Abstract
Purpose
Though C-C chemokine ligand 2 (CCL2) has been demonstrated to play a pivotal role in prostate cancer tumorigenesis and invasion, the role of inherited variation in the CCL2 gene in prostate cancer progression and metastases remains unanswered. This study is aimed to determine the influence of CCL2 germline variants on prostate cancer aggressiveness.
Experimental Design
We performed an association study between six single nucleotide polymorphisms (SNPs) in the CCL2 gene and prostate cancer clinicopathologic variables in a large hospital based Caucasian patient cohort (N =4073).
Results
Genetic variantion at CCL2 is associated with markers of disease aggressiveness. Three SNPs, each in strong linkage disequilibrium, are associated with a higher (>7) biopsy Gleason score: CCL2-1811 A/G, −2835A/C and +3726 T/C (P =0.01, 0.03 and 0.04 respectively). The CCL2 −1811 G allele is addionally associated with advanced pathologic stages in patients who underwent radical prostatectomy (P = 0.04). In haplotype analysis, we found that the frequency of a common haplotype, H5, was higher among patients with D’Amico good risk features (Ppermutation = 0.04).
Conclusions
These results support the influence of CCL2 variants on prostate cancer development and progression.
Keywords: Prostate cancer, CCL2, Single-nucleotide Polymorphisms
Introduction
Chemokines and chemokine receptors are major mediators of leukocyte trafficking into the sites of the immune response. They participate in defense against microbial infection, in Th1/Th2 polarization of the immune response, allograft rejection and angiogenesis as well as in tumorigenesis and metastasis (1, 2). C-C chemokine ligand 2 (CCL2), also known as monocyte chemoattractant protein-1 (MCP-1), is a member of the C-C beta chemokine family that is produced by macrophages, fibroblasts, and endothelial cells to stimulate chemotaxis of monocyte/macrophages and other inflammatory cells through its receptor, CCR2 (2,3). CCL2 and its receptor CCR2 have recently been shown to play key roles in promoting tumorigenesis and metastasis via distinct mechanisms (4–7). First, CCL2 has a direct promotional effect on tumor cell growth and survival. Second, CCL2 has a modulatory effect on the tumor microenvironment by promoting macrophage mobilization and infiltration into the tumor bed. Third, CCL2 can promote osteoclast maturation in the bone tumor microenvironment. Fourth, CCL2 can suppress cytotoxic T lymphocytes. The multiple roles of CCL2 in the promotion of tumorigenesis make the CCL2/CCR2 axis an attractive therapeutic target for cancer treatment.
Single-nucleotide polymorphisms (SNPs) analysis of CCL2 has suggested that this chemokine may play a role in host susceptibility to the development of cancer and/or cancer metastasis (8–10). Seven CCL2 polymorphisms have been studied in their relationship to disease susceptibility or severity (11). Five of them are in the promoter regulatory region of the CCL2 gene: −927 G/C, −1811 A/G, −2136 A/T, −2518 A/G, −2835 C/A; one in the first intron: +764 C/G and one in the 3’ flanking region: +3726 T/C.
Four of the above mentioned SNPs (−2136 T, −2518 G, −2835 A, +764 G) in the CCL2 gene were found to be associated with increased circulating levels of CCL2 protein, but any true association may be due to a single variant in the region, since these SNPs are in strong linkage disequilibrium (LD) (12).
Prostate cancer is the most commonly diagnosed malignancy in men in the United States (13). Although metastatic prostate cancer is initially treatable by castration, advanced prostate cancer remains incurable owing to the inevitable emergence of androgen-independent cells (14). So far, only a few inherited genetic variants have been associated with aggressiveness or response to treatment of prostate cancer (15–17). CCL2 may play a role in prostate cancer tumorigenesis and invasion and is highly expressed in the tumor microenvironment by human bone marrow endothelial cells. However, the role of CCL2 genetic polymorphisms in prostate cancer progression and metastases remains an open question. To date only one association study has evaluated the CCL2 −2518G/A polymorphism and prostate cancer risk and no association was observed in 296 cases and 311 controls (8).
We present an association study on CCL2 polymorphisms and prostate cancer aggressiveness in a large hospital-based Caucasian patient cohort. Our data suggests the influence of CCL2 germline variants on prostate aggressiveness.
