Abstract
The latent TGF-β binding proteins (LTBP-1 -3, and -4) assist in the secretion and localization of latent TGF-β molecules. Ltbp3−/− and Ltbp4S−/− mice have distinct phenotypes and only in the lungs does deficiency of either Ltbp-3 or Ltbp-4 cause developmental abnormalities. To determine if these two LTBPs have additional common functions, we generated mice deficient for both Ltbp-3 and Ltbp-4S. The only novel defect in Ltbp3−/−; Ltbp4S−/− mice was an early lethality compared to mice with single mutations. In addition lung abnormalities were exacerbated and the terminal air sac septation defect was more severe in Ltbp3−/−; Ltbp4S−/− mice than in Ltbp4S−/− mice. Decreased cellularity of Ltbp3−/−; Ltbp4S−/− lungs was correlated with higher rate of apoptosis in newborn lungs of Ltbp3−/−; Ltbp4S−/− animals compared to WT, Ltbp3−/−, and Ltbp4S−/− mice. No differences in the maturation of the major lung cell types were discerned between the single and double mutant mice. However, the distribution of Type 2 cells and myofibroblasts was abnormal, and myofibroblast segregation in some areas might be an indication of early fibrosis. We also observed differences in ECM composition between Ltbp3−/−; Ltbp4S−/− and Ltbp4S−/− lungs after birth, reflected in decreased incorporation of fibrillin-1 and -2 in Ltbp3−/−; Ltbp4S−/− matrix. The function of the lungs of Ltbp3−/−; Ltbp4S−/− mice after the first week of life was potentially further compromised by macrophage infiltration, as proteases secreted from macrophages might exacerbate developmental emphysema. Together these data indicate that LTBP-3 and -4 perform partially overlapping functions only in the lungs.
Keywords: TGF-β, LTBP, lung development, myofibroblasts, alveolarization
Introduction
The transforming growth factor-βs (TGF-β) are potent cytokines that have profound effects on development, cell growth, and immune cell differentiation (Massague et al., 2000). TGF-β production is ubiquitous and almost all cells express the TGF-β high affinity serine-threonine kinase receptors. Cells release TGF-β as part of a latent complex that may be directed to the extracellular matrix (ECM) and stored until mobilized. In order to signal, the TGF-β must be released from this complex. Release of TGF-β is known as latent TGF-β activation and is distinguished from the cleavage or processing of mature TGF-β from its propeptide, a process that normally occurs intracellularly. Thus the activity of this powerful growth factor is regulated not only at the levels of transcription and translation, but also at the level of its extracellular availability.
The latent TGF-β complex consists of the TGF-β dimer bound by non-covalent interactions to its cleaved propeptide. In this form, TGF-β is masked and cannot interact with its cell surface receptor (Annes et al., 2003). Accordingly, the TGF-β propeptide dimer is named the latency-associated peptide (LAP), and the complex of TGF-β and LAP is named the small latent complex (SLC). The LAP molecule in the SLC is often disulfide bound to a latent TGF-β binding protein (LTBP) (Annes et al., 2003). This tripartite complex of TGF-β, LAP, and LTBP is called the large latent complex (LLC). Within the LLC, TGF-β is the signaling molecule, LAP confers latency to TGF-β, and LTBP controls the localization of the latent TGF-β into the ECM.
The LTBPs comprise a group of four proteins structurally similar to the fibrillins (-1, -2 and -3), the major constituents of microfibrils (Ramirez and Dietz, 2009). The LTBPs are multi-domain proteins with molecular masses of 150–220 KD and are composed primarily of repeating calcium-binding EGF-like domains and domains containing eight cysteine residues, named 8-Cysteine (8-Cys) or TB domains (Gleizes et al., 1996; Saharinen et al., 1996). The 8-Cys domains are characteristic for the LTBP–fibrillin super family. Fibrillin-1, -2, and -3 have seven 8-Cys domains, and each of the LTBP isoforms has four 8-Cys domains. However, only the third 8-Cys domains of LTBP–1, -3 and -4 bind to LAP (Gleizes et al., 1996; Saharinen et al., 1996). There are multiple isoforms of each LTBP, generated as a result of differential splicing of LTBP transcripts (Hyytiainen et al., 2004). In addition, LTBP-1 and -4 are found in two forms – long (L) and short (S) -produced by the use of separate promoters (Oklu et al., 1998; Olofsson et al., 1995).
The formation of LLC is essential for several aspects of TGF-β function. For example, binding to an LTBP facilitates secretion of the SLC and activation of latent TGF-β by the integrin αvs6 requires the presence of an LTBP (Annes et al., 2004). Null mutations in Ltbp1L yield mice with an abnormal cardiac outflow tract, a developmental defect also observed in mice deficient for TGF-β receptor 2 in neural crest cells, and in some Tgfb2−/− mice (Choudhary et al., 2006; Kaartinen et al., 2004; Sanford et al., 1997; Todorovic et al., 2007)). Null mutations of Ltbp3 yield mice with skeletal and lung abnormalities, associated with decreased TGF-β action (Chen et al., 2002; Colarossi et al., 2005), and a hypomorphic mutation in Ltbp4S yields mice with a severe defect in terminal air sac septation, associated with altered TGF-β signaling (Sterner-Kock et al., 2002). We have also shown that mice with a Cys33Ser mutation in the Tgfb1 gene, which precludes covalent binding of TGF-β1 LAP with LTBP, have a phenotype consistent with decreased TGF-β signaling, further documenting the importance of LLC formation in regulating TGF-β levels, probably by directing TGF-β sequestration into the extra-cellular matrix (ECM) (Yoshinaga et al., 2008).
LTBPs interact with multiple ECM proteins, and therefore LTBPs may have functions independent of regulating TGF-β activity. LTBP -1, -2, and -4 bind to fibrillin-1 and -2 by non-covalent interactions (Rifkin, 2005), and the LTBPs have been detected in microfibrils of multiple tissues (Dallas et al., 2000; Ono et al., 2009). Sterner-Kock et al. and we documented that Ltbp4S−/− mice display an abnormal organization of the elastic fibers in their lungs, skin and blood vessels (Dabovic et al., 2008; Sterner-Kock et al., 2002). We suggested that these defects in elastogenesis are not TGF-β related, as normalization of TGF-β levels, while improving the degree of alveolar septation, did not abolish defects in elastic fiber assembly (Dabovic et al., 2008).
Although the LTBP isoforms are structurally similar, they share only 31% identity and 41% similarity of amino-acid sequence (Koli et al., 2001; Noguera et al., 2003; Todorovic et al., 2005). This may indicate overlapping functions or there may be specific functions for individual LTBPs in a single tissue. Several unique properties of individual LTBPs have been revealed by in vitro studies: 1) LTBP-1 and – 4 bind fibrillins-1 and -2, whereas LTBP-3 does not (Isogai et al., 2003); 2) LTBP-1 and -4 do not require binding to SLC for efficient secretion from cells, whereas LTBP-3 is secreted only as LLC (Chen et al., 2002; Penttinen et al., 2002); 3) LTBP-1 and -3 bind all three isoforms of TGF-β (TGF-β 1, 2 and 3), whereas LTBP-4 binds only TGF-β1, and that binding appears to be inefficient, as only a small fraction of LTBP-4S is secreted as LLC (Saharinen and Keski-Oja, 2000).
An important question is whether the different LTBP isoforms are redundant or have unique biological functions. Mice with null mutations of the genes for the different TGF-β – binding LTBPs (Ltbp1L, Ltbp3, and Ltbp4S) display distinct phenotypic abnormalities, although each Ltbp is expressed in multiple tissues and each of the three TGF-β – binding LTBPs can carry TGF-β1 (Saharinen and Keski-Oja, 2000). Only in lungs does a deficit of either Ltbp-3 or Ltbp-4 result in developmental abnormalities, i.e. defective terminal air sac septation (Colarossi et al., 2005; Sterner-Kock et al., 2002).
To address the question whether LTBP-3 and LTBP-4 have specific or/and overlapping functions, we have generated mice with null mutations in both Ltbp3 and Ltbp4S. We reasoned that if LTBP-3 and LTBP-4S have common or redundant functions, we would observe novel abnormalities in mice deficient in both Ltbp-3 and Ltbp-4S. Moreover, within the lung, this approach should demonstrate how the individual LTBPs contribute to organ development.
Materials and Methods
Antibodies
Antibodies to P-Smad2 and Smad2/3 were purchased from Cell Signaling Technology (Danvers, MA) or from Chemicon (Millipore, Billerica, MA). Antibodies against Aquaporin-5 and Pro-Surfactant Protein C were purchased from Chemicon (Millipore). Anti-Podoplanin antibody was purchased from Abcam (Cambridge, UK). PECAM-1 (CD31) antibody was purchased from BD Pharmigen (San Diego, CA). Anti α-Smooth Muscle Actin Antibody was purchased from Sigma-Aldrich, (St. Louis, MO). Ki67 antibody was purchased from Novacosta Laboratories Ltd (Newcastle Upon Tyne, UK), and Histone H3 Antibody from Cell Signaling. Antibody to mouse Elastin was purchased from Elastin Products Company, Inc (Owensville, MO). Antibodies against Fibrillin-1 and -2 and Ltbp-1, -3 and -4 were provided by L.Y.S. All secondary antibodies used for immunohistofluorescent studies were purchased from Molecular Probes, Invitrogen (Carlsbad, CA).
For immunohistochemical studies staining was revealed using biotinylated secondary antibodies and the ABC Vector Elite Kit from Vector Laboratories (Burlingame, CA).
