Abstract
Elastin enables the reversible deformation of elastic tissues and can withstand decades of repetitive forces. Tropoelastin is the soluble precursor to elastin, the main elastic protein found in mammals. Little is known of the shape and mechanism of assembly of tropoelastin as its unique composition and propensity to self-associate has hampered structural studies. In this study, we solve the nanostructure of full-length and corresponding overlapping fragments of tropoelastin using small angle X-ray and neutron scattering, allowing us to identify discrete regions of the molecule. Tropoelastin is an asymmetric coil, with a protruding foot that encompasses the C-terminal cell interaction motif. We show that individual tropoelastin molecules are highly extensible yet elastic without hysteresis to perform as highly efficient molecular nanosprings. Our findings shed light on how biology uses this single protein to build durable elastic structures that allow for cell attachment to an appended foot. We present a unique model for head-to-tail assembly which allows for the propagation of the molecule’s asymmetric coil through a stacked spring design.
Keywords: AFM, SAXS, atomic force microscopy
All mammals rely on elastin to convey extensional elasticity to their tissues. Elastin dominates the mass of the aorta where it encounters the peaks and troughs of systole and diastole over the course of two billion heartbeats in a lifetime (1). The lung expands with each intake of breath and elastically contracts on exhalation. The function of these tissues benefits from minimal energy loss during elastic return in each cycle of expansion and contraction. Additionally, elastin is required to function in an environment that relies on cellular contact (2–4) without compromising persistent elasticity. This high level of physical performance demanded of elastin vastly exceeds and indeed outlasts all human-made elastomers (5).
Elastin is constructed by the hierarchical assembly and cross-linking of many tropoelastin monomers that accumulate on a microfibrillar skeleton. Tropoelastin is encoded by a single gene in humans and predominantly laid down in utero and early childhood, providing a durable resource that is intended to elastically serve until old age. This exquisite assembly helps to generate elastic tissues as diverse as artery, lung, and skin (4). Consequences of elastolytic damage in aortic aneurysms, emphysema, and solar elastosis confirm the key roles of elastin in structure and cellular interactions (6–8). These tissues rely on this paradoxical combination of organized tissue structures built from an intrinsically unstructured protein. Tropoelastin serves as a component of rigidly organized assemblies, yet enables the formation of dynamically distensible, elastic tissues.
Tropoelastin is frequently described in the literature as an unstructured protein, mainly because models of elasticity invoke an element of disorder within the structure (4, 9, 10). While this concept appears to be the case at the fine, more subtle intramolecular level, chaos alone is unlikely to explain the ordered assembly of tropoelastin which culminates in the formation of organized mammalian elastic tissues. This balance of supramolecular order and intramolecular disorder means that at the scale of the tropoelastin protein monomer, one would anticipate sufficient order to provide a functional and usable building block which displays a defined three-dimensional solution shape.
Results and Discussion
Tropoelastin Is a Monomer in Solution.
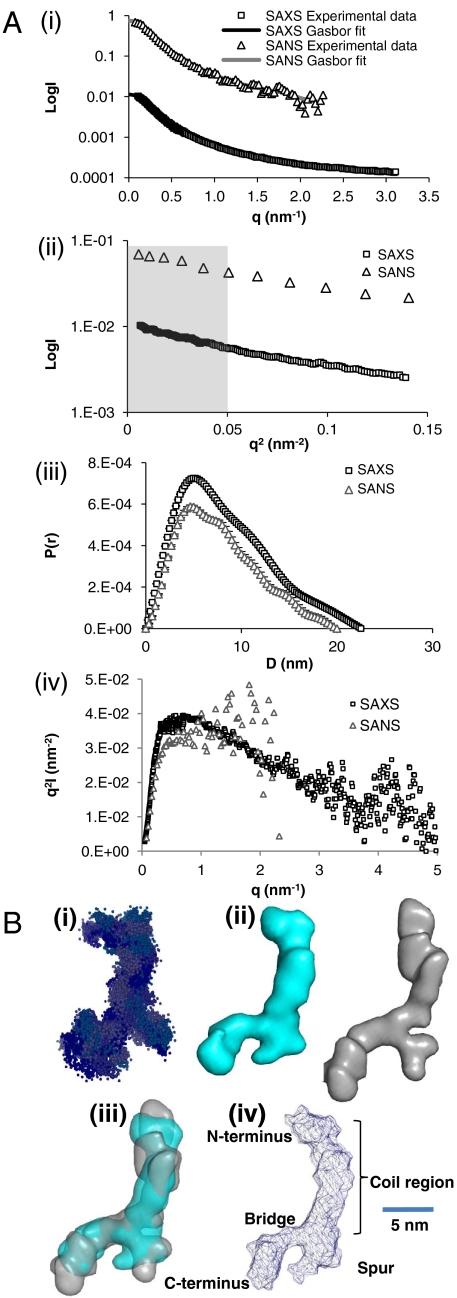
The size and oligomer status of tropoelastin (lacking domain 26A; Fig. 1) was first analyzed by multiangle laser light scattering (MALLS). Tropoelastin presented an experimental molecular mass of 59,520 Da + /-0.5% which correlates closely with the theoretical mass of 60,017 Da and is consistent with the expectation that tropoelastin is a monomer in solution (Fig. S1A). In order to determine the solution shape of human tropoelastin, X-ray and neutron scattering measurements were made on solutions of tropoelastin. Scattering data are shown in Fig. 2 A, i. These data were assessed using Guinier plots, to check that samples were free from aggregation, and the radius of gyration (Rg) and maximum particle dimension were determined (Fig. 2 A, ii, iii). The hydrodynamic radius of the monomer protein [5.09 + /-0.4 nm from MALLS (Fig. S1B)] was smaller than the Rg [6.2 nm from SAXS (small angle scattering)] indicating an elongated species. The Kratky plot traces were indicative of a nonglobular molecule with flexible linkers (11) (Fig. 2 A, iv). The X-ray and neutron scattering data were highly similar and this resulted in similar ab initio shapes from the different scattering experiments, vindicating the robustness of the shape defined.
