Abstract
The hydrodynamic radii of a series of genetically engineered monodisperse elastin like polypeptides (ELP) was determined by dynamic light scattering in aqueous solution as function of molar mass. Utilizing the known theoretical expression for the hydrodynamic radius of wormlike chains, the Kuhn statistical segment length was determined to be lk = 2.1 nm, assuming that the length of the peptide repeat unit was b = 0.365 nm, a value derived for a coiled conformation of ELP. The resulting chain stiffness is significantly larger than previously reported by force-distance curve analysis (lk < 0.4 nm). The possible occurrence of superstructures, such as hairpins or helices, would reduce the contour length of the ELP, further increasing lk. Accordingly, the value lk = 2.1 nm reported here represents a lower limit of the chain stiffness for ELP.
Keywords: elastin-like polypeptides, dynamic light scattering, kuhn length, hydrodynamic radius
Introduction
Elastin is a structural protein that is largely responsible for the elastic properties of tissues in vertebrates. It consists of alternating hydrophobic and crosslinking domains. The hydrophobic domains are dominated by hydrophobic amino acids, such as proline, valine, alanine and leucine. The crosslinking domains contain lysyl residues combined with proline-rich regions or polyalanine domains. Elastogenesis1, the biological synthesis of elastin, involves the synthesis of a soluble precursor protein, tropoelastin, that binds to galactolectin after its translation to prevent its intracellular aggregation. After secretion of the tropoelastin-galactolectin complex into the extracellular space, tropoelastin is released locally and aligned at the microfibrillar scaffold.
Elastin-like Polypeptides (ELP), are a class of artificial repetitive polypeptides of the pentameric repeat Val-Pro-Gly-Xaa-Gly (where Xaa is a guest residue that is any amino acid except Pro), and are so named because this motif recurs in the tropoelastin gene of most vertebrates. ELPs exhibit a lower critical solution temperature (LCST) phase transition in aqueous solution. The LCST behavior2 of ELPs can be precisely specified through the control of two orthogonal variables that can be completely controlled by genetically encoded synthesis of these amino acid-based biopolymers: ELP composition, notably by the type and mole fraction of the different guest residues (X), and by the chain length. ELPs are increasingly utilized for many biomedical applications, as they are non toxic, biodegradable and show good pharmacokinetics3-8.
Despite extensive studies on the biophysics of ELPs9-12, there is still some controversy regarding the origins of the LCST behavior of ELPs and their elastomeric properties. Two structural models for the remarkable elasticity of elastin have been proposed; the first “β-spiral model”, proposed by Urry9, is based on extensive studies on synthetic poly(ValProGlyValGly), and proposes that ELPs consist of a helical arrangement of type II β-spirals per pentameric peptide unit. The ProGly unit is positioned at the corner of the bend and a hydrogen bond connects the carbonyl group of the first valine with the amino residue of the fourth valine. The dipeptide segments of VG, suspended between the β-turns, exhibit large amplitude librations. The decrease of these librations upon extension results in a large decrease in entropy of the segments explaining the rubber elasticity of elastin. At variance with the classical theory of rubber elasticity, this model assumes fixed end-to-end chain lengths. Contradicting this model, molecular dynamics simulations assuming freely fluctuating ends resulted in a loss of the helical β-spiral structure13. A second model, proposed by Tamburro et al.14, involves the formation of non-recurring isolated type II β-turns for the (GlyXGlyGlyX) repeat units of the elastin sequence. The XGly or GlyGly segments build up the corners. The first and fourth glycine or the second and fifth X residue are connected via hydrogen bonds. Due to the absence of proline residues, the resulting β-turns can dynamically slide along the chain. The polypeptide can therefore freely fluctuate leading to high intrinsic entropy, which decreases upon extension. Both models have in common the presence of type II β-turns, but their dynamic interpretations are different. Molecular simulations11-13 performed by Daggett and coworkers have indicated that the hydration of the hydrophobic residues largely contributes to the gain in entropy. Upon the collapse of the polypeptide the hydration water is released thus leading to a significant increase of solvent entropy.