Mice
Ltbp3−/− mice were generated in our laboratory (Dabovic et al., 2002a); Ltbp4S−/− mice were previously described by Sterner-Kock et al. (Sterner-Kock et al., 2002) and given to us by H. von Melchner (Sterner-Kock et al., 2002). Tgfb2+/− mice were purchased from Jackson Labs (Bar Harbor, Maine). All mice were fed a normal lab diet. For staged embryos, female and male mice were housed together overnight. Noon of the following day was considered E0.5 (Embryonic day 0.5). Pregnant females were killed by CO2 asphyxiation and the embryos were collected and placed immediately in 10% buffered formalin at room temperature. All procedures were conducted according to the regulations of the NYU Langone Medical Center IACUC.
In order to increase frequency of Ltbp3−/−; Ltbp4S−/− pups, and to obtain control Ltbp3−/−, Ltbp4S−/−, and WT animals, we performed two types of crosses of animals from the same mixed genetic background: Ltbp4S−/− and WT mice were obtained from Ltbp4S+/− X Ltbp4S+/− crosses and Ltbp3−/− and Ltbp3−/−; Ltbp4S−/− mice were generated from Ltbp3+/−; Ltbp4S+/− X Ltbp3−/−; Ltbp4S+/− crosses.
Genotyping
Mice from Ltbp4S+/− X Ltbp4S+/− crosses were genotyped by PCR using reverse primers 3C7Wt: GGCTCATGCTTGAATGTTCAG and 3C7Tg: ATCATGCAAGCTGGTGGCTG specific for the WT and mutated allele, respectively, and a common forward primer P3: CCAATCTTGCTTCTTTGCTGAGC. L37F: CGTGGTGAACGTGCGTGTCCA and L38R: GCGGCAGCAAGTGCTGGGAAG for amplification of the WT allele and primers: L3G1F: CAATCCGGAGTGGCTGAACC and NeoPR: CTGCTAAAGCGCATGCTCC for the mutated allele.
Quantitative real time RT-PCR
RNA from freshly dissected lungs was extracted using Trizol (Invitrogen). Reverse transcription (RT) reactions were performed using 1 μg of RNA and Superscript III Reverse Transcriptase (Invitrogen) at 50°C for 60 minutes. The obtained cDNA samples were used for quantitative real-time RT-PCR (Q-RT-PCR) analysis (Wang et al., 2006). Q-RT-PCRs were carried out with specific primers and the Quanti Fast SYBR Green PCR Kit (Qiagen) in an iCycler Thermal Cycler (Bio-Rad). The quantity of each specific transcript was estimated using the comparative threshold cycle (TC) method and comparing the TC with that of hypoxanthine guanine phosphoribosyl transferase. The sequences of the primers are shown in Supplemental Table 4.
Histology and Immunohistochemistry
Mouse lungs were inflated with 10% buffered formalin (Sigma-Aldrich) at room temperature (for histological and immunohistochemical studies) or with 4% buffered PFA at +4oC (for in situ hybridization). Fixative was delivered through the cannulated trachea under water pressure of 25 cm for P7 and 15 cm for P0.5 and E18.5 lungs. After fixation the tissues were processed and embedded in paraffin. Five-micrometer sections were used in all studies. For histological and histomorphometric analysis the sections were stained with hematoxylin and eosin (H&E)(Sigma). Elastin was stained using the orcinol – new fuchsin technique (Sheehan and Hrapchak, 1980). Immunohistochemistry was performed as previously described (Colarossi et al., 2005; Todorovic et al., 2007).
In Situ Hybridization
Anti-sense (AS) ribo-probe for Ltbp3 was previously described (Colarossi et al., 2005) and Ltbp4 AS ribo-probe was generated from 500 bp fragment of Ltbp4 cDNA (760–1260 b of ORF) cloned in pGEM T-Easy. The probes were digoxigenin labeled using adequate primers and RNA polymerase (Roche).
In situ hybridization on paraffin sections was performed as described at http;//www.med.upenn.edu/mcrc/histology_core/nrinsitu.shtml.
Western Blot Analysis
Western blot analysis was performed on lung extracts from P7 mice. Tissue was snap-frozen in liquid nitrogen and minced using mortar and pestle and lysed for 10 min on ice in lysis buffer (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 1% TritonX-100, 1 mM glycerophosphate, 2.5 mM sodium pyrophosphate, 1 mM sodium orthovanadate) containing protease inhibitor cocktail (Roche, Indianapolis, IN). The lysates were passed through a 22-gauge syringe needle 5–10 times and centrifuged for 10 minutes at 14,000 rpm in a micro centrifuge at 4°C. The protein concentrations in the supernatants were determined using Pierce BCA kit (Thermo Scientific, Rockford, IL). Equivalent amounts of proteins from each sample were separated by SDS PAGE (Dabovic et al., 2008). Immunoreactive bands were revealed using Pierce ECL Western Blotting Substrate (Thermo Scientific). Relative intensity of the bands was evaluated using Kodak 1D 3.5.4 software (Kodak Scientific Imaging System, Rockville, MD). The ratio of the intensity of P-Smad2 versus Smad2/3 bands was normalized to the ratio calculated for the WT samples.
Histomorphometric analysis
Mean terminal sac diameter was calculated using five lung sections stained with H&E. 10–12 random fields were photographed under 20X magnification, and 2–3 horizontal lines were drawn across each photographed field in areas without large airways or vessels. Each intercept of the lines and terminal air sac walls was counted; the number of lines was multiplied by 580 (which corresponds to the length of the line connecting opposite vertices in a 20× objective microscope field in μm) and divided by the number of intercepts (Dabovic et al., 2008).
Transmission electron microscopy
(EM). For EM studies lungs were inflated with ice-cold 3% gluteraldehyde in 0.1 M cacodylate buffer (pH 7.4). The left lobe was removed, incubated in fresh fixative overnight, cut in 1.5 mm3 pieces and stained en bloc with 1% osmium tetroxide, 2% tannic acid and 2% uranyl acetate. Thereafter the samples were dehydrated and embedded in Epon as previously described (Davis, 1993). For the analysis of elastic fibers thin sections (60 nm) were put on formvar-coated grids and counterstained with 7% methanolic uranyl acetate followed by lead citrate. Sections were viewed using a Tecnai 12 transmission electron microscope at 120 kV.
Results
Phenotype of Ltbp3−/−; Ltbp4S−/− mice
In order to increase frequency of Ltbp3−/−; Ltbp4S−/− pups, and to obtain control Ltbp3−/−, Ltbp4S−/−, and WTanimals, we performed two types of crosses of animals from the same mixed genetic background: Ltbp4S−/− and WT mice were obtained from Ltbp4S+/− X Ltbp4S+/− crosses and Ltbp3−/− and Ltbp3−/−; Ltbp4S−/− mice were generated from Ltbp3+/−; Ltbp4S+/− X Ltbp3−/−; Ltbp4S+/− crosses. External examination of Ltbp3−/−; Ltbp4S−/− pups after birth revealed no obvious novel abnormalities compared to Ltbp3−/− or Ltbp4S−/− mice. Figure 1 shows the survival of WT, Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−; Ltbp4S−/− pups that were not sacrificed before 2 months of age. The most striking difference between Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−; Ltbp4S−/− mice is a shortened life span of mice deficient for both Ltbp-3 and Ltbp-4S compared to mice with single mutations (Fig. 1, Supplemental Table 1). Analysis of the distribution of different genotypes among the animals sacrificed at birth or at P7 showed that all mutant mice were born at the expected Mendelian frequency (Suppl. Table 1A); however by P7, approximately 50% of Ltbp3−/−; Ltbp4S−/− pups died (Fig. 1 and Suppl. Table 1B). Analyzes of the survival of all mice that were not sacrificed before 2 months of age and genotype distribution at P30 (Fig. 1, Suppl. Table 1C) revealed that approximately 25% of Ltbp4S−/− and 85% of Ltbp3−/−; Ltbp4S−/− pups died by P21. All Ltbp3−/− Ltbp4S−/− micethat survived after weaning age died by 1 month after birth, whereas 60–70% of Ltbp4S−/− mice lived longer than 2 months. All Ltbp3−/− mice lived longer than 2 months, as Ltbp3−/− mice have a normal lifespan [9]. Examination of dead or dying Ltbp3−/−; Ltbp4S−/− mice revealed no obvious cause of death. Because the lung septation in Ltbp3−/−; Ltbp4S−/− mice is severely affected (see below), we believe that lung defects are the major cause of premature death of Ltbp3−/−; Ltbp4S+/− animals.
Figure 1.
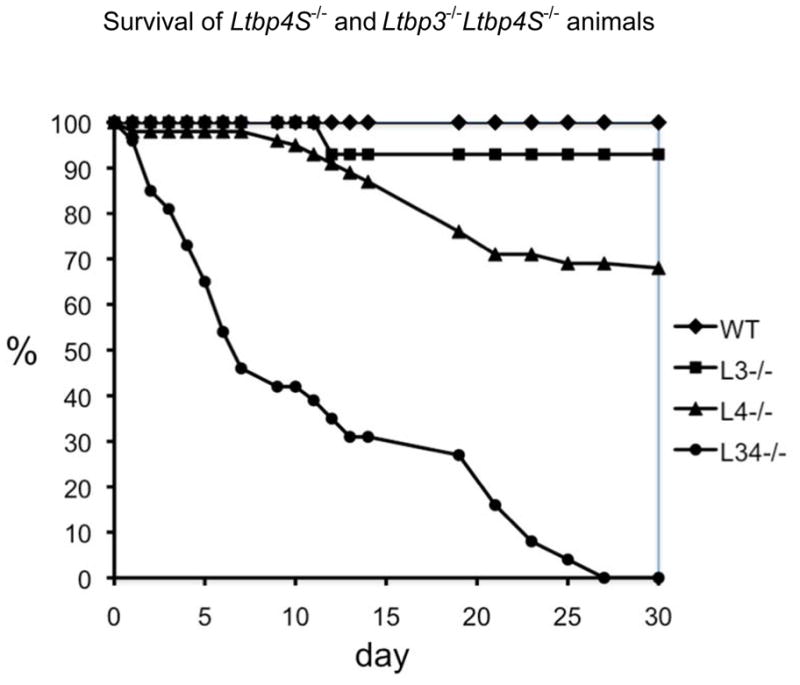
Survival of Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice. Data on mortality of WT, Ltbp3−/−, Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− mice during the first month of life were collected on the mice that were not sacrificed before 2 months of age; the initial number (N) of animals of each genotype was WT, 54; Ltbp3−/−, 14; Ltbp4S−/−, 56; Ltbp3−/−;Ltbp4S−/−, 26. 50% of Ltbp3−/−;Ltbp4S−/− mice die by P7 and the remaining 50% percent before P25. About 30% of Ltbp4S−/− mice also die by P21: however, the remaining 60–70% live more than 2 months. WT and Ltbp3−/− have normal life spans. Therefore we observed no lethality of mice during the first month after birth.