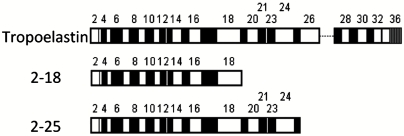
Fig. 1.
Diagrammatic representation of all the tropoelastin constructs used in this study. Black boxes represent hydrophilic domains and white boxes represent hydrophobic domains. The dotted line indicates the absence of exon 26A which is rarely present in tropoelastin.
Fig. 2.
Small angle X-ray and neutron scattering of full-length human tropoelastin. (A, i) The experimental SAXS (squares) and SANS (triangles) data are plotted as a function of q, and compared with a typical theoretical fit obtained with GASBOR (solid line). (ii) The low angle regions of the X-ray scattering data were analyzed in the form of Guinier plots (log I vs. q2), from which the radius of gyration (Rg) can be extracted from the slope (Rg2/3) of the straight line. The slope demonstrates the expected linearity for the values q ≤ 1/Rg (shaded region). (iii) Pair distribution function calculated for the SAXS (squares) and SANS (triangles) datasets. The curves show with error bars the distribution of interatomic spacings, with maxima at 22 nm (SAXS) and 20 nm (SANS). (iv) Kratky plots are shown for the SAXS (squares) and SANS (triangles) data. (B, i) GASBOR ab initio shape of full-length tropoelastin calculated from solution SAXS data, 20 models are shown superimposed. (ii) Filtered average shape of 20 individual SAXS (blue) and SANS (gray) simulations. (iii) Superimposed filtered average SAXS and SANS models. (iv) Labeled diagram of the model for full-length tropoelastin showing proposed locations of the N terminus, the spur region containing exons 20–24, and the C terminus. (Scale bar, 5 nm).
Shape of Tropoelastin.
To determine the 3D shape of tropoelastin in solution, the ab initio programs GASBOR and DAMMIN were used (12, 13). Modeling allowed us to fit the experimental data with discrepancy factors χ of between 1.3 and 2.4. A typical GASBOR fit is displayed in Fig. 2 A, i. For each program, at least twenty separate simulations were completed to determine the uniqueness of the solution. The mean normalized spatial discrepancy (NSD) between the simulated shapes was measured to establish the reliability of the solution (14). GASBOR average shapes were created separately for the SAXS and SANS (small angle neutron scattering) data, each from the multiple runs using the DAMAVER suite of programs [NSD 0.902 (SAXS), 0.927 (SANS)], in order to identify common structural features. These results are shown in Fig. 2B, where each shape is displayed as a surface representation. The ab initio shape of tropoelastin from both SAXS and SANS consistently revealed an elongated asymmetric molecule ∼20 nm along its length, 16 nm end-to-end and 3 nm wide at the narrowest point which opens to 7.5 nm at the widest point (Fig. 2 B, iv). The molecule has clearly distinct regions starting at one terminus with a long narrow region that branches to give a larger more open appearance at the opposite terminus. The narrow elongated shape persists for ∼11 nm before branching to give a “foot” at the end of the molecule. Therefore the molecule is not compact but is relatively extended.
To validate these findings, a shell model was constructed using HYDROPRO based on the ab initio SAXS model (15). This shell model was compared to hydrodynamic measurements of tropoelastin determined by sedimentation velocity analytical ultracentrifugation (16). The model fitted well with the observed hydrodynamic properties (Table S1) (theoretical sedimentation coefficient of 2.24 Svedberg vs. an experimental value of 2.3 + /-0.2 Svedberg), demonstrating that the elongated shape of the ab initio model accurately reflected the solution form of tropoelastin.
Defining the N and C Termini.
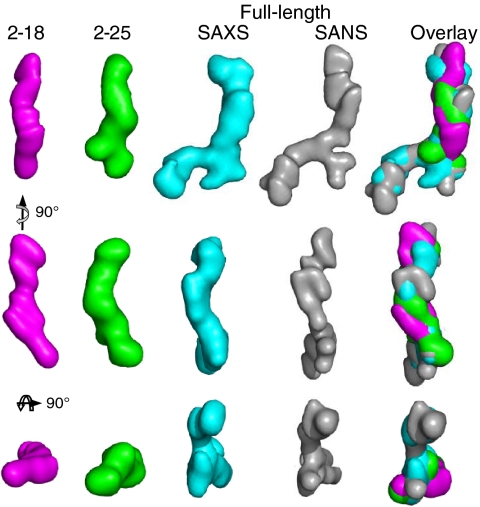
In order to determine which ends of the tropoelastin shape correspond to the N and C termini, further SAXS analysis was performed on defined fragments that extended from a fixed N terminus. Scattering was conducted on the tropoelastin fragments exons 2–18 and exons 2–25 at BioCAT and Diamond Light Source, respectively (Fig. S2). Data were processed using ab initio simulations as described above and shown in Fig. 3. The shape of exons 2–18 was a uniform elongated 14.5 nm long rod-shape that was 2–3 nm wide. This region had a slight bend at approximately 60% along its length to give an internal angle of 133°. The shape of exons 2–25 overall was remarkably similar to that of exons 2–18 (NSD of 0.7 between them); however exons 2–25 had an additional protrusion at one end. The ab initio models of exons 2–18, 2–25, and full-length tropoelastin were superimposed. Exons 2–18 and 2–25 overlay closely with the linear region of the full-length molecule, and because exons 2–18 are common between all three constructs, this suggests that the linear end corresponds to the N terminus of tropoelastin. The protrusion seen in exons 2–25 corresponds exactly to the branching region in the full-length molecule and points to branching occurring at around exon 25 and the more open region therefore corresponding to the C terminus of tropoelastin.