Materials and Methods
Study population
Details of the studied Dana-Farber Harvard Cancer Center SPORE (Gelb Center) Prostate Cancer cohort have been previously described (18, 19). Briefly, the study cohort contains a total of 4073 prostate cancer patients diagnosed between 1976 and 2007, who had been consented during 1993 to 2007 to provide information, tissue and blood samples for research purposes. To control the quality of the ethnicity information from the self-reported data, we sampled three percent of self-reported Caucasian (n =180) and performed genotyping using 26 SNPs which can distinguish Caucasian population from Non-Caucasian populations (20); the genotyping data showed that none of the tested samples were in discordance. This confirmed the reliability of self-reported Caucasian ethnicity. For all individuals who ambiguously reported their ethnicity, such as reported as “American”, or who do not have the ethnicity information, their Caucasian identity was determined by genotyping using the same set of 26 SNPs. Only reliably self-reported or SNP confirmed Caucasians were eligible for this study. Age at diagnosis was calculated from the date of the first positive biopsy. Using the D’Amico risk classification criteria, prostate cancer patients were identified as at low, intermediate or high risk of clinical recurrence after primary therapy (21). Since the original D’Amico risk classification was set to predict biochemical outcome of localized patients, in this study, patients diagnosed with N1 or M1 diseases were regarded as high D’Amico risk class. Within the entire cohort, 1716 out of 4073 patients received radical prostatectomy (RP) as the primary treatment. Pathological Gleason scores and pathological stages of RP specimen were acquired by reviewing pathology reports.
Selection of SNPs
Six SNPs in the CCL2 gene were selected for our study because of their previous associations seen in other disease types (11, 12). Three SNPs are located in the distal regulatory region of the CCL2 gene: −2835 C/A (rs2857654), −2139 A/T (rs1024610), −1811 A/G (rs3760399); one in promoter region, −927 G/C (rs3760396); one in the first intron, +764 C/G (rs2857657) and one in the 3’ flanking region: +3726 T/C (rs2530797). The LD within the CCL2 gene locus is very strong and all r2 values between selected SNPs except −1811 A/G are greater than 0.80. These 6 SNPs can cover most genetic variants information of studied region (Figure 1).
Figure 1. SNPs in the CCL2 gene locus and their linkage disequilibrium status.
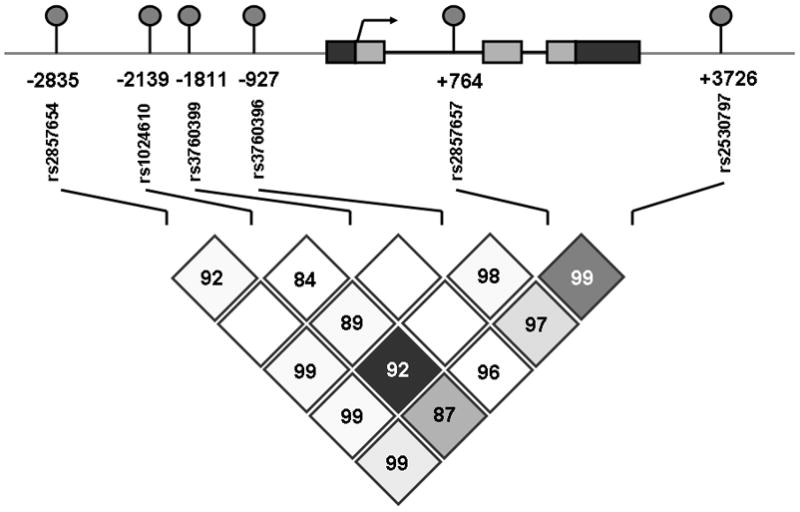
Upper part: diagram of CCL2 gene locus and selected SNPs. Light gray boxes, coding region; black boxes, untranslated regions; arrow, transcription start site; round cycles, selected SNPs. Lower part: linkage disequilibrium status between selected SNPs in our Caucasian patients cohort generated by using Haploview 4.1. Number in diamonds, r2 values between two indicated SNPs.
DNA, SNPs and Genotyping assays
All DNA samples were extracted from peripheral whole blood using QIAamp DNA Blood mini kit (QIAGEN Inc, Valencia, CA). Genotyping was performed with Sequenom iPLEX matrix-assisted laser desorption/ionization-time of flight mass spectrometry technology. For quality control, about 5% random selected duplicates were included. No discrepancy between duplicates was observed in the genotyping data of all 6 SNPs. All SNPs had greater than 99% genotype passing rates.