Ltbp3 and Ltbp4 expression
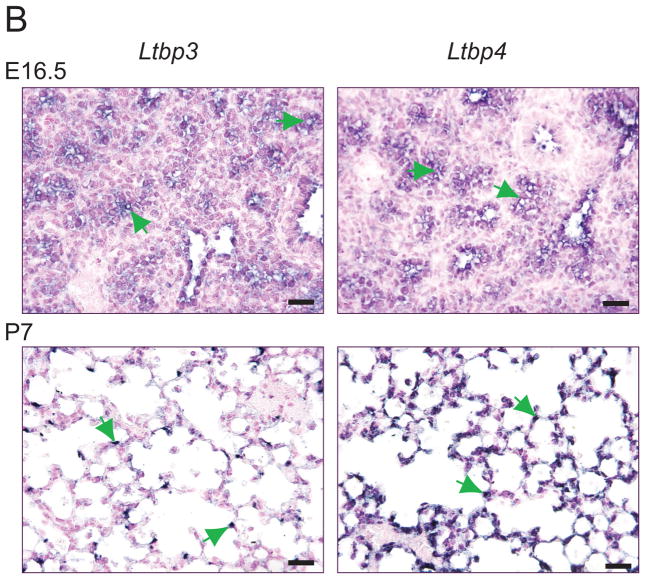
To determine which organ systems might be affected during development of Ltbp3−/−;Ltbp4S−/− mutant mice, we analyzed Ltbp3 and Ltbp4 expression in E16.5 embryos by in situ hybridization (Suppl. Fig. 1). At E16.5 Ltbp3 expression was detected in the lung, certain regions of the brain, in the cartilage, and in the brown adipose tissue. In situ hybridization with an Ltbp4 RNA probe revealed an expression pattern similar to that of Ltbp3. Thus during mouse embryogenesis, at E16.5, there was an overlap in expression of Ltbp3 and Ltbp4.
We next focused our analysis on the lungs, as both genes are expressed in the lungs and mutations in each gene yield abnormal lung development (Colarossi et al., 2005; Sterner-Kock et al., 2002). We initially examined the expression levels of the three TGF-β-binding Ltbps (1, 3, and 4) in mouse lungs from embryonic day 14.5 (E14.5) to day 7 after birth (P7) using Q-RT-PCR (Fig. 2A). The results indicate that expression of all three Ltbps can be detected in the lung as early as E14.5. The level of Ltbp1 is very low until E18.5, when there is a slight increase. This low level of expression is consistent with the observation that Ltbp1−/− mice have no obvious lung phenotype. The expression of Ltbp3 during development is somewhat (1.5 – 2 fold) higher than the expression of Ltbp1. But the major Ltbp species expressed in the embryonic and newborn lung is Ltbp4, with levels 30–40 fold higher than those of Ltbp1 or Ltbp3 at all time points examined. There is a 3–4-fold increase in expression of Ltbp1, 3 and 4 from E18.5 to P4. Thereafter, Ltbp1 and Ltbp3 expression decrease and by P7 return to the low level detected during development, whereas the expression level of Ltbp4 decreases only by 20%. These data indicate that only at P4 is there appreciable expression of both Ltbp1 and Ltbp3 compared to Ltbp4. Thus Ltbp4 is highly expressed as early as the pseudoglandular stage in lung development, whereas Ltbp3 expression is increased only later, during the saccular stage.
Figure 2.
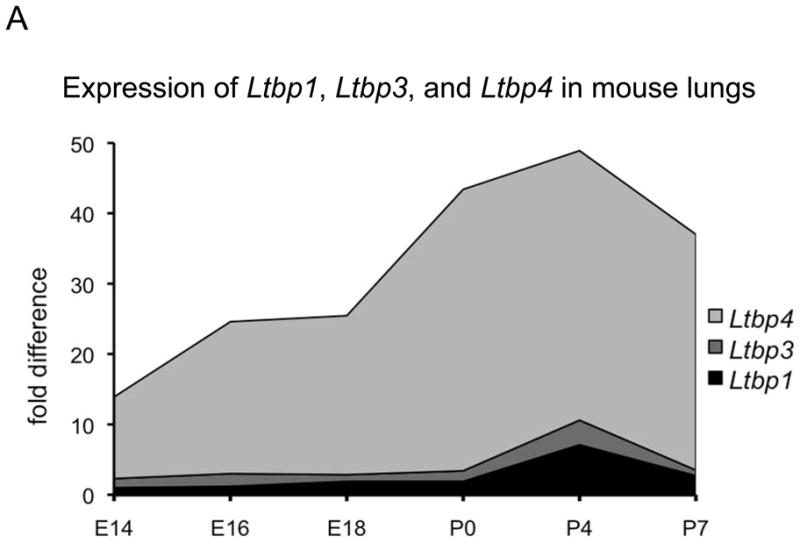
Ltbp1, Ltbp3 and Ltbp4 expression in WT mouse lungs. A. Relative expression levels of Ltbp1, Ltbp3 and Ltbp4 was assessed by Q-RT-PCR using total RNA extracted from mouse lungs as templates; 3–4 lungs were used per genotype. All samples are standardized to HPRT. Ltbp4 expression levels are 30–40 times higher than the expression levels of Ltbp3 and Ltbp1. The expression level of Ltbp3 peaks at P4 and by P7 decreases to the low levels detected during embryonic development. Ltbp4 expression is also highest at P4, but it also remains high at P7. Expression levels of Ltbp1 are very low in embryonic lungs and somewhat increased after birth at P4. However expression of Ltbp1 is lower than expression of Ltbp3 and Ltbp4 at all time points. B. ISH expression pattern of Ltbp3 and Ltbp4 in lung tissue. In embryonic lungs at E16.5 both Ltbp3 and Ltbp4 are expressed in terminal bronchioli, as indicated by arrows. P7 Ltbp4 transcripts were detected throughout the lung tissue. The expression was not uniform, and some lung cells gave stronger ISH signal (arrows). At P7 Ltbp3 transcripts were confined to a small number of lung cells (arrows). Bars: 20 μm.
Closer examination of Ltbp3 and Ltbp4 expression in developing lungs by in situ hybridization (ISH) revealed that at E14.5 (not shown) and E16.5 both genes are highly expressed in the terminal bronchioli, and at lower levels in the lung stroma (Fig. 2B) At P0.5 both Ltbp3 and Ltbp4 are expressed throughout the lung parenchyma, although the Ltbp3 ISH signal appeared much weaker, which is in accordance with Q-RT-PCR data (Fig. 2A). By P7, the Ltbp3 expression pattern changed, and RNA transcripts were detected only in a fraction of cells in the lung parenchyma (Fig. 2B). The same pattern of Ltbp3 expression is observed at P14 (data not shown). A strong ISH signal was detected throughout lung parenchyma with the Ltbp4 probe both at P7 (Fig. 2B) and P14 (data not shown), however, certain cells displayed a higher level of the transcript. The Ltbp3 and Ltbp4 ISH patterns resemble those obtained by immunostaining of lung sections with antibodies against αSMA and calponin-1, proteins characteristic for smooth muscle cells and myofibroblasts (See below Fig. 6). This suggested that both Ltbp3 and Ltbp4 are expressed in lung myofibroblasts.
Figure 6.
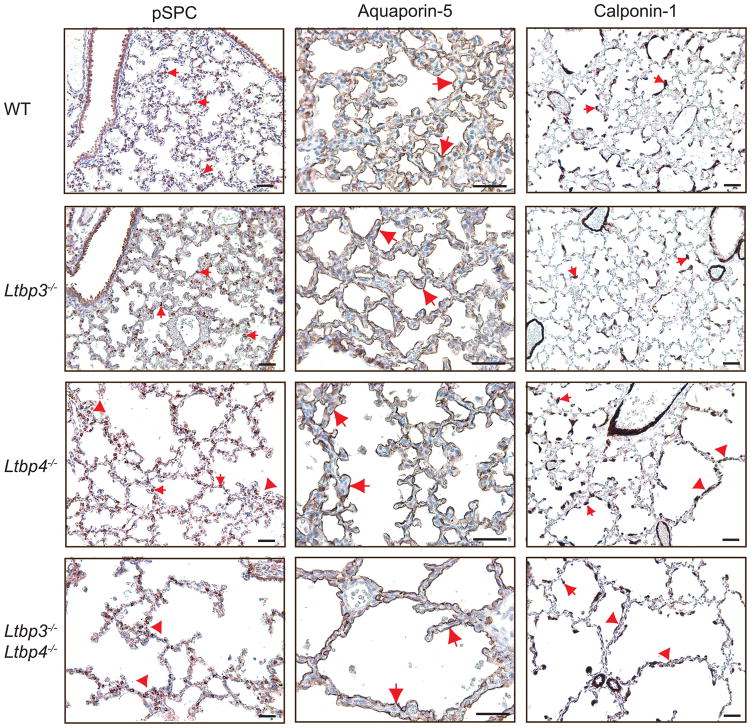
Differentiation of Type 1 and 2 epithelial cells and myofibroblasts in Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/−. Left. Type 2 cells were stained with an antibody against proSP-C. The differentiation and distribution of Type 2 cells (arrows) appear normal in WT and Ltbp3−/−lungs, whereas abnormally large clusters (arrowheads), and areas with decreased number of Type 2 cells were observed in Ltbp3−/−;Ltbp4S−/− lungs, and in the large-air sac areas in Ltbp4S−/−lungs. Center. Type 1 cells were stained with an antibody against Aquaporin-5. Aquaporin-5 is detected at the alveolar surface of all four genotypes (arrows), indicating Type-1 cell maturation in Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs. Right. SMCs and myofibroblasts (arrows) were stained with an antibody against calponin-1. In WT, Ltbp3−/− and in the areas of small, i.e. normal air sacs in Ltbp4S−/− lung parenchyma myofibroblasts (arrows) are localized on the tips of septating air sacs and in airway walls. In the areas with arrested septation in Ltbp4S−/−, as well as in Ltbp3−/−;Ltbp4S−/− lungs there was an increase in myofibroblast numbers in abnormal septae of large air sacs (arrowheads). Bars: 40 μm.