Fig. 3.
Small angle X-ray scattering of overlapping fragments of human tropoelastin. Ab initio models were calculated from SAXS data for tropoelastin constructs 2–18 (pink), 2–25 (green), and full-length tropoelastin (blue) and from SANS data for full-length tropoelastin (gray). The models are shown both individually and superimposed in three orthogonal views.
On this basis, tropoelastin is an asymmetric molecule (16) with a gradual coil along the long, spring-like axis of the molecule. This coil region accounts for most of the elasticity of tropoelastin (17). The spur region protruding from the side of the molecule corresponds to a predicted hinge region containing exons 20–24 (18, 19). Beyond the spur there is a bridge to the C-terminal region. This bridge is likely to encompass domain 26; the junction between domains 25–26 (Arg515) has the highest protease susceptibility of the molecule indicating this region is exposed (20). Between the spur and the bridge, domains 19–25 cluster such that they are enriched in cross links (21). The tropoelastin molecule terminates in a more compact “foot-like” region. This part of the molecule includes the cell-interactive C terminus of tropoelastin (22). The spur is seen in all three solved shapes that encompass this region (SAXS 2–25, SAXS tropoelastin, and SANS tropoelastin), and the foot is verified by SAXS and SANS of tropoelastin (Fig. 3). In summary, structural analysis reveals two dominant, functionally relevant parts of the molecule: the coil which contributes to elasticity and the foot that encompasses the C-terminal cell contact region.
Tropoelastin Molecules Behave as Entropic Springs.
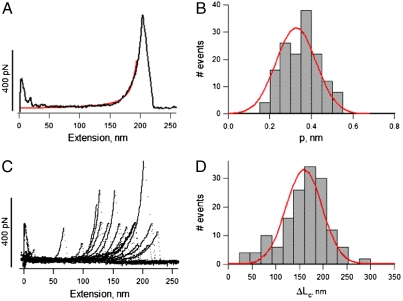
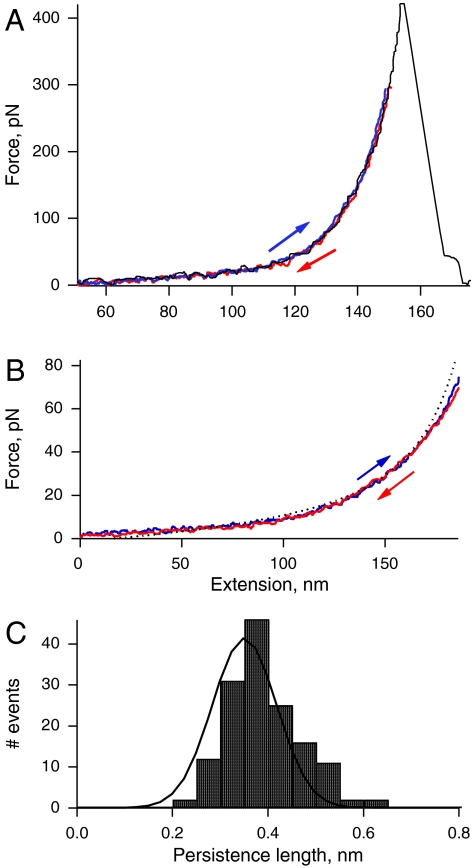
One main characteristic of elastic polymers is their ability to return to their original state after deformation. Using atomic force microscopy (AFM), we stretched and relaxed single-tropoelastin molecules several times within a time scale of approximately 500 ms and up to a force of 80 pN, depending on the length of the molecule being pulled. Tropoelastin molecules responded to pulling forces in a pattern that fits well into the worm-like chain model (red line) with some nondiscrete regions (Fig. 4A). This pattern was consistently observed for multiple single molecule analyses (Fig. 4B). However, tropoelastin does not show typical sawtooth force-extension curves that are characteristic of the unfolding of the elastic titin Ig-like or spectrin alpha-helical domains (23). The median value from the Gaussian fit of frequency of occurrence, yielded a mean persistence length of 0.36 ± 0.14 nm (n = 158) (Fig. 4C). The mean contour length was estimated to be 166 ± 49 nm (n = 158) (Fig. 4D). This value is smaller than that predicted for a 698 amino acid long polypeptide chain (∼250 nm) because the AFM tip picks molecules randomly with respect to their ends. The estimated value for the persistence length (p ∼ 0.36 nm) indicates that the molecule can be fully extended while maintaining elasticity (24–26). Using the persistence length, the Young’s Modulus of the tropoelastin molecule can be calculated as ∼3 kPa which is significantly more elastic than other extracellular matrix and elastic fiber molecules (27, 28).
Fig. 4.
