Abstract
Abnormally elevated lipid and glucose levels due to the disruption of metabolic homeostasis play causative roles in the development of metabolic diseases. A cluster of metabolic conditions, including dyslipidemia, abdominal obesity, and insulin resistance, is referred to as metabolic syndrome which has been increasing globally at an alarming rate. The primary nuclear bile acid receptor, Farnesoid X Receptor (FXR, NR1H4), plays important roles in controlling lipid and glucose levels by regulating expression of target genes in response to bile acid signaling in enterohepatic tissues. In this review, I discuss how signal-dependent FXR transcriptional activity is dynamically regulated under normal physiological conditions and how it is dysregulated in metabolic disease states. I focus on the emerging roles of post-translational modifications (PTMs) and transcriptional cofactors in modulating FXR transcriptional activity and pathways. Dysregulation of nuclear receptor transcriptional signaling due to aberrant PTMs and cofactor interactions are key determinants in the development of metabolic diseases. Therefore, targeting such abnormal PTMs and transcriptional cofactors of FXR in disease states may provide a new molecular strategy for development of pharmacological agents to treat metabolic syndrome.
1. Introduction
Metabolic syndrome affects almost one-third of adults in the United States and is increasing globally at an alarming rate [1]. It is, therefore, important to understand the molecular mechanisms that control metabolic pathways in normal and disease states, so that new strategies for therapeutic interventions can be developed. The primary nuclear bile acid receptor, Farnesoid X Receptor (FXR, NR1H4), plays an important role in controlling metabolite levels by activating or repressing groups of its target genes that regulate lipid and glucose metabolism [2–6]. Despite recent advances in understanding the biology of FXR, how FXR regulates its target genes and how FXR activity is modulated are poorly understood. Nuclear receptors (NRs), including FXR, act as biosensors for extracellular signals and transmit those signals to transcriptional machinery to alter expression of target genes [7–9]. In response to various cellular signals, including endocrine steroid/thyroid hormones, vitamins, and dietary lipids, NRs recruit distinct combinations of numerous transcriptional cofactors to effectively modulate transcription of their target genes [8, 9]. Some of these cofactors can profoundly modulate NR-mediated transcriptional responses by catalyzing post-translational modifications (PTMs) of histones at the NR target genes and also by catalyzing PTMs of non-histone proteins involved in transcriptional regulation, including the NRs and their cofactors [8, 9]. A number of excellent reviews highlighting the biological functions of FXR in health and disease have been recently published [2–6, 10–12]. Therefore, in this article, I focus on how FXR transcriptional signaling is regulated under normal conditions by transcriptional cofactors and PTMs and how FXR signaling is dysregulated in metabolic disease.
2. FXR as the primary nuclear bile acid receptor
FXR as a member of the NR superfamily
Nuclear receptors (NRs) are ligand-regulated transcription factors that function as transcriptional switches in response to lipophilic signaling molecules, including endocrine hormones, vitamins, xenobiotics, and dietary lipids [7]. They bind to specific DNA sequences and, thereby, regulate expression of target genes that are involved in every aspect of mammalian physiology. FXR is a member of the NR family [7]. FXR was originally cloned in 1995 as a novel Retinoid X Receptor (RXR, NR2B1) interacting protein (RIP14) and characterized as a NR that was weakly activated by farnesol metabolites [13, 14]. FXR is expressed mainly in liver, intestine, and kidney, but is also expressed in the adrenal gland, pancreas, and reproductive tissues [2–5, 10, 12]. Human and mouse genes encode four isoforms, FXR α1, α2, α3, α4, as a result of alternative use of two different promoters and alternative splicing between exons 5 and 6 [2–5, 10, 12]. Whether these isoforms have distinct cellular and physiological functions in the regulation of FXR target genes is unclear.
FXR ligands
FXR is a biosensor for endogenous bile acids [2–5, 10, 12]. A primary bile acid, chenodeoxy cholic acid (CDCA), is the most potent natural FXR agonist with a half maximal effective concentration (EC50) value of about 10 μM. Secondary bile acids, lithocholic acid (LCA) and deoxycholic acid (DCA), also activate FXR, but a lesser extent [2, 5, 10, 11]. Gugglesterone, a gugglipid from the mukul tree, has been reported to be a FXR antagonist [15]. Bile acids were shown to activate nuclear receptors, PXR, CAR, and vitamin D receptor in addition to FXR, and also to activate cell signaling kinase pathways [2–5, 10, 12]. Recently, a G protein-coupled receptor (GPCR), TGR5, was identified as a membrane bile acid receptor [6, 12, 16, 17]. Because bile acids can activate multiple signaling pathways, specific synthetic agonists, including GW4064 and fexaramine, and a semi-synthetic agonist, 6E-CDCA, have provided powerful tools to dissect FXR-specific transcriptional signaling [3, 4, 6, 12, 18, 19]. Structural 5 anlysis of the rat and human FXR LBD bound to synthetic or natural ligands and coactivator peptides have revealed significant insights in the mechanisms of FXR activation by its ligands [19, 20]. Amino acids involved in receptor-ligand interaction are conserved between species. Although FXR LBD contains a general organization of 12 α-helix bundles like other NRs, unexpected findings associated with bile acid binding and activation of FXR by coactivators were observed. Bile acids occupy the FXR LBD binding pocket in the reverse orientation compared to ligands for other NRs [20]. Furthermore, when bound to a ligand, FXR has two docking sites for interaction with LXXLL motifs, which results in a cooperative increase in binding affinity for coactivators which contain more than one LXXLL NR interaction domain [20].
FXR DNA binding motifs
FXR, upon heterodimerization with RXRα, binds to DNA sequences, called FXR response elements (FXREs), and activates transcription of target genes [2–5, 10, 11]. The FXRE contains two copies of a six nucleotide sequence (AGGTCA or closely related sequences) arranged as inverted repeats separated by one nucleotide, called IR1. The FXR/α RXR heterodimer also binds to and activates a variety of other FXREs, such as IR0, IR8, ER8, or DR1, but binds to the consensus IR-1 sequence with the highest affinity [2–5, 10]. In rare cases, negative FXREs have been found in FXR target genes, such as ApoA1, ApoCIII, and UGT2B4 [2–5, 10, 21]. FXR was shown to bind to the ApoA1 gene as a monomer, which is associated with repression of the gene [22]. However, whether these unusual FXREs, including the negative FXREs, play significant roles in mediating regulation of transcription by FXR and the precise molecular mechanisms by which FXR binds to its target genes as a monomer and represses their transcription are not clearly understood.
Chromatin immunoprecipitation followed by deep sequencing (ChIP–seq) is a powerful technique for the identification of direct binding sites for transcription factors in the entire genome. Guo and colleagues recently identified tissue-specific FXR binding sites in liver and intestine by Chip-seq analysis of mice treated with GW4064 [23]. Only 11% of total sites were shared between liver and intestine, demonstrating tissue-specific FXR/gene interactions. Analysis of the binding motifs revealed that a half nuclear receptor binding site was often located adjacent to the FXREs, indicating possible involvement of other nuclear receptors in modulating FXR function. Consistent with these findings, Osborne and colleagues reported that LRH-1, as a monomeric nuclear receptor partner for FXR, binds to a nuclear receptor half site near the FXRE to co-activate gene expression [24]. Consistent with previous in vitro studies [2–5, 10], the most common FXRE in these studies was the IR1 motif [23, 24]. Gene ontology analyses revealed broader biological functions of FXR than previously appreciated. FXR-binding sites were detected close to many genes involved in lipid, fatty acid, and steroid metabolism but also to other broad gene clusters related to metabolism, transport, kinase signaling, and glycolysis.