The single molecule elasticity of ELP has been experimentally investigated by force microscopy as function of temperature and ionic strength15. The force distance curves were analyzed by standard expressions for a wormlike chain model which yields the persistence length, lp, which is connected to the Kuhn statistical segment length, lk, by lp = lk/2. At T = 25 °C a very small Kuhn length was determined to lk = 0.3 nm (ELP4-120). Since it is well established that analysis of force distance curves of flexible polymers often yields values of the persistence length that are anomalously small,16 a reliable determination of the chain stiffness of ELP is still missing.
Results and Discussion
Herein, the Kuhn length of ELP was determined in dilute solution by analysis of the hydrodynamic radii (Rh) measured by dynamic light scattering (DLS) on a series of ELPs with different molecular weights. Five ELPs Gly(Val-Pro-Gly-Val-Gly)n Phe-Cys with the guest residue Valine and a single Cysteine residue at the C-terminus were investigated. Their length varied from n=20 up to n=120 (notation ELP4-n), where n is the number of pentapeptide repeats. The recombinant synthesis of each ELP construct utilized plasmid reconstruction recursive directional ligation6, and is summarized along with their purification in the Supporting Information. The samples utilized are listed in Table 1.
Table 1.
Calculated molecular weight and hydrodynamic radii determined by DLS for ELP samples.
| MW/kDa | Rh/nm | |
|---|---|---|
| ELP4-120 | 49.4 | 6.0 |
| ELP4-60 | 24.9 | 4.2 |
| ELP4-40 | 16.7 | 3.7 |
| ELP4-30 | 12.6 | 3.4 |
| ELP4-20 | 8.5 | 2.7 |
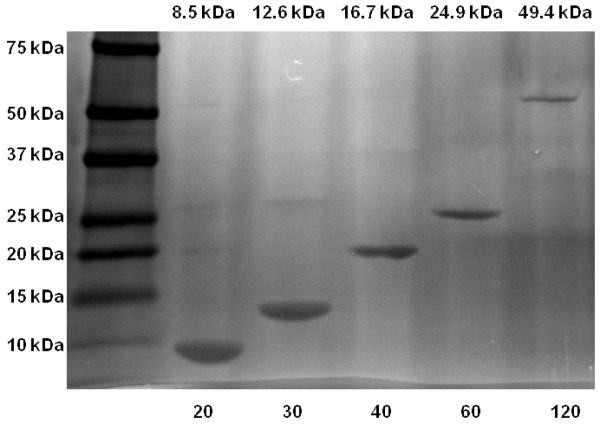
SDS-PAGE (see Figure 1) of the ELPs was carried out to verify the purity of the ELP samples utilized in this study and DLS was utilized in order to determine the Rh from the measured diffusion coefficient D by application of Stokes’ law
| (1) |
where kT is the thermal energy and η0 is the solvent viscosity. Experimental details of the DLS measurements are in the Supporting Information (SI). Briefly, the ELPs were dissolved in 20 mM NaCl, pH = 6.5, for the LS measurements at a concentration of 5 mg/mL. This small concentration effectively represents the infinite dilution limit, since Rh of sample ELP4-40 did not change when diluted to c = 0.5 g/L. All measurements were performed at a scattering angle of 30°. Due to the terminal cysteine group of the ELPs used in the present work, dimerization by formation of disulfide bridges is possible. Therefore all samples were measured directly after dissolution. Control measurements revealed that dimerization noticeably starts after 24h, only.
Figure 1.
SDS-PAGE (4-20% Tris-HCl, gradient) of purified ELPs visualized by copper staining. The left lane contains a molecular weight standard, labeled in kilodaltons (kDa). The expected molecular weight is indicated on the top of each lane. The length in pentapeptides of each ELP is labeled below each lane.