Characterization of the mechanical properties of single-tropoelastin molecules. (A) Example of a force-extension curve for a single molecule. The red line corresponds to a fit to the worm-like-chain model of polymer elasticity using 211 nm for contour length and 0.38 nm for the persistence length. (C) Superposition of several (n = 17) force-extension curves for the stretching of tropoelastin molecules. (B, D) Frequency histograms for persistence lengths, p, and contour lengths, ΔLc. The mean values of persistence and contour lengths are 0.36 ± 0.14 nm (n = 158) and 166 ± 49 nm (n = 158), respectively.
Our stretching experiments show that single-tropoelastin molecules possess ideal mechanical properties that fit the worm-like chain model and show single-chain elasticity. Tropoelastin is essentially simple in terms of its structure, consisting of two types of alternating domains: (i) hydrophilic cross-linking domains rich in Lys and Ala that tend to form α helices, and (ii) hydrophobic domains which confer elasticity, are rich in Val, Pro, Ala, and Gly and tend to form β turns. Unlike titin which is composed of domains that are characterized by well defined discrete discontinuities (sawtooth) during unfolding (23), tropoelastin shows a rather smooth curve suggestive of transient tertiary structures (29, 30). The hydrophobic domains of tropoelastin are likely to be compact amorphous structures that include distorted beta strands and fluctuating turns (31). Tropoelastin has the capacity to extend ∼8 times its resting length as the folded size of tropoelastin is ∼20 nm (Fig. 2) and it can be extended up to ∼160 nm (Fig. 4). Globular domains, such as Ig or FnIII domains, when stretched by AFM extend form a folded length of ∼4 nm to an unfolded length of ∼30 nm (32, 33), which is very similar to tropoelastin. Fig. 5 shows typical forward (blue) and reverse (red) force-extension traces after stretching and relaxing a tropoelastin molecule through multiple cycles. We observed no hysteresis in the tropoelastin molecule after repeated cycles within the time scale used. The stretch and relaxation profiles of tropoelastin impressively show it is a near perfect spring with minimal energy loss. Ideal elastic behavior clearly places tropoelastin in the same category as other natural elastomers such as rubbers and resilin (26, 34, 35).
Fig. 5.
(A) This plot shows a typical experiment where a molecule was extended (blue trace) and relaxed (red trace) two times and then it was overextended in order to detach it from the surface (black trace). This last trace has a single detachment force peak consistent with the stretching pattern of a single molecule. (B) The forward and reverse traces are well characterized by the WLC equation (dotted line) and there is no hysteresis in the time scale of the experiment (∼500 ms). (C) Histogram for the persistence length shows a mean of 0.35 ± 0.09 nm (n = 147; 26 molecules).
In summary single molecules of tropoelastin behave as perfect elastic molecules, because they can be stretched to several times the resting length and overstretching is a fully reversible process.
Implications for the Assembly and Elasticity of Vertebrate Tissue.
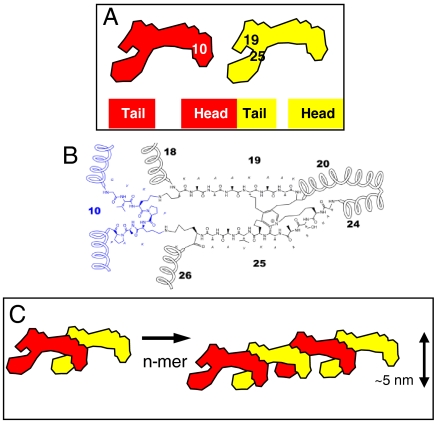
In vivo, elastin displays a prominent three-way covalent connection between domains 10, 19, and 25 that encompasses a rare lysine-specific desmosine cross link (36). The solution shape of tropoelastin places domain 19 near 25 i.e., bracketing the 20–24 hinge, whereas domain 10 is located distally in the coil part of the molecule (Fig. 6A). On the basis that the proximal 19 and 25 are donated by one tropoelastin, and 10 from a second tropoelastin molecule, a straightforward head-to-tail model ensues (Fig. 6). The model is presented simply as a string of monomers because tropoelastin propagates by rapid association via a monomer to n-mer transition, and can accommodate the participation of oligomers in the string (16). This model posits that one of the few desmosines in tropoelastin (37–39) is braced by two lysinonorleucines cross links with domain 10 (36) to establish a strong, reinforced link at the junction of conjoined tropoelastins. Furthermore, this array surrounds the junction with the longest molecular springs, which are provided by the hydrophobic domains 18, 20, 24, and 26. These elastic domains are critical to association (20, 40–42).
Fig. 6.
Head-to-tail model for nascent elastic fiber assembly. (A) Juxtaposed domains 19 and 25 on one tropoelastin molecule and domain 10 on an adjacent monomer. (B) Molecular schematic of the cross link interface. (C) Tandem assembly displaying n-mer propagation as an outcome of covalently bonded molecules.
This model mirrors the broader concept of head-to-tail assembly seen for many structural proteins such as lamin, actin, collagen I, and microfibrillar fibrillin-1. This assembly is supported by a low nanomolar binding constant (22.8 ± 2.9 nM) between associating tropoelastin monomers (Fig. S3). An attractive implication of this tandem arrangement is that the elasticity of the monomer is propagated along the extended molecule. Additionally, these fine tandem fibers would be capable of lateral association through presentation of their long, exposed hydrophobic axes to similar fibers. Lateral association is driven by hydrophobic interactions that dominate the length of the molecule, as evidenced by the self-aggregation of pure, isolated hydrophobic regions of tropoelastin (41, 43, 44).