3. Functions of FXR
FXR null mice, synthetic FXR ligands, and gene profiling analyses have been extensively used to analyze the biological functions of FXR [2–6, 10, 25]. A number of excellent articles reviewing the functions of FXR in diverse biological pathways, including metabolism, liver regeneration, anti-arthrosclerosis, tumor suppressor, and inhibition of intestinal bacterial growth, have been recently published [2–6, 10–12]. Therefore, I will briefly highlight functions of FXR in lipid and glucose metabolism, and also survey known FXR-related diseases and human polymorphisms in the FXR gene.
FXR and bile acid metabolism
FXR plays a pivotal role in maintaining bile acid homeostasis by regulating every aspect of bile acid metabolism including synthesis, transport, and refilling of the gall bladder [2–5, 10, 11, 26]. FXR senses elevated hepatic bile acid levels and inhibits hepatic bile acid biosynthesis from cholesterol by induction of a metabolic repressor, Small Heterodimer Partner (SHP) [2–5, 10]. SHP is an unusual orphan nuclear receptor that lacks a DBD [27] but acts as a bile acid-induced transcriptional corepressor [28, 29]. SHP has features of an AF-2 cofactor, i.e. that binds via LXXLL motifs to the AF-2 domain of its target NRs [30]. SHP forms non-functional heterodimers with DNA binding activators, including NRs, and inhibits their transcriptional activity [30, 31]. SHP inhibits expression of cholesterol 7α hydroxylase (CYP7A1), a rate-limiting enzyme in the bile acid biosynthetic pathway, by blocking transactivation of hepatic activators, LRH-1 and HNF-4, at the promoter. In addition, FXR protects the liver from elevated bile acid levels by regulating the expression of bile acid transporters [2, 5, 10, 11]. FXR inhibits the entry of intestinal bile acids into hepatocytes by repressing the expression of Na+-taurocholate cotransporting polypeptide (ntcp), a hepatic bile acid import transporter [32, 33]. FXR also promotes the efflux of bile acids from the liver by inducing expression of the bile salt export pump (BSEP), the multidrug resistant-associated protein 2 (MRP2), and the multidrug resistance P-glycoprotein 3 (MDR3) [32, 33]. These transporters, BSEP, MRP2, and MDR3, mediate the transport of bile acids across the canalicular membrane into the bile duct. Bile acids, together with cholesterol and phospholipids, are the major components of bile which is stored in the gallbladder. Moschetta et al.[11, 34] demonstrated that FXR plays a critical role in preventing gallstone formation by induction of bile acid transporters, BSEP and MDR2, which results in increased bile acid levels in the gall bladder and prevents cholesterol crystallization in the bile.
FXR and lipid metabolism
The function of hepatic FXR in regulating fatty acid, triglyceride, and lipoprotein metabolism is complex. Previous clinical studies have suggested an inverse relationship between the bile acid pool size and triglyceride (TG) levels [3, 5, 6, 10]. Treatment of dyslipidemic patients with bile acid binding resin resulted in decreased cholesterol levels but also in undesirable side effects of increased plasma TG levels and VLDL levels [5]. Conversely, administration of bile acids to humans or animals was shown to decrease plasma triglyceride levels and but also, undesirably, to decrease HDL levels [5, 10]. Consistent with these clinical studies, activation of FXR by treatment with FXR agonists resulted in decreased hepatic expression of SREBP-1c, a key lipogenic activator, and increased expression of PPARα which promotes fatty acid β-oxidation [2, 5, 10, 11]. FXR activation also resulted in increased expression of the VLDL receptor that promotes TG clearance. In addition, FXR activation led to increased expression of apo CII that coactivates lipoprotein lipase but decreased expression of apo CIII which functions as an inhibitor of lipoprotein lipase. Consistent with these findings, activation of FXR transcriptional signaling by synthetic or natural FXR ligands has been shown to have beneficial metabolic outcomes in some metabolic pathways, but undesirable effects in others, especially HDL biosynthesis [2–6, 10, 11]. In this regard, understanding the precise molecular mechanisms by which FXR activity is regulated in a gene-specific manner will be important in order to identify molecular targets for such selective therapeutic agents.
FXR and glucose metabolism
FXR plays an important role in controlling glucose levels [2, 5, 6, 10, 11]. Normal glucose homeostasis was disrupted in FXR-null mice with development of insulin resistance and hyperglycemia [35]. The insulin resistance and impaired insulin signaling in FXR null mice were likely due to elevated fatty acid levels [35]. Activation of FXR by treatment with an agonist resulted in repression of the gluconeogenic genes, PEPCK and glucose-6-phosphatase, and these effects were not observed in FXR- or SHP-null mice, suggesting that the FXR/SHP pathway plays a role in the regulation of hepatic glucose production [35]. Consistent with these findings, activation of FXR resulted in improved insulin sensitivity and glucose tolerance in diabetic obese mice [36]. Treatment of diabetic mice with FXR agonists or hepatic expression of constitutively active FXR-VP16 led to significant reduction of plasma glucose levels and improved insulin sensitivity [36].
FXR-related human diseases and human polymorphisms
Consistent with the roles of FXR in regulating levels of metabolites, FXR null mice exhibited a cluster of metabolic abnormalities, including dyslipidemia, cholesterol gall stone formation, hepatic steatosis, cholestasis, hyperglycemia, insulin insensitivity, and arthrosclerosis [2–6, 10, 25]. Furthermore, new functions of FXR in liver regeneration, defense mechanisms against intestinal bacteria, tumor suppression, and diabetic kidney pathology have been discovered [37–39]. Given that FXR is a critical metabolic regulator, genetic variations in FXR could underlie FXR-related diseases. Several human FXR polymorphisms have been reported [40, 41]. Intrahepatic cholestasis of pregnancy (ICP) is characterized by liver impairment, pruritus, and elevated maternal serum bile acids and can cause premature delivery and intrauterine death. Williamson and colleagues have identified 4 novel heterozygous FXR variants in ICP, three amino acid coding changes, M1V, W80R, and M173T, and one change in the mRNA at the nucleotide before the ATG translation start codon (G-1T) [41]. Using FXR expression plasmids encoding the variants, they further showed functional defects in either translation efficiency or activity for 3 of the 4 variants (G-1T, M1V, and M173T). Kim and colleagues have analyzed FXR polymorphisms in European, African, Chinese, and Hispanic-Americans populations [40]. Five polymorphisms in the coding region of FXR, including two rare single nucleotide polymorphisms (SNPs) and a common SNP, were identified in this study. The two rare SNPs within the hinge region of FXR, C643T (H215Y), and G646T (A216S), and the SNP (G-1T) were detected [40]. Expression of FXR target genes, SHP and OATP, but not BSEP, was significantly reduced in livers harboring the FXR (G-1T) mutant, which suggests that the effects of the FXR polymorphism on transcriptional responses may be gene-specific. Additional studies in human populations with different susceptibilities to metabolic disorders may provide valuable insights into FXR signaling that could be exploited to treat diseases.