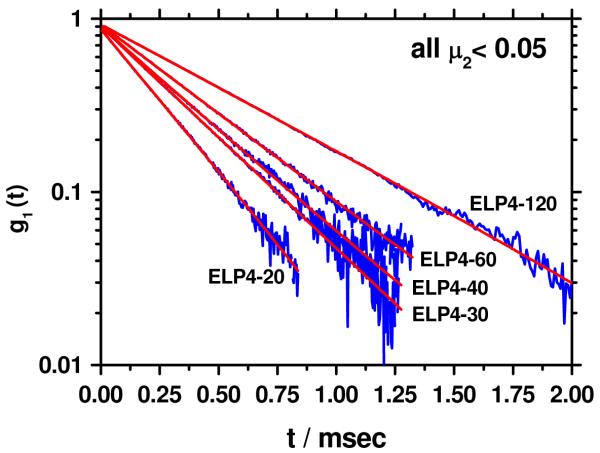
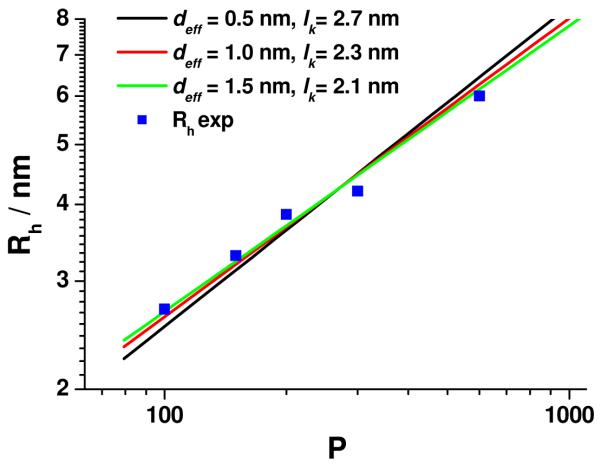
The correlation functions for all ELP samples decayed single exponentially for g1(t) > 0.2 (Figure 2). If a cumulant fit is applied, the normalized 2nd cumulant μ2, a measure for the polydispersity of the sample, is negligible, i.e. μ2< 0.05, which is within experimental error the expected range for monodisperse samples. In Figure 3 the double logarithmic plot of the hydrodynamic radius, Rh, of the ELPs versus the number of repeat units, P, is shown. A quantitative analysis of the data was performed by application of the Kratky-Porod wormlike chain model for the calculation of the hydrodynamic radius, as described in detail elsewhere.17 Briefly, the segment distribution function derived by Koyama18 was applied to numerically calculate Rh. The results are in perfect agreement with the Yamakawa-Fujii theory19,20 for wormlike cylinders.
Figure 2.
Correlation function g1 (t) of ELP4-20, ELP4-30, ELP4-40, ELP4-60 and ELP4-120 plotted vs. the correlation time t in a semi logarithmic scale. The red curves represent a single exponential fit to the data in a time regime where g1(t) > 0.2.
Figure 3.
Double logarithmic plot of the hydrodynamic radius Rh vs. the number of peptide repeat units P. The lines represent the best fits to the data assuming a repeat unit length b = 0.365 nm. Black line: deff = 0.5 nm, lk = 2.7 nm, red line: deff = 1.0 nm, lk = 2.3 nm, green line: deff = 1.5 nm, lk = 2.1 nm.