Statistical methods
We analyzed each SNP as a categorical variable with a common homozygote, a rare homozygote, and a heterozygote. Observed genotype distributions were tested for departure from Hardy-Weinberg equilibrium using Pearson’s goodness-of-fit test. No SNP violated Hardy-Weinberg equilibrium (all P value > 0.10).
To investigate the association between genotypes and biopsy Gleason grade at the time of diagnosis, we estimate Odds Ratios (ORs) and their 95% confidence intervals (CIs) using unconditional logistic regression. In a sub-cohort of patients who received RP, we also examined the association between CCL2 SNPs and RP pathologic stages with unconditional logistic regression. These analyses were adjusted for age at diagnosis. Cochran-Armitage test for trend was exploited to assess for the presence of genotype dose effect. Prostate cancer aggressiveness at diagnosis was categorized according to D’Amico risk classes (low, intermediate or high risk) with criteria described previously. Haplotypes were constructed using an accelerated EM algorithm similar to the partition/ligation method (22). Haplotype frequencies in low D’Amico risk group and intermediate or high group were estimated separately. Two-sided chi-square tests were performed to determine the association between each haplotype and risk of having intermediate or high D’ Amico risk class prostate cancer at the time of diagnosis with the low risk group as reference for comparison. One thousand permutation tests were exploited to correct multiple testing bias in association analysis of haplotypes and D’ Amico risk class.
Unconditional logistic regression tests were performed using SAS version 9.1 (SAS Institute Inc, Cary, NC) and P < 0.05 (two-sided) was considered statistically significant. The haplotype association analyses were done by using Haploview 4.1 (23). A P < 0.05 (two-sided) was considered statistically significant.
Results
Subject characteristics
Selected clinical characteristics of study participants are described previously (19). Briefly, the cohort contains 4073 patients and all participants are Caucasian. The mean age at diagnosis is 61.3 years (range: 42 to 91 years). The biopsy Gleason core was <7 in 1771 (47%), of 7 in 1272 (34%) patients and >7 in 707 (19%) patients. Among patients who had sufficient information for modified D’Amico risk classification; 1004 (30%) patients were low-risk, 1357 (40%) patients were intermediate-risk and 986 (30%) patients high-risk. One thousand seven hundred and sixteen patients underwent RP; and among them 1161 (68%) men had organ-confined (T1 or T2) disease at the time of surgery, while 475 (28%) men had extraprostatic tumor (T3 or T4) and 80 (4%) men had metastatic tumor (N1 or M1).
Correlation of CCL2 SNPs with Biopsy Gleason score
We first estimated associations between CCL2 SNPs and biopsy Gleason score (Table 1). The −2835 AA genotype had an OR of 1.42 (95% CI, 1.04–1.94) of having tumor biopsy Gleason score >7 compared to Gleason score <7. Similarly, the −1811 AG or GG genotype had an OR of 1.47 (95% CI, 1.08–2.01) for having a biopsy Gleason score >7 prostate cancer at the time of the diagnosis compared with the AA genotype. The +3726 TT genotype also had an OR of 1.33 (1.01–1.75) of having biopsy Gleason >7 tumor compared with CC genotype. Since these three SNPs are strongly correlated, the observed associations could due to one causal SNP rather than three independent results. We did not observe any statistically association when comparing genotypes of cases biopsy Gleason score of 7 with Gleason score <7.
Table 1.