This study has analyzed the shape of hydrated tropoelastin and the elastic properties of the tropoelastin monomer. We find that elastin’s extraordinary capacity for efficient elasticity resides with the monomer. The solution shape studies establish a design principle for making robust mechanical extension springs that function in an aqueous environment. Gray et al. (45) propose the oiled coil based on the elastic functionality of the hydrophobic segments, while several groups provide evidence for a compatible filamentous model (41, 46, 47). It is clear from the current findings that these models are not mutually exclusive and can be merged. Hydrophobic domains define interspersed elastic segments that can each form a large coil. The shape of tropoelastin is dominated by a condensed coil, so the molecule can now be considered as an “oiled coiled coil.” Remarkably, there are distinct regions of the molecule responsible for elasticity and cell binding, as the cell-binding C terminus is bridged elastically to the coil that dominates tropoelastin. A model of head-to-tail tandem microfibers as intermediates in elastin assembly reveals how to retain elasticity, because the assembled tropoelastin molecules are effectively propagated springs.
Methods
Expression and Purification of Tropoelastin.
Tropoelastin was expressed in bacteria and purified essentially as described previously (48). SHELΔ26A (Synthetic Human Elastin without domain 26A) corresponds to amino acid residues 27–724 (gi 182,020) and describes the 60 kDa mature form of the secreted protein following removal of the signal peptide. Subfragments of tropoelastin, the molecule 2–18 (SHEL N18) and 2–25 (M155n) were prepared as previously described (49–51).
SAXS/SANS.
Small angle X-ray solution scattering on full-length tropoelastin was carried out on ID02 at the European Synchrotron Radiation Facility, Grenoble, France, using 1 m and 5 m sample to detector distances. Measurements were taken at a protein concentration of 5 mg/mL in PBS at 10 °C. The corresponding profiles were merged so as to cover the momentum transfer interval 0.0038 Å-1 < q < 0.53 Å-1. The modulus of the momentum transfer is defined as q = 4π sin θ/λ, where 2θ is the scattering angle, and λ is the wavelength. Images were collected on a Thomson X-ray Intensifier (TH 49–427) lens coupled to a FReLoN CCD camera (2,048 × 2,048 pixels). The wavelength of X-rays used was 0.1 nm. Multiple images were obtained in time frames of 0.1 s to check for radiation damage and protein aggregation between frames. The scattering images obtained were all spherically averaged, corrections for detector artifacts and normalization to absolute scattering intensities were performed using in-house software. Buffer scattering intensities were subtracted from the sample image to remove background scattering using Primus (52). Small angle neutron solution scattering on full-length tropoelastin was carried out at the ISIS Facility. Measurements were taken at a protein concentration of 20 mg/mL in PBS at 10 °C and covered the momentum transfer interval 0.0075 Å-1 < q < 0.29 Å-1.
Synchrotron measurements of tropoelastin encoded by exons 2–18 were conducted at BioCAT. Protein was dissolved in 10 mM Tris HCl pH 7.0, 150 mM NaCl, 10 mM DTT and centrifuged at 16,000 × g for 15 min. The top 50% of the solution was used for data measurement to eliminate potential aggregates. SAXS data were collected while the sample was pumped at 10 μL/ sec through a 1.5 mm diameter quartz capillary that was maintained at 23 °C. The beam was focused on an 8 cm × 16 cm Aviex CCD area detector that was placed at a distance of 1.91 m from the sample cell to record five successive frames of 1 s exposure. The data were averaged and azimuthally integrated then background scattering from buffer was subtracted with in-house written Igor macros. The experimental configuration delivered a q range of 0.006–0.49 A-1 with an X-ray wavelength of 1.033 Å and flux 1013 photons/ sec. Guinier plot examination confirmed that radiation damage was effectively minimized.
Synchrotron measurements of tropoelastin encoded by exons 2–25 were collected using standard procedures on the I22 beamline at Diamond Light Source equipped with a photon counting detector (53, 54). The beam was focused onto the detector placed at a distance of 5 m from the sample cell and X-ray wavelength of 0.083 nm. The covered range of momentum transfer was 0.005 < q < 0.25 Å-1. The data were normalized to the intensity of the incident beam and the scattering of the buffer was subtracted using an in-house program. To check for radiation damage and aggregation during the SAXS experiment, the data were collected in 10 or 20 successive 10 s frames. For all SAXS data, the radius of gyration, forward scattering intensity, and distance distribution function p(r) were evaluated with the program GNOM (55). Particle shapes were restored from the experimental scattering profiles using ab initio modeling with GASBOR (13) and DAMMIN (12). Many GASBOR and DAMMIN simulations (∼20) were performed for each protein fragment, and these generated very similar but not identical shapes in each case. An averaged filtered shape was generated using DAMAVER and DAMFILT.
Atomic Force Microscopy.