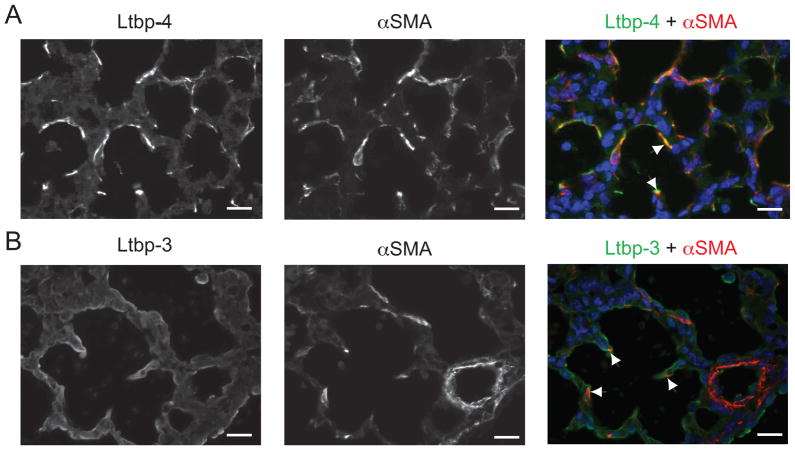
Co-immunostaining of lung sections with antibodies against LTBP-4 and αSMA revealed that myofibroblasts are the cells with the highest Ltbp4 expression level (Fig. 3). Immunofluorescent staining of lung sections with an antibody against mouse Ltbp-3 gave a relatively weak signal, consistent with the low levels of Ltbp3 mRNA, as estimated by Q-PCR. Nevertheless the co-immunostaining of lung sections with Ltbp-3 and αSMA antibodies indicated that some, although not all, lung myofibroblasts, express Ltbp-3 (Fig. 3). Therefore, our data show that both Ltbp-3 and -4are expressed by the myofibroblasts in the lungs.
Figure 3.
Lung myofibroblasts synthesize both Ltbp-3 and Ltbp-4. P0.5 WT lung sections were analyzed by immunofluorescence using antibodies against SMC/myofibroblast specific αSMA and an antibody against either Ltbp-4 (A) or Ltbp-3 (B). Nuclei were stained with DAPI. A. Double immunofluorescence of WT cells stained with both Ltbp-4 (left panel) and αSMA (middle panel). The yellow signal indicates overlap of Ltbp-4 and αSMA signals (right panel, arrowheads). B. Double immunofluorescence of WT cells stained with both Ltbp-3 (left panel) and αSMA (middle panel). A small but real number ofαSMA-positive cells (arrows) was also stained with Ltbp-3 (arrowheads), indicating that Ltbp-3 is expressed in some lung myofibroblasts. Bars: 20μm.
Defective lung development in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice
We next examined structure and cellular functions in the lungs of WT, Ltbp3, Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice to elucidate the cause of the defects in mutant lung development. Histological examination of the lungs at P0.5 indicated no differences between the WT and the Ltbp3−/− animals (Suppl. Fig. 2). Histomorphometric analysis revealed a significant increase in average terminal air sac diameter in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs compared to Ltbp3−/− and WT lungs (Suppl. Fig. 2). However, the additional loss of Ltbp-3 did not yield a more severe defect in lung septation and there was no obvious difference between the lung structure of Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice at this time point. The similarity between lung defects in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice at P0.5 was not surprising, considering the low level of Ltbp3 expression in lungs during development. We also assessed the expression levels of the three Ltbps in P0.5 mutant lungs by Q-RT-PCR (Suppl. Table 2). Our data indicate that in the absence of one or two Ltbps there was no compensatory increase in expression of other Ltbps. The fact that a small amount of Ltbp3 mRNA was measured in Ltbp3−/− lungs reflects the presence of some abnormal transcripts, as Ltbp3−/− animals were generated by an out-of-frame deletion of exons 2 and 3 (Chen et al., 2002), which introduced a stop codon 16 amino acids after 92 amino-acids coded by exon 1 (or 118 amino-acids after the start codon) and eventually caused degradation of the aberrant mRNA (Chen et al., 2002).
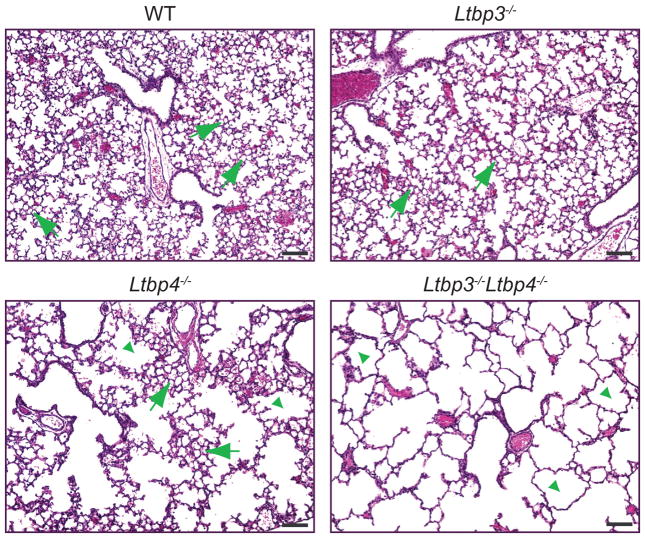
Lung development in mice proceeds after birth, with terminal air sac septation and alveolarization occurring from P0.5 to P21. At P7 the lungs from Ltbp3−/− mice were indistinguishable from WT lungs, indicating that on the mixed genetic background of the animals used in this study, lack of Ltbp-3 did not cause lung defects (Fig. 4). Histological analysis at P7 revealed that the terminal air sac septation defects in Ltbp4S−/− mice showed a great degree of variability – from relatively mild to very severe (data not shown). Yet, in lung sections from most Ltbp4S−/− mice at P7, the alveoli had a patch-like appearance that reflected uneven septation resulting in areas with both small and abnormally large air sacs (Fig. 4). At P7 lung defects in Ltbp3−/−;Ltbp4S−/− mice were more severe than those in Ltbp4S−/− mice, as there was no formation of small air sacs or alveoli during first week after birth, and Ltbp3−/−;Ltbp4S−/− lungs appeared arrested in the saccular stage, with architecture similar to that of P0.5 lungs. These data indicate that the loss of Ltbp-3 exacerbates the structural defect induced by the lack of Ltbp-4S and suggest that both Ltbp-3 and Ltbp-4 play important roles in lung development. Ltbp-4 might substitute for LTBP-3, as Ltbp3−/− mice have no lung defects, but Ltbp3 might only partially compensate for Ltbp-4, as reflected by the differences between Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/−lungs. The partial alveolarization in Ltbp4S−/− lungs is not a result of an increased expression of Ltbp3, as Q-RT-PCR analysis of lungs at P7 indicated no compensatory increase of Ltbp3 expression in Ltbp4S−/− lungs (Suppl. Table 2). In addition, there was no increase of Ltbp4 expression in Ltbp3−/− lungs.
Figure 4.
Histology of Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs at P7. Lung sections were stained with hematoxylin and eosin. WT and Ltbp3−/− lungs display normal morphology with small terminal air sacs (arrows) generated by lung septation and alveolarization after birth. In Ltbp4S−/− lungs some areas undergo alveolarization, and have clusters of small terminal air sacs (arrows). Those areas are separated by areas with large air-spaces, where lung septation is arrested (arrowheads), giving Ltbp4S−/− lungs a patch like appearance in tissue sections. In Ltbp3−/−;Ltbp4S−/−lungs all terminal air sacs are greatly enlarged (arrowheads) indicating a severe decrease in lung septation after birth. Bars: 100 μm.
Increased apoptosis in Ltbp3−/−;Ltbp4S−/− lungs
The developmental emphysema in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice may be caused by 1) decreased proliferation, 2) increased apoptosis and/or 3) defective differentiation of lung cell lineages (Bourbon et al., 2009). Therefore, we examined cell proliferation in the lung parenchyma at E16.5, E18.5, P05 and P7 by immunostaining lung sections with two markers of dividing cells: P-Histone 3 and Ki67. We also assessed the rate of apoptosis by immunostaining apoptotic cells in lung sections or by labeling double-strand DNA breaks with fluorophores using enzymatic reactions. We found no differences in cell proliferation or apoptosis comparing WT, Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs at E16.5 and E 18.5, although the defects in lung septation are apparent at E18.5 (data not shown). At P0.5 there were no differences in cell proliferation in the lungs of all four genotypes (data not shown). However, at this time a significant increase in the apoptotic index was observed in Ltbp3−/−;Ltbp4S−/− lungs compared to WT, Ltbp3−/− and Ltbp4S−/− lungs (Fig. 5). While our studies of proliferation and apoptosis in embryonic lungs did not reveal the cause of decreased cell numbers in the lung parenchyma of Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice at E18.5 and P0.5, our data do indicate that increased cell death, rather than decreased cell proliferation, as a cause of decreased cell number in the lung parenchyma of Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice at P7. This difference in apoptotic index between Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs might contribute to the differences in lung architecture observed in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice at P7.