Genotype frequencies, odds ratios and 95% CI comparing biopsy Gleason grade at diagnosis < 7, 7 and > 7
| SNPs | N (%) | OR (95% CI) * | |||||||
|---|---|---|---|---|---|---|---|---|---|
| < 7 | 7 | > 7 | 7 vs. < 7 | P value§ | > 7 vs. < 7 | P value§ | |||
| −2835 C/A (rs2857654) | |||||||||
| CC | 945 (53.4) | 683 (53.7) | 345 (48.9) | ref | 0.37 | ref | 0.02 | ||
| AC | 687 (38.8) | 511 (40.2) | 288 (40.9) | 1.01 | (0.87–1.18) | 1.12 | (0.93–1.36) | ||
| AA | 139 (7.8) | 78 (6.1) | 72 (10.2) | 0.77 | (0.57–1.04) | 1.42 | (1.04–1.94) | ||
| Total | 1771 (100.0) | 1272 (100.0) | 705 (100.0) | ||||||
| −2139 A/T (rs1024610) | |||||||||
| AA | 1096 (62.8) | 769 (61.0) | 461 (65.7) | ref | 0.41 | ref | 0.15 | ||
| AT | 573 (32.8) | 438 (34.7) | 216 (30.8) | 1.10 | (0.94–1.28) | 0.91 | (0.75–1.11) | ||
| TT | 76 (4.4) | 54 (4.3) | 25 (3.5) | 1.03 | (0.72–1.48) | 0.80 | (0.50–1.28) | ||
| Total | 1745 (100.0) | 1261 (100.0) | 702 (100.0) | ||||||
| −1811 A/G (rs3760399) | |||||||||
| AA | 1645 (93.2) | 1172 (92.6) | 636 (90.2) | ref | 0.51 | ref | 0.01 | ||
| AG+GG | 120 (6.8) | 94 (7.4) | 69 (9.8) | 1.10 | (0.83–1.46) | 1.47 | (1.08–2.01) | ||
| Total | 1765 (100.0) | 1266 (100.0) | 705 (100.0) | ||||||
| −927 G/C (rs3760396) | |||||||||
| GG | 1158 (65.6) | 791 (62.3) | 452 (64.1) | ref | 0.09 | ref | 0.46 | ||
| CG | 538 (30.5) | 426 (33.5) | 223 (31.6) | 1.16 | (0.99–1.36) | 1.06 | (0.88–1.29) | ||
| CC | 69 (3.9) | 53 (4.2) | 30 (4.3) | 1.12 | (0.77–1.61) | 1.10 | (0.70–1.71) | ||
| Total | 1765 (100.0) | 1270 (100.0) | 705 (100.0) | ||||||
| +764 C/G (rs2857657) | |||||||||
| CC | 1063 (63.5) | 745 (62.0) | 453 (66.3) | ref | 0.47 | ref | 0.20 | ||
| CG | 547 (32.7) | 411 (34.2) | 207 (30.3) | 1.08 | (0.92–1.27) | 0.89 | (0.74–1.09) | ||
| GG | 64 (3.8) | 46 (3.8) | 23 (3.4) | 1.05 | (0.71–1.55) | 0.84 | (0.51–1.37) | ||
| Total | 1674 (100.0) | 1202 (100.0) | 683 (100.0) | ||||||
| +3726 T/C (rs2530797) | |||||||||
| CC | 274 (15.8) | 199 (16.0) | 94 (13.5) | ref | 0.93 | ref | 0.01 | ||
| CT | 833 (48.1) | 595 (47.9) | 310 (44.4) | 0.97 | (0.78–1.20) | 1.06 | (0.81–1.39) | ||
| TT | 625 (36.1) | 448 (36.1) | 294 (42.1) | 0.96 | (0.77–1.20) | 1.33 | (1.01–1.75) | ||
| Total | 1732 (100.0) | 1242 (100.0) | 698 (100.0) | ||||||
Adjusted by age at diagnosis.
Cochran-Armitage test for trend.
Correlation of SNPs with Pathologic stages
In the patients who underwent RP, we classified men as either having evidence of extraprostatic (T3/T4) or metastatic (N1 or M1) disease or localized disease (T1/T2) at prostatectomy. When analyzing the association of CCL2 genotypes and pathologic stages in RP patients (Table 2), we only found that the −1811 AG or GG genotype was significantly correlated with the development of extraprostatic or metastatic prostate cancer (OR, 1.50; 95% CI, 1.03–2.18, P = 0.04), compared with AA genotype. All other five investigated genotypes were not found to be associated with pathologic stages in RP patients.
Table 2.