The Kuhn statistical length lk is obtained by fitting the calculated Rh to the measured Rh values. The parameters needed for the fit are the length per repeat unit b and the effective chain cross-section deff. The calculated fits were obtained by assuming b = 0.365 nm, which was estimated from the well-known peptide bond lengths and angles21 and by assuming different values for the effective chain cross-section deff of 0.5 nm, 1.0 nm and 1.5 nm, which are impossible to exactly calculate for non rigid particles. Given the different side chain size of the amino acids in ELPs, the chosen d values include the extremes of a very small hydrodynamic effect of the side chain (deff = 0.5 nm) and a maximum effect reflecting the maximum extension of the largest amino acid side chain (Valine). The data are fitted best by deff = 1.5 nm and lk = 2.1 nm, although the experimental data would also be compatible with deff = 1 nm and lk = 2.3 nm (see separate plots and linear presentation of the data in the Supporting Information). This somewhat large value of deff could be either due to an additional hydration shell or could be caused by thicker chain regions originating from intramolecular superstructures like hair pins. The latter is unlikely as revealed by a recent simulation study combined with NMR investigations which have highlighted the role of proline for the conformational change of ELP with temperature.22 At T = 283 °K no evidence for intramolecular interactions are found whereas at T = 303 °K βII-type turns may emerge. The data in the present work were recorded at T = 293 °K, where ELP most probably exhibits an “unstructured” conformation.
It should be noted that lk = 2.1 nm is much larger than the previously reported lk ≤ 0.4 nm derived from force-distance curve analysis in AFM measurements of single ELP molecules15. This discrepancy is not entirely unexpected, as AFM has been reported to yield extremely small Kuhn lengths that are physically anomalous.16
Conclusion
The present work has investigated the chain stiffness of ELP at temperatures well below the UCST based on the interpretation of the hydrodynamic radii by the Yamakawa-Fujii theory of wormlike chains. The value for the Kuhn statistical segment length, lk = 2.1 nm, compares well with those reported for other water soluble flexible polymers such as PEO23 (lk = 2 nm), single stranded RNA24 (lk = 2 nm) or other coiled polypeptides like poly-sodium-L-glutamat25 (lk = 1.2 nm) or denatured bovine A1 basic protein26 (lk = 3.1 nm). As discussed above the contour length utilized in the present analysis ignores possible secondary structures of ELP such as helices, hair pins etc. Since such effects would shorten the contour length, leading to an increase of the Kuhn length, and accordingly, the value reported herein constitutes a minimum value of the Kuhn length of ELPs.
Unfortunately, the present results do not provide a better understanding of the LCST behavior of ELP as function of temperature or pH nor is the presented wormlike chain analysis applicable to folded or even compact rigid ELP conformations which are expected to form close to the LCST transition. Although hydrodynamic theories are well developed for instance for rigid and flexible rods27,28 it would be difficult to consistently derive meaningful values for the contour length and the thickness, given the huge number of possible rigid conformations.
Supplementary Material
Acknowledgement
This work was supported by the DFG Priority Programm SPP 1259 (M.S.), the Graduate School of Excellence MAINZ (M.S.), and by the National Institutes of Health through grant # GM61232 (A.C.).
Footnotes