Figure 5.
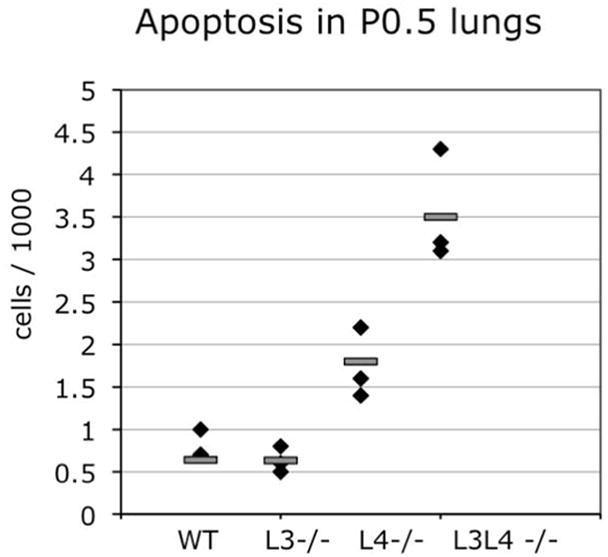
Increased apoptosis in P0.5 Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs. Apoptotic cells were revealed by labeling DNA brakes with fluorophores using in situ oligo ligation (ISOL) assay (Chemicon). Very few apoptotic cells were detected in WT and Ltbp3−/− lungs, an increased number of apoptotic cells was detected in Ltbp4S−/− lungs (P≤0.044), and a further increase in cell death was observed in Ltbp3−/−;Ltbp4S−/− lungs (P≤0.003). In addition there is a statistically significant difference between the apoptotic index in P0.5 Ltbp3−/−;Ltbp4S−/− compared to Ltbp4S−/−lungs (P≤0.0.23).
Cell differentiation in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs
The lung parenchyma is composed of two types of epithelial cells named Type 1 and Type 2 cells. Type 1 cells, which line terminal air sacs, are involved in oxygen exchange and comprise 90% of lung epithelial cells. Type 2 cells, which produce surfactant proteins B and C that are essential for lung function, comprise 5–10% of lung epithelial cells. Because 25% of Ltbp4S−/− and 80% of Ltbp3−/−;Ltbp4S−/− pups die by P12 presumably because of severe impairment of lung function, we looked for defects in Type 1 and Type 2 cell differentiation in the lungs of the mutant animals at P7. The presence of functional Type 2 cells was examined by immunostaining of lung sections with proSurfactant-C antibody, and the presence of functional Type 1 cells was assessed by staining lung sections with antibodies against Aquaporin-5. Our results revealed that differentiation of both Type 1 and Type 2 cells occurred in the absence of either or both Ltbp-3 or Ltbp-4S (Fig. 6). Yet, the distribution of Type 2 cells is abnormal in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs. We observed abnormally large clusters of Type 2 cells in some areas, and decreased numbers in others (Fig. 6A). Staining of P7 lung sections with an antibody against calponin-1 revealed defects in localization of myofibroblasts in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs (Fig. 6). In WT and Ltbp3−/− lungs myofibroblasts are found at the tips of the growing air sac septae and underneath airway epithelium, and myofibroblasts are evenly distributed through the lung tissue. However in P7 Ltbp4S−/− lungs, in the areas with defective septation and large air sacs, as well as throughout Ltbp3−/−;Ltbp4S−/− lungs, there is an increased number of calponin-1 and αSMA- producing cells in aberrant air sac septae, an indication of lung fibrosis (Araya and Nishimura, 2010).
As the lungs perform air-blood O2 and CO2 exchange, the lung is a highly vascularized tissue and blood-vessel formation is critical for proper function. Therefore we examined Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs for blood vessel formation and number. We stained lung sections from E16.5, E18.5 and P0.5 animals with an antibody against PECAM, a marker of endothelial cells. We detected no differences in blood-vessel formation or number between Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs (data not shown).
In summary, our data suggest normal differentiation of all major lung cell types, epithelial, endothelial, and smooth muscle cells in Ltbp3−/−, Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs. However, abnormalities in the distribution of myofibroblasts are present in the abnormal air sac septae.
Elastogenesis in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs
Lung function relies on tissue elasticity, which in animals with a closed circulation is provided by elastin (Wagenseil and Mecham, 2007). Two major components of elastic fibers are the fibrillins (-1 and -2) and elastin. Fibrillins are initially organized in microfibers, and elastin interaction with microfibers is essential for proper elastic fiber formation, the process designated as elastogenesis. Elastogenesis is a complex, multistep process, which requires interactions of a number of ECM proteins that are involved in binding of microfibers to cell surface molecules (integrins αV, α1, s3, s5, s6), cross-linking of tropoelastin monomers by lysyl oxidases LOX, LOXL-1, and/or direct interaction of elastin with microfibrils, composed primarily of (fibrillins-1 and -2, fibulins – 4 and -5, and mAGP-1 and 2. LTBP-1, -2, and -4 associate with microfibrils, and LTBP-4 plays an important role in elastogenesis, as Ltbp4S−/− mice display abnormal elastic fibers in their lungs, skin and blood vessels (Dabovic et al., 2008; Sterner-Kock et al., 2002). To address the potential role of defective elastogenesis in the lungs of Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice, we analyzed levels of mRNA transcripts for several proteins involved in elastogenesis. Our Q-RT-PCR data indicate that expression of Eln, Fbn1, Fbln4 and 5 are not decreased in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs (Suppl. Table 3) at P0.5 and P7 suggesting that abnormal elastogenesis in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs cannot be attributed to decreased production of major elastic fiber proteins. However, at P0.5 we detected a 3–5 fold decrease in Lox and a 5–10 fold increase in Loxl1 expression in Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− compared to WT lungs. Conversely, at P7 expression of Lox1 is increased 1.7–2.5 fold in Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− compared to WT lungs. The expression of Loxl1 at P7 is slightly decreased in Ltbp3−/−, Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− compared to WT lungs. A striking change in Loxl1 expression was observed in Ltbp3−/−;Ltbp4S−/− lungs, where the Loxl1 expression level is only about 10% of that detected in WT lungs. The mechanism underlying deregulation of the expression of Lox and Loxl1 in the absence of Ltbp3 and Ltbp4S in lung tissue is not clear, but decreased expression of the enzymes that catalyze cross-linking of elastin monomers might contribute to the defects in elastic fiber assembly observed in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice.
Electron microscopic (EM) studies of lung elastic fibers revealed no differences between Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice, both at P0.5 and P7 (Suppl. Fig. 3). The abnormal elastogenesis observed in the lungs of Ltbp4S−/− mice was not exacerbated in Ltbp3−/−;Ltbp4S−/− animals. Therefore we suggest that Ltbp-3 does not contribute to elastic fiber assembly and that regulation of elastic fiber assembly is a specific function of LTBP-4.
Microfibril Composition
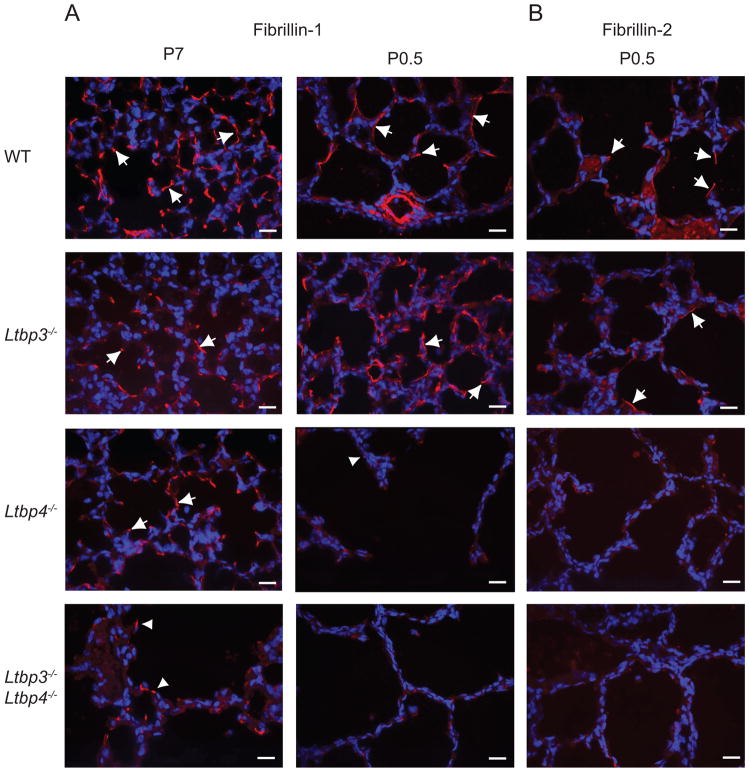
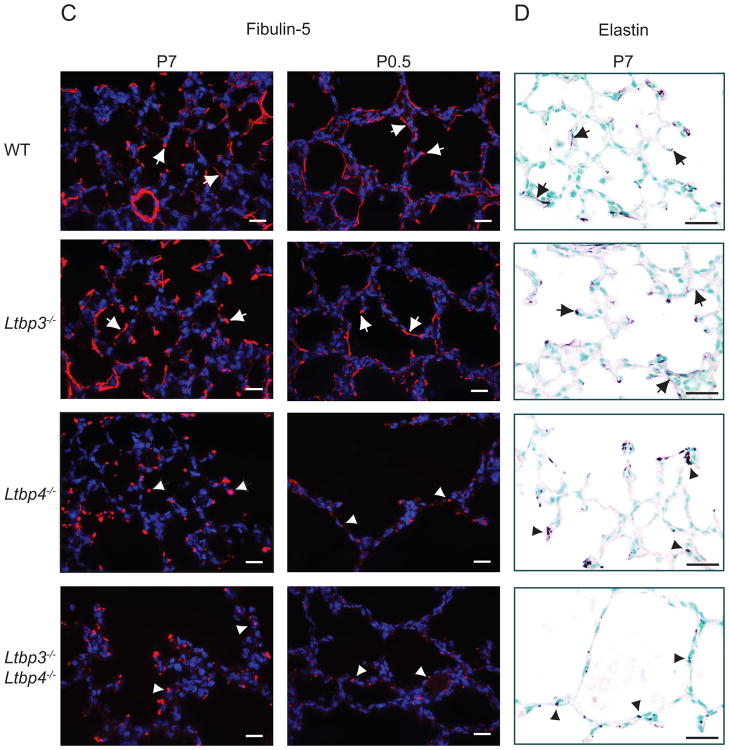
The EM analysis of elastic fibers in Ltbp4S−/− lungs indicated that the elastin incorporation in the microfibril bundles is defective in the absence of Ltbp-4. The major proteins in the microfibrils are fibrillin-1 and -2, and Fbn1−/−/Fbn2−/− mice also display defective elastogenesis in lung and blood vessels (Charbonneau et al., 2010a; Charbonneau et al., 2010b). In addition, in vitro studies have shown that LTBP-4 interacts with fibrilin-1 and -2, and immunohistochemical analysis has indicated LTBP-4 association with microfibrils in multiple tissues (Isogai et al., 2003; Ono et al., 2009). However, the defect of elastic fiber assembly in Ltbp4S−/− animals resembles most closely the defect reported for fibulin-5 deficient (Fbln5−/−) mice that have abnormally large elastin aggregates deposited next to microfibrils (Choi et al., 2009). Therefore, to examine whether the lack of LTBP-4 affects the distribution of fibrillin-1 and -2 and fibulin-5, we examined the ECM of lungs from WT, Ltbp3−/−, Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− animals at P0.5 or P7 by immunofluorescence using antibodies against fibrillin-1 and -2 and fibulin-5 (Fig. 7, Suppl. Fig. 4).