Genotype frequencies, odds ratios and 95% CI comparing Pathologic Stage in RP patients
| SNPs | N (%) | OR (95%CI) * | P value§ | ||
|---|---|---|---|---|---|
|
T1 or T2 |
T3 or T4 or N1 or M1 |
||||
| −2835 C/A (rs2857654) | |||||
| CC | 622 (53.6) | 291 (52.4) | ref | 0.59 | |
| AC | 458 (39.4) | 222 (40.0) | 1.02 | (0.82–1.26) | |
| AA | 81 (7.0) | 42 (7.6) | 1.12 | (0.75–1.67) | |
| Total | 1161 (100.0) | 555 (100.0) | |||
| −2139 A/T (rs1024610) | |||||
| AA | 705 (61.5) | 320 (58.3) | ref | 0.28 | |
| AT | 389 (33.9) | 203 (37.0) | 1.14 | (0.91–1.42) | |
| TT | 53 (4.6) | 26 (4.7) | 1.06 | (0.65–1.75) | |
| Total | 1147 (100.0) | 549 (100.0) | |||
| −1811 A/G (rs3760399) | |||||
| AA | 1080 (93.5) | 503 (90.8) | ref | 0.04 | |
| AG+GG | 75 (6.5) | 51 (9.2) | 1.50 | (1.03–2.18) | |
| Total | 1155 (100.0) | 554 (100.0) | |||
| −927 G/C (rs3760396) | |||||
| GG | 729 (63.1) | 369 (66.6) | ref | 0.26 | |
| CG | 380 (32.9) | 161 (29.1) | 0.82 | (0.65–1.03) | |
| CC | 47 (4.0) | 24 (4.3) | 1.05 | (0.62–1.79) | |
| Total | 1156 (100.0) | 554 (100.0) | |||
| +764 C/G (rs2857657) | |||||
| CC | 696 (63.1) | 321 (60.7) | ref | 0.49 | |
| CG | 358 (32.5) | 187 (35.3) | 1.12 | (0.89–1.40) | |
| GG | 48 (4.4) | 21 (4.0) | 0.92 | (0.54–1.58) | |
| Total | 1102 (100.0) | 529 (100.0) | |||
| +3726 T/C (rs2530797) | |||||
| CC | 184 (16.1) | 78 (14.2) | ref | 0.61 | |
| CT | 541 (47.5) | 271(49.5) | 1.14 | (0.84–1.55) | |
| TT | 415 (36.4) | 199 (36.3) | 1.07 | (0.78–1.48) | |
| Total | 1140 (100.0) | 548 (100.0) | |||
Adjusted by the age at diagnosis.
Cochran-Armitage test for trend.
Correlation of Haplotypes with D’Amico risk classification
D’Amico risk classifications system uses integrated clinical information to estimate the prostate cancer aggressiveness. Since the analysis on the association of individual CCL2 SNP with D’Amico risk classifications was null (data not shown), to more fully understand the extent of CCL2 genetic variation effect on prostate cancer aggressiveness, we performed a haplotype-based association analysis of D’Amico risk classifications. We found that the CCL2 gene was encompassed in one haplotype block in our participants (Figure 1). Six CCL2 SNPs delineate 7 common haplotypes (H1 through H7) that accounted for 98% of all haployptes in our studied Caucasian patients (Table 3). Haplotypes were then constructed in patients having low D’Amico risk and intermediate or high D’Amico risk class tumors, respectively. One major CCL2 haplotype (H5, CAAGCT), which can only defined by all six SNPs, was significantly associated with a reduced risk of having more aggressive prostate cancer at the time of diagnosis when compared all other haplotypes (P =0.01). This result indicated that interaction between CCL2 genetic variants may exist. The frequency of H5 was higher in low D’Amico risk group (10.9 %) than in intermediate or high risk group (8.3%). The H5 carriers had a reduced risk of having intermediate or high risk prostate cancer when compared with non-carriers (OR =0.80, 95% CI 0.66–0.96). This result remains significant after 1000 permutation tests (adjusted P =0.04). No other haplotype was significantly associated with D’Amico risk classification.
Table 3.
Frequency Distributions of Constructed Haplotypes with D’ Amico Risk Classifications
| Haplotypes* | All subjects | Low Risk | Intermediate or High Risk | P Value† | P value§ | |
|---|---|---|---|---|---|---|
| H1 | AAAGCT | 0.271 | 0.275 | 0.271 | 0.68 | 0.99 |
| H2 | CAACCT | 0.196 | 0.185 | 0.200 | 0.18 | 0.67 |
| H3 | CTAGGC | 0.189 | 0.195 | 0.190 | 0.64 | 0.98 |
| H4 | CAAGCC | 0.186 | 0.188 | 0.181 | 0.52 | 0.96 |
| H5 | CAAGCT | 0.089 | 0.102 | 0.083 | 0.01Ψ | 0.04 |
| H6 | CAGGCT | 0.039 | 0.032 | 0.042 | 0.05 | 0.35 |
| H7 | CAAGGC | 0.012 | 0.008 | 0.015 | 0.03 | 0.18 |
| Overall | 0.04 |
The SNPs order is the same as given if Figure 1.