4. Transcriptional Cofactors and Post-Translational Modifications (PTMs)
Despite recent advances in understanding of the biology of FXR, the molecular basis of how FXR regulates expression of its target genes and how the transcriptional activity of FXR is regulated are relatively unexplored. In this section, I survey the transcriptional cofactors (also called transcriptional coregulators) and PTMs that potentially modulate FXR transcriptional signaling pathways. Unliganded NRs have been shown to interact with SMRT/N-CoR corepressor complexes and, thereby, repress their target genes [9, 42]. Likewise, unliganded or antagonist-bound FXR may be associated with these transcriptional corepressors at its target genes although this area is not well studied for FXR. In response to diverse cellular signals including endocrine hormones and dietary lipids, nuclear receptors such as FXR, recruit distinct combinations of transcriptional cofactors to effectively regulate transcription of their target genes [7, 9, 43]. These cofactors often profoundly modulate nuclear receptor transcriptional signaling pathways by catalyzing PTMs at two levels, first, by modifying histones at NR target genes, and second, modifying non-histone regulatory proteins including NRs and the cofactors themselves [9, 44]. Transcriptional cofactors, therefore, not only catalyze PTMs but also serve as targets of PTMs in response to cellular signals.
PTMs of histones
There are two classes of chromatin modifying cofactors that alter chromatin structure and gene activity [44] . The first class includes histone modifying cofactors which catalyze PTMs of histones [45]. PTMs of core histones serve as a code that increases or decreases transcription by modulating transcriptional factor access to DNA via altered local chromatin structure. Histone acetyltransferases (HATs) or histone deacetylases (HDACs) covalently modify histones by adding or removing, respectively, acetyl groups on lysine residues in core histones[44, 46]. Histone acetylation and deacetylation generally correlate with gene activation and repression, respectively [44, 46]. In contrast to acetylation, histone methylation may result in either gene activation or repression depending on the amino acid residues that are methylated and type of methylation [44, 47]. For instance, methylation of histone H3 at Lys 4 (H3K4) is associated with gene activation, whereas gene repression is associated with methylation of H3K9 [48–51]. The functional importance of histone modifying transcriptional cofactors in FXR/SHP-mediated feedback regulation of hepatic bile acid synthesis was recently demonstrated [50, 52, 53]. In response to bile acid signaling, SHP coordinately recruits chromatin modifying cofactors, including HDACs and G9a lysine methyltransferase, to the promoter of the key bile acid biosynthetic CYP7A1 gene, resulting in histone modification and gene repression [50, 52]. Importantly, blocking endogenous G9a methyltransferase activity in liver by expression of a G9a dominant mutant reversed bile acid-mediated CYP7A1 inhibition which resulted in an enlarged gall bladder and elevated bile acid pools [50].
The second type of transcriptional cofactors modulating chromatin structure and gene activity is ATP-dependent chromatin remodeling complexes such as Swi/Snf [54]. Using energy from ATP hydrolysis, these chromatin remodeling complexes alter nucleosome structure by disrupting DNA and histone interactions, resulting in either transcriptionally activated accessible or transcriptionally repressive closed chromatin configurations [55]. A Brg-1-containing Swi/Snf complex was recently shown to interact with bile acid-activated FXR and to be recruited to the SHP promoter, resulting in chromatin remodeling to an open chromatin configuration and gene activation [56]. These studies demonstrate that chromatin remodeling cofactors as well as cofactors catalyzing PTMs have a role in regulation of bile acid biosynthesis.
PTMs of non-histone regulatory proteins
In addition to PTM of histones, numerous studies have shown that transcriptional cofactors, such as HATs and HDACs, also modulate activity of non-histone proteins by post-translational acetylation and deacetylation, respectively [8, 9, 57]. Transactivation by important metabolic regulators including FXR, LXR, SREBP-1, Foxo-1, HNF-4, p53, and PGC-1α , were substantially modulated by their acetylation status [58-64]. Acetylation and deacetylation of these proteins can increase or decrease their transcriptional activity by altering DNA binding, interaction with other cofactors, cellular localization, or protein stability[58–64]. For instance, acetylation of p53, SREBP-1c, and HNF-4 increased their ability to bind to the DNA [59, 62, 64], whereas acetylation of Foxo-1 and FXR decreased their DNA binding and transactivation ability [58, 63]. Interestingly, acetylation of p53 at different sites selectively affected biological pathways by targeting different subsets of p53 target genes [65]. Since FXR also activates or inhibits subsets of target genes, it will be interesting to test whether PTMs of FXR also contribute to such different gene-specific functional outcomes.
5. FXR and Transcriptional Cofactors
Bile acids as signaling molecules
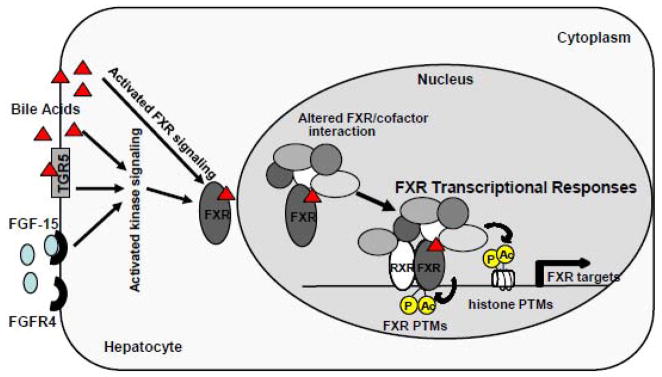
It is now clear that bile acid signaling pathways in hepatocytes are complex (Fig. 1). Bile acids facilitate digestion of lipid-soluble nutrients but also function as signaling molecules that mediate integrative metabolic regulation by activating both cellular signaling kinases and nuclear receptors, primarily FXR [2, 6, 17, 66]. In addition, recent studies have identified a GPCR, TGR5, as a membrane bile acid receptor [6, 16]. An intriguing role of FXR as an integrator of a gut/liver endocrine regulatory axis was also demonstrated [67]. FXR activation induces expression of FGF-15 in the small intestine. The secreted FGF-15 binds to a liver transmembrane receptor tyrosine kinase, FGF-15 receptor 4 (FGFR4), and triggers cellular signaling kinase cascades in hepatocytes. Activation of bile acid receptors and cell signaling pathways, such as ERK, JNK, PKB (Akt), and PKC, results in altered expression of hepatic genes involved in the regulation of lipid and glucose metabolism [2, 17, 66]. In response to these multiple genomic and non-genomic bile acid-activated signaling pathways, FXR differentially interacts with its transcriptional cofactors, resulting in the recruitment of distinct combinations of transcriptional cofactor complexes to its target genes. FXR cofactors catalyze PTMs of histones and FXR itself, to effectively modulate transcription of target genes (Fig. 1).
Fig. 1. FXR transcriptional responses by PTMs and cofactor interaction in response to bile acid signaling.
Bile acids activate cellular kinases (JNK, ERK, PKB, PKC), nuclear receptors, primarily FXR, and a membrane GPCR TGR5. Intestinal FGF-15 (FGF-15 is the mouse homologue of human FGF-19), upon induction by bile acid-activated FXR, binds to a liver membrane receptor tyrosine kinase, FGFR4, and triggers cellular kinase signaling cascades in hepatocytes. In response to these multiple bile acid-activated cellular signaling pathways, interaction of FXR with transcriptional cofactors is changed, which results in altered PTMs of FXR as well as histones to effectively modulate expression at target genes.