Figure 7.
Microfibril composition in Ltbp3−/−, Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs. Distribution of fibrillin-1, fibrillin-2 and fibulin-5 analyzed by immunofluorescence. (Elastin was revealed by resorcinol- new fuchsin staining. A. Fibrillin-1 distribution in Ltbp3−/−, Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs. At P7 in WT and Ltbp3−/− lungs fibrillin-1 is detected at the tips of the growing septae and in the walls of terminal air sacs, as indicated by arrows. In Ltbp4S−/− lungs fibrillin-1 fibers were detected in the areas with small, i.e. normal, air sacs (arrows), whereas very few short fibers were detected in the areas with large air sacs (arrowheads). In Ltbp3−/−;Ltbp-4S−/−lung parenchyma the fibrillin-1 staining intensity was greatly decreased. B. Fibrillin-2 distribution in P0.5 lungs. The intensity of immunofluorescent staining obtained using a fibrillin-2 antibody was weak; however, fibrillin-2 fibers were detected in WT and Ltbp3−/− lungs (arrows), but not in Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs. C. Fibulin-5 distribution in Ltbp3−/−, Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs. Fibulin-5 distribution in WT and Ltbp3−/− lungs at P0.5 and P7 is similar to fibrillin-1 distribution. Fibulin-5 staining was in terminal air sac walls and in the tips of growing septae (arrows). In Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs fibulin-5 antibody revealed no fibers, and fibulin-5 deposits appeared globular (arrowheads). In addition the staining signal appeared weaker in Ltbp3−/−;Ltbp-4S−/− than in Ltbp4S−/− lungs. D. Distribution of elastin in P7 Ltbp3−/−, Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs. The elastin staining pattern appeared similar to fibulin-5 distribution. Elastin fibers were detected in WT and Ltbp3−/− lungs (arrows), and globular deposits in Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs (arrowheads). Bars: 20 μm.
At P7 large amounts of fibrillin-1 were detected in the walls of pulmonary blood vessels and subjacent to the airway epithelium (Suppl. Fig. 4) in WT, Ltbp3−/−, Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs. Fibrillin-1 fibers were observed in the walls of terminal air sacs/alveoli in WT, Ltbp3−/−, and Ltbp4S−/− animals, but were almost undetectable in the air sac walls in Ltbp3−/−;Ltbp4S−/− lungs (Fig. 7A). Therefore, at P7 the major differences in fibrillin-1 amount and distribution between Ltbp3−/−;Ltbp4S−/− and Ltbp4S−/− mice were observed in lung parenchyma. In Ltbp4S−/− lungs fibrillin-1 fibers were detected in the walls of terminal air sacs in the areas with small, i.e. normal, air sacs, whereas very few short fibers were detected in the areas with large air sacs. In Ltbp3−/−;Ltbp4S−/− lung parenchyma the fibrillin-1 staining intensity was greatly decreased, very few short fibrils were observed in air sac walls, and the staining signal at the tips of the growing septae appeared decreased compared to Ltbp4S−/− lungs. At P0.5 the fibrillin-1 staining intensity was much weaker in the terminal air sacs in Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− lungs than in the WT and Ltbp3−/− lungs. These data indicated that fibrillin-1 incorporation into the ECM was defective in Ltbp4S−/− and Ltbp3−/;Ltbp4S−/− lungs. This decrease in fibrillin-1 incorporation in lung ECM might contribute to the defects observed in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs.
Fibrillin-2 distribution is similar to fibrilin-1 distribution at P0.5, although the intensity of immunofluorescent staining obtained using a fibrillin-2 antibody was much weaker compared to fibrillin-1 staining in WT and Ltbp3−/− lungs (Fig. 7B). The fibrillin-2 staining intensity was greatly decreased in Ltbp4S−/−, and Ltbp3−/−;Ltbp4S−/− airways compared to WT and Ltbp3−/− mice, indicating defective incorporation of fibrillin-2 in lung ECM in the absence of Ltbp-4 or Ltbp-3 and Ltbp-4.
Defects in fibulin-5 distribution in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs were obvious both at P0.5 and P7. Immunofluorescent staining of the lungs from WT and Ltbp3−/− mice at P0.5 revealed fibulin-5 in the lamellae between SMCs in blood vessel walls, in the fibers subjacent to the airway epithelium, at the tips of the growing septae, and in the walls of terminal air sacs (Fig. 7C and Suppl. Fig. 4). In the lungs of Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice, fibulin-5 was not organized in fibers and appeared as dot-like aggregates in lung parenchyma, around the airways, and between the fragmented lamellae between the SMCs in blood vessels. At P7 the fibulin-5 staining pattern in WT and mutant lungs was similar to the pattern observed at P0.5, whereas, the fibulin-5 aggregates in the parenchyma and around the airways in the lungs of Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− mice appeared larger, and only few short and dotted fibers were detected in the areas where septation occurred in Ltbp4S−/− lungs. The overall staining pattern obtained with the fibulin-5 antibody resembled the pattern observed with elastin staining of lung sections from WT, Ltbp3−/−, Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− animals at P0.5 and P7. Therefore, our data indicate that in the absence of Ltbp-4 fibulin-5 cannot interact with microfibrils, but fibulin-5 does bind to elastin. These results suggest that LTBP-4 may facilitate fibulin-5 binding to microfibrils, which is essential for elastogenesis.
Inflammation in the lungs of Ltbp3−/−;Ltbp4S−/− mice
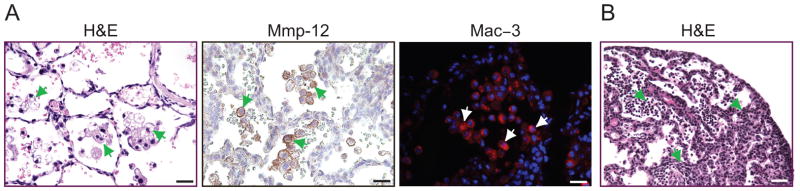
Starting from P6 we observed inflammation in Ltbp3−/−;Ltbp4S−/− lungs. Immunohistochemistry with the macrophage-specific antibody Mac-3 showed an increased number of macrophages in both Ltbp3+/−;Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs (Fig. 8A and data not shown). This inflammatory response could be induced by defective elastogenesis, as elastin that is not incorporated in elastic fibers is prone to degradation and elastin degradation products are chemo-attractant for macrophages (Guo et al., 2006; Senior et al., 1980). In addition large infiltrations of lymphocytes were also observed in some samples at P12 (Fig. 8B). This inflammation might further compromise lung function and contribute to the premature death of Ltbp3−/−;Ltbp4S−/− animals.
Figure 8.
Inflammation in Ltbp3−/−;Ltbp4S−/− lungs. A. Macrophage infiltration in Ltbp3−/−; Ltbp4S−/− lungs. Clusters of large, activated macrophages were observed in air sacs of Ltbp3−/−;Ltbp4S−/− mice (arrows). The activated macrophages were positive for metallo-protease MMP-12 and for a macrophage-specific Mac-3 (arrows). B. Large lymphocyte infiltrations (arrows) were observed by H&E staining in some Ltbp3−/−;Ltbp4S−/− animals at P10-P21. Bars: A −20 μm, B − 40 μm.
Discussion
LTBP-1, -3 and -4 bind latent TGF-β and through interactions with extracellular proteins, such as fibrillins and fibronectin, target LLC to the ECM. However, the biological functions of LTBPs are not fully understood. We generated mice deficient for both Ltbp-3 and Ltbp-4 in an attempt to reveal possible biological processes regulated by both proteins, i.e. common functions of Ltbp-3 and Ltbp-4.