Two-sided chi-square test, each haplotype compared with all other haplotypes.
After 1000 permutation test.
OR for carriers versus non-carries, 0.80 (95% CI, 0.66–0.96).
Discussion
Chemokines and their receptors have been detected in most tumors (1, 2). CCL2 remains one of the best studied chemokines. It has been demonstrated that CCL2 may play a role in prostate cancer tumorigenesis and metastasis (4, 5). CCL2 is not only involved in inflammatory responses, but also may stimulate prostate cancer cell chemoattraction, proliferation and survival (6, 7). Over-expression of CCL2 in human prostate cancer cells significantly increased local tumor burden in vivo (24, 25). Other in vivo data further demonstrated that CCL2-neutralizing antibodies effectively inhibit prostate cancer growth (26). SNPs located in the regulatory region of the gene may increase or decrease transcriptional activity and polymorphisms affecting the gene encoding proteins may affect the function of encoded protein, potentially leading to the association with disease susceptibility or severity (15, 27). Therefore, it is biologically plausible that genetic polymorphisms that increase CCL2 expression may be associated with prostate cancer aggressiveness, reflected by clinicopathologic traits such as biopsy Gleason score, tumor grade or an integrated risk estimator such as D’Amico classification. In the current study, we correlated genetic variations of the CCL2 gene to three clinical traits in a large hospital based prostate cancer patient cohort.
We found that SNP −1811A/G, which occurs in the promoter region of CCL2 gene, was associated with higher biopsy Gleason grade (>7) at diagnosis in all patients, and also associated with advanced pathologic stage in RP patients. Another promoter region polymorphism, −2835 C/A, and a downstream, SNP +3726T/C, are associated with higher biopsy Gleason grade. Because of the tight LD between these three SNPs, it is very difficult to separate their influence in our genetic association study.
Variation at the CCL2 promoter has been reported to affect transcriptional binding sites, consequently, influencing expression (12, 28–30). Binding of Interferon Regulatory Factor-1 (IRF-1) and/or Prep1/Pbx 2 transcription factor complex is influenced by theCCL2-2518 A/G polymorphism (28, 31). Another functional study showed that the −927 C SNP he −927 C SNP is associated with PARP-1 and ARNT binding, and the −362 G is associated with a STAT binding site. CCL2 −2518, −927 and −362 polymorphisms are associated with altered transcriptional activity in vitro (32, 33). Intriguingly, both of the two promoter SNPs which are associated with prostate cancer aggressiveness in current study, CCL2 −1811 A/G and CCL2 −2835 C/A, could disrupt a potential transcription factor Sp1 binding site respectively, predicted by the web software TRANSFAC 4.0 (34), though the hypothesis remains to be validated.
Another possibility is that the observed association between CCL2 polymorphisms and CCL2 aggressiveness is due to linkage with another as-yet unknown polymorphisms. For example, the SNP rs4430796, which located in the same chromosome region as the CCL2 gene, 17q12, was correlated to prostate cancer risk and aggressiveness recently (35–38). However, there is more than 3.5M bps distance between rs4430796 and CCL2 gene locus. More comprehensive resequencing and functional studies are needed to explore functional sites.
To better evaluate the influence of CCL2 genetic variants on prostate cancer aggressiveness, we augmented our analyses with haplotypes and D’Amico risk classification to examine more comprehensively. The haplotype frequency distributions in our participants are in consistent with previous heart study in a population of European ancestry (12). It showed that one common haplotype H5 was associated with D’Amico risk classification. Since H5 is defined by all six studied SNPs, only interaction of all six SNPs together identifies H5 and its correlation with D’Amico risk. The association of H5 remained significant after 1000 times permutation tests.
In conclusion, our results indicate that inherited variants in the CCL2 gene are associated with prostate cancer aggressiveness in Caucasian patients. This finding also provides evidence that CCL2 may be involved in prostate cancer tumorigenesis and metastasis. Additional follow-up of our sample, further validation in other larger set of prostate cancer samples and functional study will help to clarify the role of CCL2 genotype in prostate cancer aggressiveness.