Potential FXR cofactors
About a dozen transcriptional cofactors that can potentially modulate FXR transactivation ability have been reported (Table 1). In general, interactions of FXR with coactivators, including Brg-1, p300, CARM1, PRMT1, and ASCOM, are increased after treatment with FXR agonists, and the coactivators are recruited to FXR target genes, which results in histone modification or chromatin remodeling and increased gene transcription. Interestingly, some of these cofactors, including p300 and SIRT1, acetylate and deacetylate not only histones at FXR target genes but also FXR itself [58, 69]. Although not much studies on corepressors for FXR were done, based on other NRs such as PPAR, LXR and TR [42], this is a likely possibility.
Table 1.
Reported Transcriptional cofactors of FXR
| Cofactors | Cellular function | Regulation of FXR signaling | References |
|---|---|---|---|
| PGC-1α | a key metabolic regulator mitochondria function oxidative phosphorylation | a coactivator of FXR during fasting increases FXR transactivation increases expression of the FXR gene | [68] |
| p300 | Transcriptional integrator Lys acetylase | acetylates H3K9/14 at the FXR targets acetylates FXR at K217 dynamic transcription of FXR targets | [58, 69] |
| SRC-1 | general NR AF2 coactivator Lys acetylase | FXR agonists increases SRC-1 interaction w/ FXR LBD, increases FXR transactivation | [70, 71] |
| SIRT1 | NAD+-dependent deacetylase A life-longevity protein | deacetylates H3K9/14 at the FXR targets deacetylates FXR dynamic transcription of FXR targets | [58, 69] |
| Brg-1 | an ATPase of Swi/Snf complexes | FXR agonist increases interaction w/FXR increases FXR transactivation mediates chromatin remodeling at FXR targets | [72] |
| CARM1 | Arg methyltransferase | FXR agonist increases interaction w/FXR increases FXR transactivation methylates H3R17 at the BSEP promoter | [48] |
| PRMT1 | Arg methyltransferase | FXR agonist increases interaction w/FXR increases FXR transactivation methylates H4R3 the BSEP and SHP promoters | [49] |
| ASCOM | a coactivator complex containing ASC-2 and H3K4 Lys methyltransferases (MLL3/4) | FXR agonist increases ASCOM recruitment to targets tri-methylates H3K4 and gene activation impaired FXR signaling and disrupted BA levels by mice expressing inactive MLL3 mutant | [51] |
| GPS2 | a subunit of the N-coR corepressor complex | interacts with FXR increases the CYP8B1 expression by activated FXR by promoting the enhancer and promoter interaction | [73] |
| Ku proteins | DNA-dependent kinase catalytic subunits | FXR corepressor interacts with the hinge region of FXR decreases FXR activity of the BSEP gene | [74] |
| DRIP205 | transcription mediator | FXR agonist increases interaction w/FXR increases FXR transactivation binding w/ RXR is important for DRP205 action | [75] |
| TRRAP | transcription mediator | FXR agonist increases interaction w/FXR increases FXR transactivation | [76] |
a. PGC-1 coactivator
Edwards and colleagues reported that PGC-1α , a critical metabolic regulator in mitochondrial biogenesis and function, increases both FXR levels and its transactivation activity [68]. PGC-1α functions as a transcriptional coactivator of FXR that directly interacts with FXR and enhance its transactivation activity. PGC-1α also increases transcription of the FXR gene by coactivation of PPARγ and HNF-4 resulting in increased FXR mRNA levels. Hepatic expression of PGC-1α and FXR was increased during fasting, which 14 results in decreased plasma TG levels and increased fatty acid β-oxidation needed to supply energy demands during fasting [68].
b. P300 acetylase and SIRT1 deacetylase
P300 catalyzes covalent modification of acetylation at lysine residues in histones and transcriptional factors/cofactors. Recently, Fang et al., showed that p300 acts as a coactivator for FXR in SHP gene induction by catalyzing acetylation of K9/K14 of histone H3 which are activating histone modifications [69]. P300 was also shown to acetylate FXR. However, unexpectedly, acetylation of FXR by p300 decreased the transactivation potential of FXR by inhibiting DNA binding of the FXR/RXRα heterodimer [58] (see next section). Interestingly, down-regulation of p300 altered expression of SHP and other metabolic FXR target genes involved in lipoprotein and glucose metabolism, such that beneficial lipid and glucose profiles would be expected [69].
SIRT1 is a NAD+-dependent deacetylase that removes acetyl groups from modified lysines in both histones and non-histone regulatory proteins [77, 78]. SIRT1 senses cellular nutrient levels and functions as a key metabolic regulator by modulating activity of metabolic regulators via protein deacetylation. Our group recently reported SIRT1 profoundly modulates FXR transcriptional signaling by deacetylation of both histones and FXR [58]. On one hand, SIRT1 increases the transactivation potential of FXR by deacetylating FXR because heterodimerization of FXR with RXRα and binding to the DNA is impaired by acetylation of FXR. On the other hand, SIRT1 deacetylates histones at the SHP promoter, which is expected to maintain transcriptional silencing. These paradoxical effects of SIRT1 and p300 on dynamic FXR transcriptional signaling will be discussed in detail in following sections.
c. Brg-1, an ATPase of Swi/Snf complexes
A recent study using in vivo chromatin remodeling assays and functional studies along with siRNA approaches has identified Brg-1, an ATPase in Swi/Snf complexes, as a FXR coactivator [72]. Interestingly, Miao et al. [72] recently showed that Brm and Brg-1, two ATPases of Swi/Snf complexes, have distinct roles in FXR/SHP-mediated feedback regulation of hepatic bile acid biosynthesis. While Brg-1 is a FXR coactivator for SHP gene induction, Brm is a critical component of the SHP complexes, which inhibit expression of both the CYP7A1 gene and the SHP gene. Activation of FXR by agonists in HepG2 cells or mice in vivo increased the interaction of FXR with Brg-1 and increased occupancy of FXR and Brg-1 at the SHP promoter.
d. Histone methyltransferases, CARM1, PRMT1, and ASCOM
Ananthanarayanan and colleagues originally reported that CARM1 interacts with FXR and enhances the transactivation ability of FXR [48]. In response to treatment with a FXR agonist, occupancy of CARM1 and FXR was increased at the promoter of BSEP, a known FXR target gene that functions as an ATP-dependent canalicular bile acid transporter. Increased occupancy of CARM1 at the BSEP promoter resulted in increased methylation at Arg-17 of histone H3 and gene activation. Fiorucci and colleagues have also identified PRMT1 as a FXR coactivator [49]. Treatment with a semi-synthetic FXR agonist, 6E-CDCA, resulted in increased interaction of FXR with PRMT1 and increased mRNA levels of the FXR target genes, BSEP and SHP. After treatment with 6E-CDCA, both the occupancy of PRMT1 and histone H4 methylation at the promoters of the BSEP and SHP genes were increased. Lee and colleagues recently demonstrated that a coactivator complex ASCOM that contains activating signal cointegrator-2 (ASC-2), the histone H3 K4 methyltransferases, MLL3 and MLL4, is a coactivator of FXR [51]. ASC-2, MLL3, and MLL4 were recruited to FXR target genes in a ligand-dependent manner, resulting in H3K4 trimethylation and gene activation. Importantly, expression of FXR target genes was partially impaired in mice expressing a catalytically inactive MLL3 mutant and these mice exhibited disrupted bile acid homeostasis.