The mechanical properties of single-tropoelastin molecules were studied using a home-built single molecule AFM (32, 56, 57) that consists of a detector head (Digital Instruments) mounted on top of a single axis piezoelectric positioner with a strain gauge sensor (P841.10, Physik Instrumente). The P841 has a total travel of 15 μm and is attached to two piezoelectric positioners (P280.10A, Physik Instrumente) that are used to control the x and y position. This system has a z-axis resolution of a few nanometers and can measure forces in the range of 10–10,000 piconewtons (pN). The monitoring of the force is reported by the cantilever, and the control of the movement of the piezoelectric positioners, are achieved by means of two data acquisition boards (PCI 6052E, PCI 6703, National Instruments), and controlled by custom-written software (LabView; National Instruments and Igor, Wavemetrics). For AFM pulling experiments we used square-based pyramidal silicon-nitride tips with a nominal tip radius ∼20 nm on V-shaped 200-μm-long silicon-nitride cantilevers with a nominal spring constant of ∼50 pN/nm (MLCT-AUHW: silicon-nitride gold-coated cantilevers; Veeco Metrology Group). The spring constant of each individual cantilever was calculated using the equipartition theorem (58). The rms force noise (1-kHz bandwidth) was ∼10 pN. Unless noted, the pulling speed of the different force–distance curves was in the range of 0.1–0.5 nm/ms.
Single-Tropoelastin Mechanics.
In a typical experiment, a small aliquot of the purified tropoelastin (≤ 50 μL, 10 μg/mL) was allowed to adsorb to a clean glass coverslip (for ∼10 min) and then rinsed with PBS pH 7.4. Random segments of tropoelastin molecules were then picked up by adsorption to the cantilever tip. The probability of picking up a single molecule was typically kept low (less than one in 50 attempts) by controlling the concentration of tropoelastin used to prepare the coverslips. We found that most pulls (> 70%) did not show any force peaks indicating that no molecules were picked up by the AFM tip. The remaining force-extension curves show a pattern consistent with the stretching of one or more molecules. For example, Fig. 4A shows a force-extension curve that we attribute to the stretching of a single molecule; this can be nicely fitted to the worm-like-chain equation (red line). We also observed other patterns that had multiple peaks in their force-extension curves; these we attribute to the stretching of several molecules in parallel. Single molecule recordings account to only about 1% of the force-extension dataset.
Analysis of Force-Extension Curves.
The extensibility of tropoelastin molecules were analyzed using the worm-like chain model of polymer elasticity (59, 60):
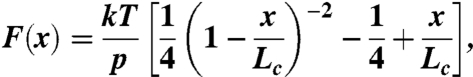 |
where F is force, p is the persistence length, x is end-to-end length, Lc is contour length of the stretched polymer. The adjustable parameters are the persistence length, p, and the contour length, Lc.
Calculation of Young’s Modulus.
The persistence length, p, is proportional to the Young’s modulus, E (61):
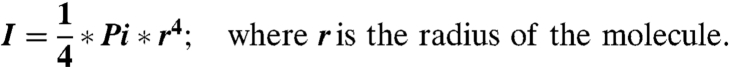 |
Assuming a wet sphere, the radius of the molecule can be taken as 5 nm which yields a Young’s modulus of E = 2.9 kPa.
Supplementary Material
Acknowledgments.
We thank Theyencheri Narayanan of the European Synchrotron Radiation Facility, Grenoble, France. The support of the European Synchrotron Radiation Facility, Advanced Photon Source, and Diamond Light Source is gratefully acknowledged. We thank Dr. Tom Jowitt from the Biomolecular Analysis Facility, University of Manchester for assistance with MALLS data collection and Dr. Adam Huffman for computing support. We thank Hugh Beedie and James Osborne (Cardiff University Information Services) for implementation of distributed software. A.S.W. acknowledges funding support from the Australian Research Council, National Health and Medical Research Council, National Heart Research Fund, University of Sydney, and the Australian Institute of Nuclear Science and Engineering. The AFM study was funded by the John Sealy Memorial Endowment Fund for Biomedical Research and in part by National Institutes of Health (NIH) Grant R01DK073394.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014280108/-/DCSupplemental.
References
- 1.Li DY, et al. Elastin is an essential determinant of arterial morphogenesis. Nature. 1998;393:276–280. doi: 10.1038/30522. [DOI] [PubMed] [Google Scholar]
- 2.Niklason LE, et al. Functional arteries grown in vitro. Science. 1999;284:489–493. doi: 10.1126/science.284.5413.489. [DOI] [PubMed] [Google Scholar]
- 3.Niklason LE, Langer R. Prospects for organ and tissue replacement. Jama J Am Med Assoc. 2001;285:573–576. doi: 10.1001/jama.285.5.573. [DOI] [PubMed] [Google Scholar]
- 4.Vrhovski B, Weiss AS. Biochemistry of tropoelastin. Eur J Biochem. 1998;258:1–18. doi: 10.1046/j.1432-1327.1998.2580001.x. [DOI] [PubMed] [Google Scholar]
- 5.Urry DW, et al. Elastin: a representative ideal protein elastomer. Philos T Roy Soc B . 2002;357:169–184. doi: 10.1098/rstb.2001.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campa JS, Greenhalgh RM, Powell JT. Elastin degradation in abdominal aortic aneurysms. Atherosclerosis. 1987;65:13–21. doi: 10.1016/0021-9150(87)90003-7. [DOI] [PubMed] [Google Scholar]
