 1984;29:451–471. [Google Scholar]
- 10.Franklin RB, Mills AL. In: The Spatial Distribution of Microbes in the Environment. 1st Ed. Franklin RB, Mills AL, editors. The Netherlands: Springer, Dordrecht; 2007. pp. 31–60. [Google Scholar]
- 11.Arndt H. Rotifers as predators on components of the microbial web (bacteria, heterotrophic flagellates, ciliates)—a review. Hydrobiologia. 1993;255/256:231–246. [Google Scholar]
- 12.Pejler B. Relation to habitat in rotifers. Hydrobiologia. 1995;313/314:267–278. [Google Scholar]
- 13.Kutikova LA. Bdelloid rotifers (Rotifera, Bdelloidea) as a component of soil and land biocenoses. Biol Bull. 2003;30:271–274. [PubMed] [Google Scholar]
- 14.Ramette A, Tiedje JM. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc Natl Acad Sci USA. 2007;104:2761–2766. doi: 10.1073/pnas.0610671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt SK, et al. Phylogeography of microbial phototrophs in the dry valleys of the high Himalayas and Antarctica. Proc Roy Soc B. 2010;278:702–708. doi: 10.1098/rspb.2010.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 17.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 19.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenetics and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- 20.Webb CO. Exploring the phylogenetic structure of ecological communities: An example for rain forest trees. Am Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 21.De Meester L, Gomez A, Okamura B, Schwenk K. The Monopolozation Hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecol. 2002;23:121–135. [Google Scholar]
- 22.Darling KF, Kucera M, Pudsey CJ, Wade CM. Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proc Natl Acad Sci USA. 2004;101:7657–7662. doi: 10.1073/pnas.0402401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foissner W. Biogeography and dispersal of microorganisms: A review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- 24.Telford RJ, Vandvik V, Birks HJB. Dispersal limitations matter for microbial morphospecies. Science. 2006;312:1015. doi: 10.1126/science.1125669. [DOI] [PubMed] [Google Scholar]
- 25.Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. Eukaryotic microbes, species recognition and the geographic limits of species: Examples from the kingdom Fungi. Philos Trans R Soc Lond B Biol Sci. 2006;361:1947–1963. doi: 10.1098/rstb.2006.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 27.Fenchel T, Finlay BJ. The ubiquity of small species: Patterns of local and global diversity. Bioscience. 2004;54:777–784. [Google Scholar]
- 28.King AJ, et al. Biogeography and habitat modelling of high-alpine bacteria. Nature Commun. 2010;1:53. doi: 10.1038/ncomms1055. [DOI] [PubMed] [Google Scholar]
- 29.Kaya M, Herniou EA, Barraclough TG, Fontaneto D. Inconsistent estimates of diversity between traditional and DNA taxonomy in bdelloid rotifers. Org Divers Evol. 2009;9:3–12. [Google Scholar]
- 30.Ricci C. Dormancy patterns in rotifers. Hydrobiologia. 2001;446/447:1–11. [Google Scholar]
- 31.Gladyshev E, Meselson M. Extreme resistance of bdelloid rotifers to ionizing radiation. Proc Natl Acad Sci USA. 2008;105:5139–5144. doi: 10.1073/pnas.0800966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins D, Underwood M. Zooplankton may not disperse readily in wind, rain, or waterfowl. Hydrobiologia. 1998;387:15–21. [Google Scholar]
- 33.Mark Welch DB, Mark Welch JL, Meselson M. Evidence for degenerate tetraploidy in bdelloid rotifers. Proc Natl Acad Sci USA. 2008;105:5145–5149. doi: 10.1073/pnas.0800972105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- 35.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin AP. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol. 2002;68:3673–3682. doi: 10.1128/AEM.68.8.3673-3682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birky CW. Workshop on barcoded DNA: Application to rotifer phylogeny, evolution, and systematics. Hydrobiologia. 2007;593:175–183. [Google Scholar]
- 38.Barraclough TG, Birky CW, Jr, Burt A. Diversification in sexual and asexual organisms. Evolution. 2003;57:2166–2172. doi: 10.1111/j.0014-3820.2003.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 39.Birky CW., Jr Heterozygosity, heteromorphy, and phylogenetic trees in asexual eukaryotes. Genetics. 1996;144:427–437. doi: 10.1093/genetics/144.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratnasingham S, Herbert PDN. The Barcoding of Life data system. Mol Ecol Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 42.Mark Welch D, Meselson M. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- 43.Robeson MS2nd, et al. Environmental DNA sequencing primers for eutardigrades and bdelloid rotifers. BMC Ecol. 2009;9:25. doi: 10.1186/1472-6785-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su XZ, Wu Y, Sifri CD, Wellems TE. Reduced extension temperatures required for PCR amplification of extremely A+T-rich DNA. Nucleic Acids Res. 1996;24:1574–1575. doi: 10.1093/nar/24.8.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A, Ludwig T, Meier H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 47.Keane TM, Naughton TJ, McInerney JO. MultiPhyl: A high-throughput phylogenomics webserver using distributed computing. Nucleic Acids Res. 2007;35(Web Server issue):W33–37. doi: 10.1093/nar/gkm359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huson DH, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birky CW, Jr, Adams J, Gemmel M, Perry J. Using population genetic theory and DNA sequences for species detection and identification in asexual organisms. PLoS ONE. 2010;5:e10609. doi: 10.1371/journal.pone.0010609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levenberg K. A method for the solution of certain non-linear problems in least squares. Q Appl Math. 1944;2:164–168. [Google Scholar]
- 53.Marquardt SW. An algorithm for least-squares estimation of nonlinear parameters. J Soc Ind Appl Math. 1963;11:431–441. [Google Scholar]
- 54.Bailey TC, Gatrell AC. Interactive Spatial Data Analysis. Edinburgh Gate, England: Prentice Hall; 1995. [Google Scholar]
- 55.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2010;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.