e. GPS2, DRIP205, TRRAP, and Ku proteins
Treuter and colleagues recently identified GPS2, a stoichiometric subunit of the N-CoR corepressor complex, as a novel FXR interacting protein in the regulation of a major bile acid biosynthetic gene, CYP8B1 [73]. From chromatin immunoprecipitation and functional studies using siRNA, they found that the CYP8B1 gene was a direct FXR target gene and that GPS2 augments expression of the CYP8B1 gene by the activated FXR via facilitating physical interaction between the CYP8B1 enhancer and promoter regions [73]. DRIP205 and TRRAP were also identified as transcriptional coactivators of FXR from in vitro interaction studies and cell-based functional reporter studies [75, 76]. Treatment with a FXR agonist including bile acids increased the interaction of FXR with these cofactors. Gain or loss of function studies showed that these cofactors augment the transactivation ability of FXR. Cell-based reporter assays using a RXRα heterodimerization-deficient FXR mutant (L433R) revealed that FXR heterodimerization with RXR is important for coactivation of FXR activity by DRIP205 [75]. DNA-dependent protein kinase catalytic subunits, Ku80, and Ku70, were also identified as FXR-interacting proteins that function as corepressors [74]. The Ku proteins interacted with the hinge region of FXR and expression of these proteins decreased the promoter activity of the BSEP gene. These interactios suggest that phosphorylation of FXR may also be an important regulatory PTM. In this regard, it will be interesting to determine if kinase signaling activated by bile acids or FGF-15 targets FXR. Most of these cofactors have been analyzed in vitro or in cultured cells, so it will be important to examine whether they modulate FXR transcriptional activity in vivo.
6. Acetylation of FXR in Health and Disease
Recent studies have shown that PTMs of FXR profoundly modulate its transactivation ability in response to cellular signals (Fig. 2). Fang et al., recently reported that p300 acetylates FXR in vitro and in cells [69]. By examination of the acetylation of FXR in vivo in normal and disease model mice, our group further obtained evidence for biological functions of acetylation of FXR in normal and disease conditions [58]. FXR acetylation was normally dynamically regulated by p300 acetylase and SIRT1 deacetylase in response to bile acid signaling or fasting/feeding cycles in normal mice [58]. In contrast, FXR acetylation was constitutively highly elevated in livers of diet-induced obese mice and ob/ob mice, two mouse models of the metabolic syndrome [58]. Most previous gene regulation studies have been focused on either PTMs of histones or PTMs of non-histone regulatory proteins, but these studies showed that FXR transcriptional signaling was modulated both by acetylation of FXR and acetylation of histones at the target gene.
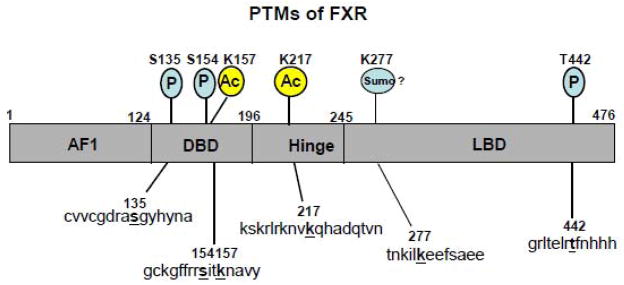
Fig. 2. Reported PTMs of FXR.
FXR is a target of PTMs, including acetylation (Ac), phosphorylation (P), and potentially sumoylation (Sumo), in response to diverse cellular signals. 29 FXR was also shown to be ubiquitinated but target sites are not known. Amino acid targets of PTMs are indicated above the schematic diagram of human FXR and the sequence motifs are shown below. The indicated sequence motifs are conserved in mouse FXR sequences.
Dynamic FXR acetylation in normal animals
Lys-217 in the hinge region of FXR was identified by tandem mass spectrometry analysis as a major acetylation target site for p300. Acetylation at Lys-157 in the DNA binding domain was also detected. Consistent with roles of the NR hinge region in protein flexibility to allow for simultaneous receptor dimerization and DNA binding [7, 43], acetylation of FXR at Lys-217 inhibited heterodimerization with RXRα and binding of FXR/RXR heterodimers to DNA/chromatin [58]. Consistent with these observations, functional studies showed that FXR acetylation, especially at Lys-217, leads to decreased FXR transactivation ability. As summarized in Fig. 3, in response to FXR activating signals, including bile acids, feeding, or synthetic agonists, interaction of FXR with p300 is increased, whereas that of SIRT1 is decreased. Subsequently, occupancy of FXR and p300 is increased at the SHP promoter, whereas that of SIRT1 is decreased, resulting in acetylation of histones. Acetylation of histones H3 at K9/K14 by p300 is associated with gene activation and is probably the major factor in the transcriptional activation of the SHP gene. However, p300 also catalyzes acetylation of FXR and inhibits heterodimerization with RXRα and DNA binding. The fate of FXR, presumably dissociation from the promoter by this mechanism, is not clear. This apparently paradoxical effect may be an important mechanism to terminate transcriptional responses of FXR to a stimulus, such as feeding or bile acid signaling, which is essential in a dynamically regulated system to maintain homeostasis.
Fig. 3. Dynamic acetylation/deacetylation of FXR in normal physiology.
In response to low bile acid levels during fasting, occupancy of SIRT1 at the SHP promoter is increased and histones are deacetylated, so that gene expression remains at a low basal level. In response to FXR activation signals such as bile acids after feeding, interaction of FXR with p300 and SIRT1 is altered and subsequently, occupancy of FXR and p300 is increased at the SHP promoter, resulting in acetylation of histones H3 at K9/K14 and gene activation. P300 also catalyzes acetylation of FXR and inhibits heterodimerization with RXR which then results in dissociation from the promoter and transcriptional termination. The fate of the dissociated FXR has not been clearly determined. FXR may be deacetylated by SIRT1 and undergo ubiquitination-proteasomal degradation or rebind to the SHP promoter. Acetylation and deacetylation of FXR, therefore, appear to be a dynamic process in response to fluctuating bile acid levels during fasting and feeding cycles.
During the fasting state which results in increased SIRT1 levels and activity, FXR may be deacetylated by SIRT1 and undergo ubiquitination-proteasomal degradation or rebind to the SHP promoter (Fig. 3). In the fasting state, expression of FXR may be increased by PGC-1α [68] but occupancy of SIRT1 at the SHP promoter is increased and histones are deacetylated, so that gene expression remains at a low basal level. Therefore, acetylation and deacetylation of FXR appear to be a dynamic process in response to fluctuating bile acid levels during fasting and feeding cycles and are tightly balanced by the opposing actions of p300 and SIRT1. This scenario is analogous to the modulation of PGC-1α activity by opposing actions of GCN5 acetylase and SIRT1 deacetylase [60, 61]. Although GCN5 potentially increases transcription of PGC-1α target genes by acetylating histone H3, GCN5 also directly acetylates PGC-1α at multiple lysine residues and negatively regulates its coactivator potential. Conversely, SIRT1 deacetylates and increases the coactivator potential of PGC-1α.