- 7.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 8.Chen VL, Fleischmajer R, Schwartz E, Palaia M, Timpl R. Immunochemistry of elastotic material in sun-damaged skin. J Invest Dermatol. 1986;87:334–337. doi: 10.1111/1523-1747.ep12524421. [DOI] [PubMed] [Google Scholar]
- 9.Urry DW. Entropic elastic processes in protein mechanisms. Ii. simple (passive) and coupled (active) development of elastic forces. J Protein Chem. 1988;7:81–114. doi: 10.1007/BF01025240. [DOI] [PubMed] [Google Scholar]
- 10.Debelle L, Tamburro AM. Elastin: molecular description and function. Int J Biochem Cell Biol. 1999;31:261–272. doi: 10.1016/s1357-2725(98)00098-3. [DOI] [PubMed] [Google Scholar]
- 11.Mertens HD, Svergun DI. Structural characterization of proteins and complexes using small-angle x-ray solution scattering. J Struct Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from x-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toonkool P, Regan DG, Kuchel PW, Morris MB, Weiss AS. Thermodynamic and hydrodynamic properties of human tropoelastin. analytical ultracentrifuge and pulsed field-gradient spin-echo Nmr studies. J Biol Chem. 2001;276:28042–28050. doi: 10.1074/jbc.M103391200. [DOI] [PubMed] [Google Scholar]
- 17.Holst J, et al. Substrate elasticity provides mechanical signals for the expansion of hemopoietic stem and progenitor cells. Nat Biotechnol. 2010;28:1123–1128. doi: 10.1038/nbt.1687. [DOI] [PubMed] [Google Scholar]
- 18.Kumashiro KK, Ho JP, Niemczura WP, Keeley FW. Cooperativity between the hydrophobic and cross-linking domains of elastin. J Biol Chem. 2006;281:23757–23765. doi: 10.1074/jbc.M510833200. [DOI] [PubMed] [Google Scholar]
- 19.Dyksterhuis LB, Baldock C, Lammie D, Wess TJ, Weiss AS. Domains 17-27 of tropoelastin contain key regions of contact for coacervation and contain an unusual turn-containing crosslinking domain. Matrix Biol. 2007;26:125–135. doi: 10.1016/j.matbio.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Jensen SA, Vrhovski B, Weiss AS. Domain 26 of tropoelastin plays a dominant role in association by coacervation. J Biol Chem. 2000;275:28449–28454. doi: 10.1074/jbc.M004265200. [DOI] [PubMed] [Google Scholar]
- 21.Wise SG, Mithieux SM, Raftery MJ, Weiss AS. Specificity in the coacervation of tropoelastin: solvent exposed lysines. J Struct Biol. 2005;149:273–281. doi: 10.1016/j.jsb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Bax DV, Rodgers UR, Bilek MM, Weiss AS. Cell adhesion to tropoelastin is mediated via the C-terminal grkrk motif and integrin alphavbeta3. J Biol Chem. 2009;284:28616–28623. doi: 10.1074/jbc.M109.017525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberhauser AF, Carrion-Vazquez M. Mechanical biochemistry of proteins one molecule at a time. J Biol Chem. 2008;283:6617–6621. doi: 10.1074/jbc.R700050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin G, et al. Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for the assembly of microfibrils. J Biol Chem. 2002;277:50795–50804. doi: 10.1074/jbc.M210611200. [DOI] [PubMed] [Google Scholar]
- 25.Li H, et al. Multiple conformations of Pevk proteins detected by single-molecule techniques. Proc Natl Acad Sci USA. 2001;98:10682–10686. doi: 10.1073/pnas.191189098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lv S, et al. Designed biomaterials to mimic the mechanical properties of muscles. Nature. 2010;465:69–73. doi: 10.1038/nature09024. [DOI] [PubMed] [Google Scholar]
- 27.Kielty CM, Sherratt MJ, Marson A, Baldock C. Fibrillin microfibrils. Adv Protein Chem. 2005;70:405–436. doi: 10.1016/S0065-3233(05)70012-7. [DOI] [PubMed] [Google Scholar]
- 28.Kielty CM, Sherratt MJ, Shuttleworth CA. Elastic fibres. J Cell Sci. 2002;115:2817–2828. doi: 10.1242/jcs.115.14.2817. [DOI] [PubMed] [Google Scholar]
- 29.Muiznieks LD, Weiss AS. Flexibility in the solution structure of human tropoelastin. Biochemistry. 2007;46:8196–8205. doi: 10.1021/bi700139k. [DOI] [PubMed] [Google Scholar]
- 30.Dyksterhuis LB, Carter EA, Mithieux SM, Weiss AS. Tropoelastin as a thermodynamically unfolded premolten globule protein: the effect of trimethylamine N-oxide on structure and coacervation. Arch Biochem Biophys. 2009;487:79–84. doi: 10.1016/j.abb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Samouillan V, Andre C, Dandurand J, Lacabanne C. Effect of water on the molecular mobility of elastin. Biomacromolecules. 2004;5:958–964. doi: 10.1021/bm034436t. [DOI] [PubMed] [Google Scholar]
- 32.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 33.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by Afm. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 34.Elvin CM, et al. Synthesis and properties of crosslinked recombinant pro-resilin. Nature. 2005;437:999–1002. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 35.Cordier P, Tournilhac F, Soulie-Ziakovic C, Leibler L. Self-healing and thermoreversible rubber from supramolecular assembly. Nature. 2008;451:977–980. doi: 10.1038/nature06669. [DOI] [PubMed] [Google Scholar]