Supporting Information Available: Synthesis of ELP, expression and purification of ELP, SDS-Page analysis, light scattering characterization and various plots of the data shown in Fig. 3. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Debelle L, Tamburro AM. Int. J. Biochem. Cell Biol. 1999;31:261–272. doi: 10.1016/s1357-2725(98)00098-3. [DOI] [PubMed] [Google Scholar]
- (2).Meyer DE, Chilkoti A. Biomacromolecules. 2004;5:846–851. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- (3).MacKay JA, Chen MN, McDaniel JR, Liu WG, Simnick AJ, Chilkoti A. Nat. Mater. 2009;8:993–999. doi: 10.1038/nmat2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Chow D, Nunalee ML, Lim DW, Simnick AJ, Chilkoti A. Mater. Sci. Eng., R. 2008;62:125–155. doi: 10.1016/j.mser.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mackay JA, Chilkoti A. Int. J. Hyperth. 2008;24:483–495. doi: 10.1080/02656730802149570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).McDaniel JR, MacKay JA, Quiroz FG, Chilkoti A. Biomacromolecules. 2010;11:944–952. doi: 10.1021/bm901387t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Meyer DE, Chilkoti A. Biomacromolecules. 2002;3:357–367. doi: 10.1021/bm015630n. [DOI] [PubMed] [Google Scholar]
- (8).Dandu R, Von Cresce A, Briber R, Dowell P, Cappello J, Ghandehari H. Polymer. 2009;50:366–374. [Google Scholar]
- (9).Urry DW, Parker TM. J. Muscle Res. Cell Motil. 2002;23:543–559. [PubMed] [Google Scholar]
- (10).Tamburro AM. Nanomedicine. 2009;4:469–487. doi: 10.2217/nnm.09.18. [DOI] [PubMed] [Google Scholar]
- (11).Li B, Alonso DOV, Daggett V. J. Mol. Biol. 2001;305:581–592. doi: 10.1006/jmbi.2000.4306. [DOI] [PubMed] [Google Scholar]
- (12).Li B, Daggett V. J. Muscle Res. Cell Motil. 2002;23:561–573. doi: 10.1023/a:1023474909980. [DOI] [PubMed] [Google Scholar]
- (13).Li B, Alonso DOV, Bennion BJ, Daggett V. J. Am. Chem. Soc. 2001;123:11991–11998. doi: 10.1021/ja010363e. [DOI] [PubMed] [Google Scholar]
- (14).Tamburro AM, Guantieri V, Pandolfo L, Scopa A. Biopolymers. 1990;29:855–870. doi: 10.1002/bip.360290419. [DOI] [PubMed] [Google Scholar]
- (15).Valiaev A, Lim DW, Schmidler S, Clark RL, Chilkoti A, Zauscher S. J. Am. Chem. Soc. 2008;130:10939–10946. doi: 10.1021/ja800502h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Janshoff A, Neitzert M, Oberdorfer Y, Fuchs H. Angew. Chem., Int. Ed. 2000;39:3213–3237. doi: 10.1002/1521-3773(20000915)39:18<3212::aid-anie3212>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- (17).Schmidt M. Macromolecules. 1984;17:553–560. [Google Scholar]
- (18).Koyama R. J. Phys. Soc. Jpn. 1973;34:1029–1038. [Google Scholar]
- (19).Yamakawa H, Fujii M. Macromolecules. 1973;6:407–415. [Google Scholar]
- (20).Yamakawa H, Fujii M. Macromolecules. 1974;7:649–654. doi: 10.1021/ma60041a020. [DOI] [PubMed] [Google Scholar]
- (21).Voet D, Voet JG. Biochemie. VCH; Weinheim: 1994. 1. korr. Nachdr. der 1. Aufl.. ed. [Google Scholar]
- (22).Glaves R, Baer M, Schreiner E, Stoll R, Marx D. ChemPhysChem. 2008;9:2759–2765. doi: 10.1002/cphc.200800474. [DOI] [PubMed] [Google Scholar]
- (23).Kawaguchi S, Imai G, Suzuki J, Miyahara A, Kitano T. Polymer. 1997;38:2885–2891. [Google Scholar]
- (24).Caliskan G, Hyeon C, Perez-Salas U, Briber RM, Woodson SA, Thirumalai D. Phys. Rev. Lett. 2005;95 doi: 10.1103/PhysRevLett.95.268303. [DOI] [PubMed] [Google Scholar]
- (25).Shimizu S, Muroga Y, Hyono T, Kurita K. J. Appl. Crystallogr. 2007;40:S553–S557. [Google Scholar]
- (26).Krigbaum WR, Hsu TS. Biochemistry. 1975;14:2542–2546. doi: 10.1021/bi00682a038. [DOI] [PubMed] [Google Scholar]
- (27).Adamczyk Z, Sadlej K, Wajnryb E, Ekiel-Jezewska ML, Warszynski P. J. Colloid Interface Sci. 2010;347:192–201. doi: 10.1016/j.jcis.2010.03.066. [DOI] [PubMed] [Google Scholar]
- (28).Mansfield ML, Douglas JF. Macromolecules. 2008;41:5412–5421. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.