Elevated FXR acetylation in disease
In high fat diet-induced obese mice, ob/ob mice, or mice depleted of hepatic SIRT1 by adenoviral expressed siRNA, FXR acetylation levels were constitutively and highly elevated with deleterious gene expression patterns and metabolic outcomes [58]. Interaction of FXR with p300 was constitutively highly elevated, whereas FXR interaction with SIRT1 and its heterodimer partner, RXRα, was dramatically decreased [58]. Heterodimerization with RXRα and subsequently binding of the heterodimer to the DNA were impaired, which may result in aberrant FXR transcriptional pathways in these metabolic disease conditions (Fig. 4). Whether elevated FXR acetylation is a cause or consequence of deleterious metabolic effects remains unclear. Low activity and/or levels of SIRT1 in these metabolic disease model mice may contribute to elevated FXR acetylation so that FXR acetylation is a consequence of the metabolic abnormalities. Alternatively, aberrant expression of metabolic target genes due to dysregulated FXR transcriptional pathways may underlie metabolic abnormalities, which is consistent with elevated FXR acetylation levels in both diet-induced obese and ob/ob mice. However, a primary role for highly elevated FXR acetylation does not rule out a vicious positive feedback loop in which detrimental metabolic outcomes result in enhancement of the elevated FXR acetylation. These findings provide an intriguing correlation between elevated FXR acetylation, decreased FXR transactivation ability, and deleterious effects in metabolic disease states. Dynamic acetylation and deacetylation of FXR in normal mice may be required for activation of FXR target genes, while continuously elevated FXR acetylation in the diseased states blocks activation. An interesting question is whether acetylation affects general FXR transcriptional activity and pathways on all its target genes or whether acetylation has gene-specific effects.
Fig. 4. Constitutively highly elevated FXR acetylation in pathophysiology.

In diet-induced obese (DIO) mice and leptin-deficient ob/ob mice, FXR acetylation levels are constitutively and highly elevated with deleterious gene expression patterns and metabolic outcomes. Interaction of FXR with p300 is highly elevated, whereas FXR interaction with SIRT1 and its heterodimer partner, RXRα, is dramatically decreased. Aberrant PTMs and inappropriate FXR/cofactor complexes contribute to dysregulation of FXR transcriptional activity and pathways in metabolic disease states.
Remaining questions
There are important remaining questions about the molecular regulation of FXR activity by PTMs. Does FXR acetylation at Lys-157 or Lys-217 leads to gene-specific changes in FXR transcriptional responses by forming complexes with different combinations of transcriptional cofactors, resulting in different functional consequences? Also, an interesting still unresolved question is whether elevated FXR acetylation at K217 is a cause or consequence of metabolic diseases. Transgenic knock-in mice expressing acetylation mimic or defective mutants of Lys-217 or transient expression of these mutants using adenoviral delivery would help address these questions. It will be important to determine whether kinases downstream of the bile acid signaling pathways influence FXR acetylation and identify which abnormal cellular kinase signaling pathways underlie the elevated acetylation of FXR in disease conditions. While most studies of FXR have focused on the liver, it will be also interesting to examine whether elevated FXR acetylation is detected in other FXR-related diseases such as cancer and aging, and in other tissues.
7. FXR and Other Post-Translational Modifications
FXR Phosphorylation
Staels and colleagues recently reported that calcium-dependent PKC kinase phosphorylates FXR at Ser-135 and Ser-154, in the DBD (Fig. 2) [79]. Using pharmacological activators or inhibitors of PKC, they showed that phosphorylation of FXR by PKC promoted FXR interaction with PGC-1α and increased its transactivation activity. Consistent with these results, a PKC phosphorylation-defective FXR mutant exhibited decreased ligand-dependent FXR transactivation [79]. Since bile acids activate PKC, it will be interesting to examine whether bile acids increase PKC-mediated phosphorylation of FXR. Enhanced activity of FXR as a result of post-translational phosphorylation was also demonstrated by Schneider and colleagues who showed that the membrane protein ATPase class 1 type 8B member (called familial intrahepatic cholestasis 1 (FIC1) protein) activates FXR via protein phosphorylation [80]. FIC1 is critical for normal bile acid transport, and mutation of this gene leads to a spectrum of liver diseases including intrahepatic cholestasis [33]. Using phosphorylation-defective or mimic FXR mutants, along with pharmacological inhibitors and siRNA approaches, they showed that PKC zeta phosphorylates FXR at Thr-442, which is important for the FIC1 effect (Fig. 2) and that phosphorylation of FXR at Thr-442 augments transactivation ability of FXR and increases its nuclear localization. These findings show that the membrane bound FIC1 signals to FXR via PKC zeta and that FIC1-related liver disease is likely associated with downstream effects of decreased FXR activity in the regulation of bile acid homeostasis.
FXR ubiquitination and sumoylation
Compared to FXR acetylation and phosphorylation studies, post-translational ubiquitination and sumoylation of FXR have not been thoroughly examined. Our group recently showed that treatment with MG132, a proteasome inhibitor, markedly increased FXR levels, suggesting that FXR may undergoes proteosomal degradation [58]. Furthermore, FXR was robustly ubiquitinated in vitro and in cells. However, the residue(s) in FXR that are ubiquitinated is not known. The half-lives of FXR were determined to be 5–6 hr for ligand-activated FXR wild type and about 2 hr for acetylation defective mutants [58], indicating that FXR acetylation increases its stability. Phosphorylation of FXR in response to bile acid signaling may modulate ubiquitination and proteasomal degradation of FXR. Supporting such a possibility, we recently reported that in response to treatment with bile acid or FGF-19, activated ERK kinase phosphorylates SHP at Ser-26, which increases stability of SHP by inhibiting ubiquitination at Lys-122 and Lys-123 and proteasomal degradation [56].
A recent study reported that Lys-279 in the LBD of FXR is a potential sumoylation site in the regulation of intestinal innate immunity (Fig. 2) [81]. FXR contains at least two potential sumoylation consensus sites (WKXE/D), Lys-122 or Lys-277, where W indicates any hydrophobic amino acid and X indicates any amino acid. Overexpression of the K277R mutant impaired inhibition of the expression of the TNFα gene caused by synthetic FXR agonist 6E-CDCA (also called INT-747), whereas FXR wild type showed full trans-repression activity [81]. Additional studies will be required to unequivocally demonstrate that Lys-277 is the major sumoylation site in FXR and to directly demonstrate the functional roles of sumoylation in FXR transcriptional signaling pathways.
8. Concluding Remarks
In response to constant nutrient and hormonal fluctuations, the body must maintain metabolic homeostasis by altering expression of genes in metabolic tissues. The inability to sense and adapt to these fluctuations which leads to over accumulation of metabolites, plays a crucial causative role in the development of metabolic disorders. The nuclear bile acid receptor FXR senses elevated bile acid levels and connects bile acid signaling with altered gene activity to maintain lipid and glucose homeostasis. Dysregulation of FXR activity and transcriptional pathways due to aberrant PTMs and inappropriate FXR/cofactor complexes could be a key determinant in the development of metabolic disease. Therefore, targeting these abnormal PTMs and transcriptional cofactors of FXR in disease states to correct altered transcriptional signaling may represent new molecular strategies for development of pharmacological agents to treat the metabolic syndrome. For example, FXR synthetic ligands that cause differential interaction with specific groups of transcriptional cofactors and alter FXR PTMs might be developed into therapeutic agents. Furthermore, inhibitors of histone modifying enzymes such as histone deacetylases and methylases have emerged as effective therapeutic options [82–84]. Therefore, combinatorial use of FXR ligands together with these epigenomic drugs that specifically target histone modifying enzymes may provide new strategies for developing next generation drugs for the treatment of FXR-related diseases such as metabolic syndrome and cancer.