The major phenotype in Ltbp4S−/− mice is a developmental defect in lung septation observed as early as E18.5 (Dabovic et al., 2008). We have shown that the absence of Ltbp-4S synthesis in Ltbp4S−/− mouse lungs causes excessive TGF-β signaling, and that decreasing TGF-β expression or signaling in vivo either by pharmacological or genetic intervention meliorated Ltbp4S−/− lung septation (Dabovic et al., 2008).
The major pathological changes in Ltbp3−/− mice are observed in the skeletal system, and manifest as fusion of the bones in the skull base resulting in severe cranio-facial malformations by P8, and defects in bone turnover resulting in osteopetrosis – like defects in endochondral bones by 1 month after birth. Ltbp3−/− mice also display post-natal abnormalities in lung development, i.e. decreased alveologenesis (Choudhary et al., 2006; Dabovic et al., 2008). The defective terminal air sac septation in Ltbp3−/− mice is dependent upon the genetic background, as 100% of Ltbp3−/− mice on a 129SvEv genetic background but only 0–5% on a mixed genetic background have impaired alveologenesis.
Another obvious difference between Ltbp-3 and Ltbp-4 function, revealed by our studies of the phenotypes of Ltbp3−/− and Ltbp4S−/− mice, as well as by the analysis of the effects of LTBP3 and LTBP4 mutations in humans, is a function of Ltbp-4S in elastin assembly. LTBP4 deficiency results in severe impairment of elastic fiber formation in all elastic tissues, whereas no elastogenesis defect was observed in LTBP-3 deficient mice or humans. In addition, Ltbp1S−/−, Ltbp1L−/−, and Ltbp3−/− mice have no defect in elastin assembly, suggesting that modulation of elastic fiber formation is a characteristic function of LTBP-4 (Annes et al., 2004; Chen et al., 2002; Dabovic et al., 2008; Drews et al., 2008; Sterner-Kock et al., 2002; Todorovic et al., 2007). Ltbp4S−/− mice also display an abnormality in lung elastogenesis apparent as early as E14.5–16.5 during embryonic development. Defective elastin assembly may affect lung septation after birth. Thus both elevated TGF-β levels and abnormal elastogenesis may contribute to lung defects observed in Ltbp4S−/− mice at P7.
The phenotype of Ltbp3−/−Ltbp4S−/− mice is more severe than the phenotypes of either Ltbp3−/− or Ltbp4S−/− mice, as Ltbp3−/−;Ltbp4S−/− mice die by P21, whereas Ltbp3−/− mice have a normal life span, and about 55% of Ltbp4S−/− mice live more than 2 months. However, we did not find additional developmental defects in any tissue other than the lung in Ltbp3−/−;Ltbp4S−/− mice, compared to Ltbp3−/− and Ltbp4S−/− animals. Therefore, we conclude that these two Ltbps do not have major overlapping activities other than in the lung.
The impairment of lung septation in Ltbp3−/−;Ltbp4S−/− mice was more severe than that observed in Ltbp4S−/− lungs and this may have resulted in the early death of the double mutant mice. As the Ltbp3−/− mice analyzed in this study did not have lung defects, we suggest that, although Ltbp-3 may play a role in alveolarization of the lungs, Ltbp-3 function is not essential, and in some genetic backgrounds may be compensated by Ltbp-4. The nature of the modifying genes responsible for the differences in penetrance of the Ltbp3−/− phenotype is unknown. Nevertheless, in Ltbp3−/−;Ltbp4S−/− lungs, Ltbp-3 deficiency exacerbates the lung defects. Ltbp3−/−;Ltbp4S−/− lung development is arrested in saccular stage, whereas terminal air sac septation in the lungs of Ltbp4S−/− animals may continue after birth. Because in individual Ltbp4S−/− mice we observed a great variability in lung septation at P7 and in some animals (10–20%) the lungs appeared as severely affected as the lungs of Ltbp3−/−;Ltbp4S−/− mice, we suggest that LTBP-4 function in lung development is essential. LTBP-4 might contribute initially to lung development by modulating both TGF-β levels and elastic fiber assembly. We also suggest that LTBP-3 contributes to the regulation of terminal air sac septation after birth, but the mechanism of LTBP-3 action is not yet understood.
We examined a number of parameters of lung development and structure in order to clarify why the Ltbp3−/−;Ltbp4S−/− phenotype is stronger than the Ltbp3−/− phenotype. There were no differences in the elastogenesis defect, in the maturation of cell types, or in cell proliferation at all time points studied. We found an increase in apoptotic index and in the number or distribution of myofibroblasts in the lung parenchyma after birth; however the mechanisms underlying morphological abnormalities seen at birth or shortly thereafter are unclear. We did observe decreased incorporation of fibrillin-1 and fibrillin-2 in Ltbp3−/−;Ltbp4S−/− lungs compared to Ltbp4S−/− lungs, which might indicate an Ltbp-3 function in lung ECM assembly, either through regulation of TGF-β levels (which, in turn, may change expression levels of many ECM proteins and affect ECM structure) or through interaction with ECM proteins. The loss of fibrillin and defective elastin incorporation may also be responsible for the heightened level of inflammation observed in Ltbp3−/−;Ltbp4S−/− lungs compared to Ltbp4S−/− lungs, as the unincorporated fibrillin and elastin might be degraded and it is known that fibrillin and elastin degradation products are chemoattractants for macrophages (Houghton et al., 2006).
The contribution of the inflammatory response to the morbidity of Ltbp3−/−;Ltbp4S−/− animals is unclear at this time, but it would be interesting to examine the effect of anti-inflammatory agents on the lung phenotype. Decreased levels of active TGF-β might also yield enhanced inflammation, as TGF-β is a powerful suppressor of the inflammatory response, yet TGF-β signaling is increased in Ltbp4S−/− and Ltbp3−/−;Ltbp4S−/− lungs (20) and therefore we hypothesize that matrix degradation, rather than decreased TGF-β levels, is a cause for the macrophage infiltration in the lungs of Ltbp3−/−; Ltbp4S−/− mice.
The interpretation of the lung phenotype in Ltb4S−/− or Ltbp3−/−;Ltbp4S−/− animals is confounded by the fact that Ltbp-4 deficiency has a profound effect on elastogenesis. It has been reported that defective elastogenesis may cause developmental emphysema, and mice deficient for fibrillin-1 and -2, elastin, fibulin-4 or -5 display defective terminal air sac septation (Hucthagowder et al., 2006; McLaughlin et al., 2006; Nakamura et al., 2002; Neptune et al., 2003; Urban et al., 2005; Yanagisawa et al., 2002). However, aberrant organization of elastic microfibers may have a subsequent effect on TGF-β activities and TGF-βs, in turn, affects matrix synthesis and turnover (ref). It might be possible to dissect out TGF-β binding activities of Ltbp-4 from its matrix organizing functions by generating mice in which the TGF-β binding residues in Ltbp-4 are altered to eliminate formation of the Ltbp-TGF-β complex but not destroy the matrix forming capacity of the Ltbp-4. Experiments are in progress to test this hypothesis.
Supplementary Material
Acknowledgments
Contract grant sponsor: NIH; Contract grant numbers: CA034282, AR49698 to DBR.
Contract grant sponsor: Canadian Institutes of Health; Contract grant numbers: MOP57663, MOP86713 to ECD. ECD is a Canada Research Chair
The authors thank other members of the Rifkin lab for their contributions to this work.