- 36.Brown-Augsburger P, Tisdale C, Broekelmann T, Sloan C, Mecham RP. Identification of an elastin cross-linking domain that joins three peptide chains. possible role in nucleated assembly. J Biol Chem. 1995;270:17778–17783. doi: 10.1074/jbc.270.30.17778. [DOI] [PubMed] [Google Scholar]
- 37.Foster JA, et al. Isolation and characterization of cross-linked peptides from elastin. J Biol Chem. 1974;249:6191–6196. [PubMed] [Google Scholar]
- 38.Lonky SA, Gochman N, Smith S, Bergeron-Lynn G, Jacobs K. Amino acid analysis of elastin hydrolysates using a lithium citrate gradient: quantification of elastin from whole lung. Clin Chim Acta. 1981;110:227–233. doi: 10.1016/0009-8981(81)90351-x. [DOI] [PubMed] [Google Scholar]
- 39.Daamen WF, Hafmans T, Veerkamp JH, Van Kuppevelt TH. Comparison of five procedures for the purification of insoluble elastin. Biomaterials. 2001;22:1997–2005. doi: 10.1016/s0142-9612(00)00383-5. [DOI] [PubMed] [Google Scholar]
- 40.Toonkool P, Jensen SA, Maxwell AL, Weiss AS. Hydrophobic domains of human tropoelastin interact in a context-dependent manner. J Biol Chem. 2001;276:44575–44580. doi: 10.1074/jbc.M107920200. [DOI] [PubMed] [Google Scholar]
- 41.Tamburro AM, Bochicchio B, Pepe A. Pathol Biol. Vol. 53. Paris: 2005. The dissection of human tropoelastin: from the molecular structure to the self-assembly to the elasticity mechanism; pp. 383–389. www.ncbi.nlm.nih.gov/pubmed/16085114. [DOI] [PubMed] [Google Scholar]
- 42.Keeley FW, Bellingham CM, Woodhouse KA. Elastin as a self-organizing biomaterial: use of recombinantly expressed human elastin polypeptides as a model for investigations of structure and self-assembly of elastin. Philos T Roy Soc B. 2002;357:185–189. doi: 10.1098/rstb.2001.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostuni A, Bochicchio B, Armentano MF, Bisaccia F, Tamburro AM. Molecular and supramolecular structural studies on human tropoelastin sequences. Biophys J. 2007;93:3640–3651. doi: 10.1529/biophysj.107.110809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao M, et al. Sequence and structure determinants for the self-aggregation of recombinant polypeptides modeled after human elastin. J Biol Chem. 2003;278:48553–48562. doi: 10.1074/jbc.M308465200. [DOI] [PubMed] [Google Scholar]
- 45.Gray WR, Sandberg LB, Foster JA. Molecular model for elastin structure and function. Nature. 1973;246:461–466. doi: 10.1038/246461a0. [DOI] [PubMed] [Google Scholar]
- 46.Cox BA, Starcher BC, Urry DW. Coacervation of alpha-elastin results in fiber formation. Biochim Biophys Acta. 1973;317:209–213. doi: 10.1016/0005-2795(73)90215-8. [DOI] [PubMed] [Google Scholar]
- 47.Urry DW, Long MM. On the conformation, coacervation and function of polymeric models of elastin. Adv Exp Med Biol. 1977;79:685–714. doi: 10.1007/978-1-4684-9093-0_59. [DOI] [PubMed] [Google Scholar]
- 48.Martin SL, Vrhovski B, Weiss AS. Total synthesis and expression in Escherichia coli of a gene encoding human tropoelastin. Gene. 1995;154:159–166. doi: 10.1016/0378-1119(94)00848-m. [DOI] [PubMed] [Google Scholar]
- 49.Vrhovski B, Jensen S, Weiss AS. Coacervation characteristics of recombinant human tropoelastin. Eur J Biochem. 1997;250:92–98. doi: 10.1111/j.1432-1033.1997.00092.x. [DOI] [PubMed] [Google Scholar]
- 50.Wu WJ, Vrhovski B, Weiss AS. Glycosaminoglycans mediate the coacervation of human tropoelastin through dominant charge interactions involving lysine side chains. J Biol Chem. 1999;274:21719–21724. doi: 10.1074/jbc.274.31.21719. [DOI] [PubMed] [Google Scholar]
- 51.Wu WJ, Weiss AS. Deficient coacervation of two forms of human tropoelastin associated with supravalvular aortic stenosis. Eur J Biochem. 1999;266:308–314. doi: 10.1046/j.1432-1327.1999.00891.x. [DOI] [PubMed] [Google Scholar]
- 52.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. Primus: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 53.Berry A, et al. The Rapid2 X-ray detection system. Nucl Instrum Methods. 2003;513:260–263. [Google Scholar]
- 54.Lewis RA, et al. The rapid high rate large area X-ray detector system. Nucl Instrum Methods. 1997;392:32–41. [Google Scholar]
- 55.Semenyuk AV, Svergun DI. Gnom—a program package for small-angle scattering data processing. J Appl Crystallogr. 1991;24:537–540. [Google Scholar]
- 56.Bullard B, Garcia T, Benes V, Leake MC, Linke WA, Oberhauser AF. The molecular elasticity of the insect flight muscle proteins projectin and kettin. Proc Natl Acad Sci USA. 2006;103:4451–4456. doi: 10.1073/pnas.0509016103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller E, Garcia T, Hultgren S, Oberhauser AF. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys J. 2006;91:3848–3856. doi: 10.1529/biophysj.106.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Florin E-L, et al. Sensing specific molecular interactions with the atomic force microscope. Biosensors and Bioelectronics. 1995;10:895–901. http://dx.doi.org/10.1016/0956-5663(95)99227-C. [Google Scholar]
- 59.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 60.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 61.Graham JS, Vomund AN, Phillips CL, Grandbois M. Structural changes in human type I collagen fibrils investigated by force spectroscopy. Exp Cell Res. 2004;299:335–342. doi: 10.1016/j.yexcr.2004.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.