Research Highlights.
FXR has important roles in controlling lipid and glucose levels.
FXR activity is regulated by transcriptional cofactors which catalyze PTMs of FXR as well as histones.
Acetylation of FXR is normally dynamically regulated, but is dysregulated in metabolic disease states.
Aberrant FXR cofactors and PTMs may be targets for development of new therapeutic agents to treat metabolic disease.
Acknowledgments
Because of space constraints, some of primary references could not be cited directly, instead, other review articles were included in the references. I thank Byron Kemper for critical reading 23 of the manuscript. This work was supported by NIH DK062777, NIH DK80032, and ADA basic research award to J.K.K.
Abbreviations
- FXR
Farnesoid X Receptor
- PTM
post-translational modification
- NR
nuclear receptors
- AF-1
activation function 1
- AF-2
activation function 2
- DBD
DNA binding domain
- LBD
ligand binding domain
- CDCA
chenodeoxycholic acid
- FXRE
FXR response element
- IR1
inverted repeat 1
- ChIP
chromatin immunoprecipitation
- CYP7A1
cholesterol 7 hydroxylase
- FGF- 15/19
fibroblast growth factor 15/19
- TG
triglyceride
- SHP
Small Heterodimer Partner
- SNP
single nucleotide polymorphism
- ICP
intrahepatic cholestasis of pregnancy
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- NAD
nicotine amide dinucleotide
- PKC
protein kinase C
- FIC1
familial intrahepatic cholestasis 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grundy SM, Brewer HB, Jr, Cleeman JI, Smith SC, Jr, Lenfant C. Definition of metabolic syndrome: Report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation. 2004;109:433–8. doi: 10.1161/01.CIR.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 2.Lee FY, Lee H, Hubbert ML, Edwards PA, Zhang Y. FXR, a multipurpose nuclear receptor. Trends Biochem Sci. 2006;31:572–80. doi: 10.1016/j.tibs.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Cariou B, Staels B. FXR: a promising target for the metabolic syndrome? Trends Pharmacol Sci. 2007;28:236–43. doi: 10.1016/j.tips.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Edwards PA. FXR signaling in metabolic disease. FEBS Lett. 2008;582:10–8. doi: 10.1016/j.febslet.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Staels B, Kuipers F. Bile acid sequestrants and the treatment of type 2 diabetes mellitus. Drugs. 2007;67:1383–92. doi: 10.2165/00003495-200767100-00001. [DOI] [PubMed] [Google Scholar]
- 6.Fiorucci S, Mencarelli A, Palladino G, Cipriani S. Bile-acid-activated receptors: targeting TGR5 and farnesoid-X-receptor in lipid and glucose disorders. Trends Pharmacol Sci. 2009;30:570–80. doi: 10.1016/j.tips.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 8.Lonard DM, O'Malley WB. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–28. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 10.Fiorucci S, Rizzo G, Donini A, Distrutti E, Santucci L. Targeting farnesoid X receptor for liver and metabolic disorders. Trends Mol Med. 2007;13:298–309. doi: 10.1016/j.molmed.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Modica S, Moschetta A. Nuclear bile acid receptor FXR as pharmacological target: are we there yet? FEBS Lett. 2006;580:5492–9. doi: 10.1016/j.febslet.2006.07.082. [DOI] [PubMed] [Google Scholar]
- 12.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–91. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 13.Seol W, Choi HS, Moore DD. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol Endocrinol. 1995;9:72–85. doi: 10.1210/mend.9.1.7760852. [DOI] [PubMed] [Google Scholar]
- 14.Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–93. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 15.Urizar NL, Liverman AB, Dodds DT, Silva FV, Ordentlich P, Yan Y, Gonzalez FJ, Heyman RA, Mangelsdorf DJ, Moore DD. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science. 2002;296:1703–6. doi: 10.1126/science.1072891. [DOI] [PubMed] [Google Scholar]
- 16.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–77. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–93. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 18.Willson TM, Jones SA, Moore JT, Kliewer SA. Chemical genomics: functional analysis of orphan nuclear receptors in the regulation of bile acid metabolism. Med Res Rev. 2001;21:513–22. doi: 10.1002/med.1023. [DOI] [PubMed] [Google Scholar]
- 19.Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, Rosenfeld JM, Alvarez JG, Noel JP, Nicolaou KC, Evans RM. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell. 2003;11:1079–92. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mi LZ, Devarakonda S, Harp JM, Han Q, Pellicciari R, Willson TM, Khorasanizadeh S, Rastinejad F. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol Cell. 2003;11:1093–100. doi: 10.1016/s1097-2765(03)00112-6. [DOI] [PubMed] [Google Scholar]
- 21.Barbier O, Torra IP, Sirvent A, Claudel T, Blanquart C, Duran-Sandoval D, Kuipers F, Kosykh V, Fruchart JC, Staels B. FXR induces the UGT2B4 enzyme in hepatocytes: a potential mechanism of negative feedback control of FXR activity. Gastroenterology. 2003;124:1926–40. doi: 10.1016/s0016-5085(03)00388-3. [DOI] [PubMed] [Google Scholar]
- 22.Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, Fruchart JC, Dallongeville J, Hum DW, Kuipers F, Staels B. Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J Clin Invest. 2002;109:961–71. doi: 10.1172/JCI14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas AM, Hart SN, Kong B, Fang J, Zhong XB, Guo GL. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 51:1410–9. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong HK, Infante AM, Seo YK, Jeon TI, Zhang Y, Edwards PA, Xie X, Osborne TF. Genome-wide interrogation of hepatic FXR reveals an asymmetric IR-1 motif and synergy with LRH-1. Nucleic Acids Res. doi: 10.1093/nar/gkq397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinal C, Tohkin M, Miyata M, Ward J, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 26.Choi M, Moschetta A, Bookout AL, Peng L, Umetani M, Holmstrom SR, Suino-Powell K, Xu HE, Richardson JA, Gerard RD, Mangelsdorf DJ, Kliewer SA. Identification of a hormonal basis for gallbladder filling. Nat Med. 2006;12:1253–5. doi: 10.1038/nm1501. [DOI] [PubMed] [Google Scholar]
- 27.Seol W, Choi H, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 28.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–15. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 29.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Wilson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Molecular Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 30.Bavner A, Sanyal S, Gustafsson JA, Treuter E. Transcriptional corepression by SHP: molecular mechanisms and physiological consequences. Trends Endocrinol Metab. 2005;16:478–88. doi: 10.1016/j.tem.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Chanda D, Park JH, Choi HS. Molecular basis of endocrine regulation by orphan nuclear receptor Small Heterodimer Partner. Endocr J. 2008;55:253–68. doi: 10.1507/endocrj.k07e-103. [DOI] [PubMed] [Google Scholar]
- 32.Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev. 2003;83:633–71. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- 33.Suchy FJ, Ananthanarayanan M. Bile salt excretory pump: biology and pathobiology. J Pediatr Gastroenterol Nutr. 2006;43(Suppl 1):S10–6. doi: 10.1097/01.mpg.0000226385.71859.5f. [DOI] [PubMed] [Google Scholar]
- 34.Moschetta A, Bookout AL, Mangelsdorf DJ. Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat Med. 2004;10:1352–8. doi: 10.1038/nm1138. [DOI] [PubMed] [Google Scholar]