References
- Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 2004;165(5):723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFbeta activation. J Cell Sci. 2003;116(Pt 2):217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- Araya J, Nishimura SL. Fibrogenic reactions in lung disease. Annu Rev Pathol. 2010;5:77–98. doi: 10.1146/annurev.pathol.4.110807.092217. [DOI] [PubMed] [Google Scholar]
- Bourbon JR, Boucherat O, Boczkowski J, Crestani B, Delacourt C. Bronchopulmonary dysplasia and emphysema: in search of common therapeutic targets. Trends Mol Med. 2009;15(4):169–179. doi: 10.1016/j.molmed.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Charbonneau NL, Carlson EJ, Tufa S, Sengle G, Manalo EC, Carlberg VM, Ramirez F, Keene DR, Sakai LY. In vivo studies of mutant fibrillin-1 microfibrils. J Biol Chem. 2010a doi: 10.1074/jbc.M110.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau NL, Jordan CD, Keene DR, Lee-Arteaga S, Dietz HC, Rifkin DB, Ramirez F, Sakai LY. Microfibril structure masks fibrillin-2 in postnatal tissues. J Biol Chem. 2010b;285(26):20242–20251. doi: 10.1074/jbc.M109.087031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dabovic B, Annes JP, Rifkin DB. Latent TGF-beta binding protein-3 (LTBP-3) requires binding to TGF-beta for secretion. FEBS Lett. 2002;517(1–3):277–280. doi: 10.1016/s0014-5793(02)02648-0. [DOI] [PubMed] [Google Scholar]
- Choi J, Bergdahl A, Zheng Q, Starcher B, Yanagisawa H, Davis EC. Analysis of dermal elastic fibers in the absence of fibulin-5 reveals potential roles for fibulin-5 in elastic fiber assembly. Matrix Biol. 2009;28(4):211–220. doi: 10.1016/j.matbio.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary B, Ito Y, Makita T, Sasaki T, Chai Y, Sucov HM. Cardiovascular malformations with normal smooth muscle differentiation in neural crest-specific type II TGFbeta receptor (Tgfbr2) mutant mice. Dev Biol. 2006;289(2):420–429. doi: 10.1016/j.ydbio.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Colarossi C, Chen Y, Obata H, Jurukovski V, Fontana L, Dabovic B, Rifkin DB. Lung alveolar septation defects in Ltbp-3-null mice. Am J Pathol. 2005;167(2):419–428. doi: 10.1016/S0002-9440(10)62986-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabovic B, Chen Y, Choi J, Vassallo M, Dietz HC, Ramirez F, von Melchner H, Davis EC, Rifkin DB. Dual functions for LTBP in lung development: LTBP-4 independently modulates elastogenesis and TGF-beta activity. J Cell Physiol. 2008 doi: 10.1002/jcp.21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas SL, Keene DR, Bruder SP, Saharinen J, Sakai LY, Mundy GR, Bonewald LF. Role of the latent transforming growth factor beta binding protein 1 in fibrillin-containing microfibrils in bone cells in vitro and in vivo. J Bone Miner Res. 2000;15(1):68–81. doi: 10.1359/jbmr.2000.15.1.68. [DOI] [PubMed] [Google Scholar]
- Davis EC. Smooth muscle cell to elastic lamina connections in developing mouse aorta. Role in aortic medial organization. Lab Invest. 1993;68(1):89–99. [PubMed] [Google Scholar]
- Drews F, Knobel S, Moser M, Muhlack KG, Mohren S, Stoll C, Bosio A, Gressner AM, Weiskirchen R. Disruption of the latent transforming growth factor-beta binding protein-1 gene causes alteration in facial structure and influences TGF-beta bioavailability. Biochim Biophys Acta. 2008;1783(1):34–48. doi: 10.1016/j.bbamcr.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Gleizes PE, Beavis RC, Mazzieri R, Shen B, Rifkin DB. Identification and characterization of an eight-cysteine repeat of the latent transforming growth factor-beta binding protein-1 that mediates bonding to the latent transforming growth factor-beta 1. J Biol Chem. 1996;271:29891–29896. doi: 10.1074/jbc.271.47.29891. [DOI] [PubMed] [Google Scholar]
- Guo G, Booms P, Halushka M, Dietz HC, Ney A, Stricker S, Hecht J, Mundlos S, Robinson PN. Induction of macrophage chemotaxis by aortic extracts of the mgR Marfan mouse model and a GxxPG-containing fibrillin-1 fragment. Circulation. 2006;114(17):1855–1862. doi: 10.1161/CIRCULATIONAHA.105.601674. [DOI] [PubMed] [Google Scholar]
- Houghton AM, Quintero PA, Perkins DL, Kobayashi DK, Kelley DG, Marconcini LA, Mecham RP, Senior RM, Shapiro SD. Elastin fragments drive disease progression in a murine model of emphysema. J Clin Invest. 2006;116(3):753–759. doi: 10.1172/JCI25617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucthagowder V, Sausgruber N, Kim KH, Angle B, Marmorstein LY, Urban Z. Fibulin-4: a novel gene for an autosomal recessive cutis laxa syndrome. Am J Hum Genet. 2006;78(6):1075–1080. doi: 10.1086/504304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyytiainen M, Penttinen C, Keski-Oja J. Latent TGF-beta binding proteins: extracellular matrix association and roles in TGF-beta activation. Crit Rev Clin Lab Sci. 2004;41(3):233–264. doi: 10.1080/10408360490460933. [DOI] [PubMed] [Google Scholar]
- Isogai Z, Ono RN, Ushiro S, Keene DR, Chen Y, Mazzieri R, Charbonneau NL, Reinhardt DP, Rifkin DB, Sakai LY. Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J Biol Chem. 2003;278(4):2750–2757. doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Dudas M, Nagy A, Sridurongrit S, Lu MM, Epstein JA. Cardiac outflow tract defects in mice lacking ALK2 in neural crest cells. Development. 2004;131(14):3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- Koli K, Saharinen J, Hyytiainen M, Penttinen C, Keski-Oja J. Latency, activation, and binding proteins of TGF-beta. Microsc Res Tech. 2001;52:354–362. doi: 10.1002/1097-0029(20010215)52:4<354::AID-JEMT1020>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- McLaughlin PJ, Chen Q, Horiguchi M, Starcher BC, Stanton JB, Broekelmann TJ, Marmorstein AD, McKay B, Mecham R, Nakamura T, Marmorstein LY. Targeted disruption of fibulin-4 abolishes elastogenesis and causes perinatal lethality in mice. Mol Cell Biol. 2006;26(5):1700–1709. doi: 10.1128/MCB.26.5.1700-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Lozano PR, Ikeda Y, Iwanaga Y, Hinek A, Minamisawa S, Cheng CF, Kobuke K, Dalton N, Takada Y, Tashiro K, Ross J, Jr, Honjo T, Chien KR. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature. 2002;415(6868):171–175. doi: 10.1038/415171a. [DOI] [PubMed] [Google Scholar]
- Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, Ramirez F, Sakai LY, Dietz HC. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33(3):407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- Noguera I, Obata H, Gualandris A, Cowin P, Rifkin DB. Molecular cloning of the mouse Ltbp-1 gene reveals tissue specific expression of alternatively spliced forms. Gene. 2003;308:31–41. doi: 10.1016/s0378-1119(03)00463-3. [DOI] [PubMed] [Google Scholar]
- Oklu R, Metcalfe JC, Hesketh TR, Kemp PR. Loss of a consensus heparin binding site by alternative splicing of latent transforming growth factor-beta binding protein-1. FEBS Lett. 1998;425(2):281–285. doi: 10.1016/s0014-5793(98)00257-9. [DOI] [PubMed] [Google Scholar]
- Olofsson A, Ichijo H, Moren A, ten Dijke P, Miyazono K, Heldin CH. Efficient association of an amino-terminally extended form of human latent transforming growth factor-beta binding protein with the extracellular matrix. J Biol Chem. 1995;270(52):31294–31297. doi: 10.1074/jbc.270.52.31294. [DOI] [PubMed] [Google Scholar]
- Ono RN, Sengle G, Charbonneau NL, Carlberg V, Bachinger HP, Sasaki T, Lee-Arteaga S, Zilberberg L, Rifkin DB, Ramirez F, Chu ML, Sakai LY. Latent transforming growth factor beta-binding proteins and fibulins compete for fibrillin-1 and exhibit exquisite specificities in binding sites. J Biol Chem. 2009;284(25):16872–16881. doi: 10.1074/jbc.M809348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penttinen C, Saharinen J, Weikkolainen K, Hyytiainen M, Keski-Oja J. Secretion of human latent TGF-beta-binding protein-3 (LTBP-3) is dependent on co-expression of TGF-beta. J Cell Sci. 2002;115(Pt 17):3457–3468. doi: 10.1242/jcs.115.17.3457. [DOI] [PubMed] [Google Scholar]
- Ramirez F, Dietz HC. Extracellular microfibrils in vertebrate development and disease processes. J Biol Chem. 2009;284(22):14677–14681. doi: 10.1074/jbc.R900004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin DB. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J Biol Chem. 2005;280(9):7409–7412. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- Saharinen J, Keski-Oja J. Specific sequence motif of 8-Cys repeats of TGF-beta binding proteins, LTBPs, creates a hydrophobic interaction surface for binding of small latent TGF-beta. Mol Biol Cell. 2000;11(8):2691–2704. doi: 10.1091/mbc.11.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-beta with an eight cysteine repeat of its binding protein LTBP-1. EMBO J. 1996;15(2):245–253. [PMC free article] [PubMed] [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124(13):2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior RM, Griffin GL, Mecham RP. Chemotactic activity of elastin-derived peptides. J Clin Invest. 1980;66(4):859–862. doi: 10.1172/JCI109926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. Theory and practice of histotechnology. St. Louis: Mosby; 1980. [Google Scholar]
- Sterner-Kock A, Thorey IS, Koli K, Wempe F, Otte J, Bangsow T, Kuhlmeier K, Kirchner T, Jin S, Keski-Oja J, von Melchner H. Disruption of the gene encoding the latent transforming growth factor-beta binding protein 4 (LTBP-4) causes abnormal lung development, cardiomyopathy, and colorectal cancer. Genes Dev. 2002;16(17):2264–2273. doi: 10.1101/gad.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorovic V, Frendewey D, Gutstein DE, Chen Y, Freyer L, Finnegan E, Liu F, Murphy A, Valenzuela D, Yancopoulos G, Rifkin DB. Long form of latent TGF-{beta} binding protein 1 (Ltbp1L) is essential for cardiac outflow tract septation and remodeling. Development. 2007;134(20):3723–3732. doi: 10.1242/dev.008599. [DOI] [PubMed] [Google Scholar]
- Todorovic V, Jurukovski V, Chen Y, Fontana L, Dabovic B, Rifkin DB. Latent TGF-beta binding proteins. Int J Biochem Cell Biol. 2005;37(1):38–41. doi: 10.1016/j.biocel.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Urban Z, Gao J, Pope FM, Davis EC. Autosomal dominant cutis laxa with severe lung disease: synthesis and matrix deposition of mutant tropoelastin. J Invest Dermatol. 2005;124(6):1193–1199. doi: 10.1111/j.0022-202X.2005.23758.x. [DOI] [PubMed] [Google Scholar]
- Wagenseil JE, Mecham RP. New insights into elastic fiber assembly. Birth Defects Res C Embryo Today. 2007;81(4):229–240. doi: 10.1002/bdrc.20111. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006;39(4):356–362. doi: 10.1016/j.ymeth.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Davis EC, Starcher BC, Ouchi T, Yanagisawa M, Richardson JA, Olson EN. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature. 2002;415(6868):168–171. doi: 10.1038/415168a. [DOI] [PubMed] [Google Scholar]
- Yoshinaga K, Obata H, Jurukovski V, Mazzieri R, Chen Y, Zilberberg L, Huso D, Melamed J, Prijatelj P, Todorovic V, Dabovic B, Rifkin DB. Perturbation of transforming growth factor (TGF)-beta1 association with latent TGF-beta binding protein yields inflammation and tumors. Proc Natl Acad Sci U S A. 2008;105(48):18758–18763. doi: 10.1073/pnas.0805411105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.