- 35.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116:1102–9. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103:1006–11. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–6. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 38.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–5. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang T, Wang XX, Scherzer P, Wilson P, Tallman J, Takahashi H, Li J, Iwahashi M, Sutherland E, Arend L, Levi M. Farnesoid X receptor modulates renal lipid metabolism, fibrosis, and diabetic nephropathy. Diabetes. 2007;56:2485–93. doi: 10.2337/db06-1642. [DOI] [PubMed] [Google Scholar]
- 40.Marzolini C, Tirona RG, Gervasini G, Poonkuzhali B, Assem M, Lee W, Leake BF, Schuetz JD, Schuetz EG, Kim RB. A common polymorphism in the bile acid receptor farnesoid X receptor is associated with decreased hepatic target gene expression. Mol Endocrinol. 2007;21:1769–80. doi: 10.1210/me.2007-0025. [DOI] [PubMed] [Google Scholar]
- 41.Van Mil SW, Milona A, Dixon PH, Mullenbach R, Geenes VL, Chambers J, Shevchuk V, Moore GE, Lammert F, Glantz AG, Mattsson LA, Whittaker J, Parker MG, White R, Williamson C. Functional variants of the central bile acid sensor FXR identified in intrahepatic cholestasis of pregnancy. Gastroenterology. 2007;133:507–16. doi: 10.1053/j.gastro.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Lazar MA. Nuclear receptor corepressors. Nucl Recept Signal. 2003;1:e001. doi: 10.1621/nrs.01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci. 2004;29:317–24. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 44.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 46.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–59. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–60. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 48.Ananthanarayanan M, Li S, Balasubramaniyan N, Suchy FJ, Walsh MJ. Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J Biol Chem. 2004;279:54348–57. doi: 10.1074/jbc.M410021200. [DOI] [PubMed] [Google Scholar]
- 49.Rizzo G, Renga B, Antonelli E, Passeri D, Pellicciari R, Fiorucci S. The methyl transferase PRMT1 functions as co-activator of farnesoid X receptor (FXR)/9-cis retinoid X receptor and regulates transcription of FXR responsive genes. Mol Pharmacol. 2005;68:551–8. doi: 10.1124/mol.105.012104. [DOI] [PubMed] [Google Scholar]
- 50.Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol. 2007;27:1407–24. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim DH, Lee J, Lee B, Lee JW. ASCOM controls farnesoid X receptor transactivation through its associated histone H3 lysine 4 methyltransferase activity. Mol Endocrinol. 2009;23:1556–62. doi: 10.1210/me.2009-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kemper J, Kim H, Miao J, Bhalla S, Bae Y. Role of a mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol Cell Biol. 2004;24:7707–7719. doi: 10.1128/MCB.24.17.7707-7719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boulias K, Talianidis I. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 2004;32:6096–103. doi: 10.1093/nar/gkh947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–79. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 56.Miao J, Xiao Z, Kanamaluru D, Min G, Yau PM, Veenstra TD, Ellis E, Strom S, Suino-Powell K, Xu HE, Kemper JK. Bile acid signaling pathways increase stability of Small Heterodimer Partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev. 2009;23:986–96. doi: 10.1101/gad.1773909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23:425–33. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu S, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metabolism. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 60.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–38. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 61.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 62.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–51. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 63.Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102:11278–83. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, Wu SY, Chiang CM, Veenstra TD, Kemper JK. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 285:33959–70. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–39. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 66.Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–20. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–25. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18:157–69. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fang S, Tsang S, Jones R, Ponugoti B, Yoon H, Wu SY, Chiang CM, Willson TM, Kemper JK. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem. 2008;283:35086–95. doi: 10.1074/jbc.M803531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lew JL, Zhao A, Yu J, Huang L, De Pedro N, Pelaez F, Wright SD, Cui J. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J Biol Chem. 2004;279:8856–61. doi: 10.1074/jbc.M306422200. [DOI] [PubMed] [Google Scholar]
- 71.Wang S, Lai K, Moy FJ, Bhat A, Hartman HB, Evans MJ. The nuclear hormone receptor farnesoid X receptor (FXR) is activated by androsterone. Endocrinology. 2006;147:4025–33. doi: 10.1210/en.2005-1485. [DOI] [PubMed] [Google Scholar]
- 72.Miao J, Fang S, Lee J, Comstock C, Knudsen KE, Kemper JK. Functional specificities of Brm and Brg-1 Swi/Snf ATPases in the feedback regulation of hepatic bile acid biosynthesis. Mol Cell Biol. 2009;29:6170–81. doi: 10.1128/MCB.00825-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanyal S, Bavner A, Haroniti A, Nilsson LM, Lundasen T, Rehnmark S, Witt MR, Einarsson C, Talianidis I, Gustafsson JA, Treuter E. Involvement of corepressor complex subunit GPS2 in transcriptional pathways governing human bile acid biosynthesis. Proc Natl Acad Sci U S A. 2007;104:15665–70. doi: 10.1073/pnas.0706736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohno M, Kunimoto M, Nishizuka M, Osada S, Imagawa M. Ku proteins function as corepressors to regulate farnesoid X receptor-mediated gene expression. Biochem Biophys Res Commun. 2009;390:738–42. doi: 10.1016/j.bbrc.2009.10.040. [DOI] [PubMed] [Google Scholar]
- 75.Pineda Torra I, Freedman LP, Garabedian MJ. Identification of DRIP205 as a coactivator for the Farnesoid X receptor. J Biol Chem. 2004;279:36184–91. doi: 10.1074/jbc.M405126200. [DOI] [PubMed] [Google Scholar]
- 76.Unno A, Takada I, Takezawa S, Oishi H, Baba A, Shimizu T, Tokita A, Yanagisawa J, Kato S. TRRAP as a hepatic coactivator of LXR and FXR function. Biochem Biophys Res Commun. 2005;327:933–8. doi: 10.1016/j.bbrc.2004.12.095. [DOI] [PubMed] [Google Scholar]
- 77.Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol Endocrinol. 2007;21:1745–55. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- 78.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gineste R, Sirvent A, Paumelle R, Helleboid S, Aquilina A, Darteil R, Hum DW, Fruchart JC, Staels B. Phosphorylation of farnesoid X receptor by protein kinase C promotes its transcriptional activity. Mol Endocrinol. 2008;22:2433–47. doi: 10.1210/me.2008-0092. [DOI] [PubMed] [Google Scholar]
- 80.Frankenberg T, Miloh T, Chen FY, Ananthanarayanan M, Sun AQ, Balasubramaniyan N, Arias I, Setchell KD, Suchy FJ, Shneider BL. The membrane protein ATPase class I type 8B member 1 signals through protein kinase C zeta to activate the farnesoid X receptor. Hepatology. 2008;48:1896–905. doi: 10.1002/hep.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 2009;183:6251–61. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 82.Kramer OH, Gottlicher M, Heinzel T. Histone deacetylase as a therapeutic target. Trends Endocrinol Metab. 2001;12:294–300. doi: 10.1016/s1043-2760(01)00438-6. [DOI] [PubMed] [Google Scholar]
- 83.Bruneau BG. Epigenetic regulation of the cardiovascular system: introduction to a review series. Circ Res. 107:324–6. doi: 10.1161/RES.0b013e3181f17dfe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–63. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]