Translational Relevance.
The chemokine CCL2 may play a pivotal role in prostate cancer tumorigenesis and invasion. Our study analyzes the role of six common polymorphisms in the CCL2 gene, as well as the haplotypes they composed, on the risk of developing aggressive prostate cancer. We found that two promoter region SNPs, −1811A/G and −2835 A/C, a 3’ UTR SNP, +3726 T/C, and a common haplotype are associated with more aggressive prostate cancer. These findings support the relevance of CCL2 in the development of aggressive prostate cancer.
Acknowledgments
This work was supported by a SPORE in Prostate Cancer 2 P50 CA090381-06, SPORE in prostate Cancer 2 P50 CA69568-06, the Prostate Cancer Foundation and Department of Defense (DoD) Prostate Cancer Training Award W81XWH-09-1-0372.
Abbreviations
- RP
Radical Prostatectomy
- SNPs
Single Nucleotide Polymorphisms
- ORs
Odds Ratios
- CIs
Confidence Intervals
- LD
Linkage Disequilibrium
Footnotes
Competing interest statement: The authors declare no conflict of interests.
Referrences
- 1.Hippe A, Homey B, Mueller-Homey A. Chemokines. Recent Results Cancer Res. 2010;180:35–50. doi: 10.1007/978-3-540-78281-0_4. [DOI] [PubMed] [Google Scholar]
- 2.Lazennec G, Richmond A. Chemokines and chemokine receptors: new insights into cancer-related inflammation. Trends Mol Med. 2010;16:133–44. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–26. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conti I, Rollins BJ. CCL2 (monocyte chemoattractant protein-1) and cancer. Semin Cancer Biol. 2004;14:149–54. doi: 10.1016/j.semcancer.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Craig MJ, Loberg RD. CCL2 (Monocyte Chemoattractant Protein-1) in cancer bone metastases. Cancer Metastasis Rev. 2006;25:611–9. doi: 10.1007/s10555-006-9027-x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Lu Y, Pienta KJ. Multiple roles of chemokine (C-C motif) ligand 2 in promoting prostate cancer growth. J Natl Cancer Inst. 2010;102:522–8. doi: 10.1093/jnci/djq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Patel L, Pienta KJ. CC chemokine ligand 2 (CCL2) promotes prostate cancer tumorigenesis and metastasis. Cytokine Growth Factor Rev. 2010;21:41–8. doi: 10.1016/j.cytogfr.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sáenz-López P, Carretero R, Cózar JM, et al. Genetic polymorphisms of RANTES, IL1-A, MCP-1 and TNF-A genes in patients with prostate cancer. BMC Cancer. 2008;8:382. doi: 10.1186/1471-2407-8-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tse KP, Tsang NM, Chen KD, et al. MCP-1 Promoter Polymorphism at 2518 is associated with metastasis of nasopharyngeal carcinoma after treatment. Clin Cancer Res. 2007;13:6320–6. doi: 10.1158/1078-0432.CCR-07-1029. [DOI] [PubMed] [Google Scholar]
- 10.Ghilardi G, Biondi ML, La Torre A, Battaglioli L, Scorza R. Breast cancer progression and host polymorphisms in the chemokine system: role of the macrophage chemoattractant protein-1 (MCP-1) -2518 G allele. Clin Chem. 2005;51:452–5. doi: 10.1373/clinchem.2004.041657. [DOI] [PubMed] [Google Scholar]
- 11.Navratilova Z. Polymorphisms in CCL2&CCL5 chemokines/chemokine receptors genes and their association with diseases. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150:191–204. doi: 10.5507/bp.2006.028. [DOI] [PubMed] [Google Scholar]
- 12.McDermott DH, Yang Q, Kathiresan S, et al. CCL2 polymorphisms are associated with serum monocyte chemoattractant protein-1 levels and myocardial infarction in the Framingham Heart Study. Circulation. 2005;112:1113–20. doi: 10.1161/CIRCULATIONAHA.105.543579. [DOI] [PubMed] [Google Scholar]
- 13.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 14.Singer EA, Golijanin DJ, Miyamoto H, Messing EM. Androgen deprivation therapy for prostate cancer. Expert Opin Pharmacother. 2008;9:211–28. doi: 10.1517/14656566.9.2.211. [DOI] [PubMed] [Google Scholar]
- 15.Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross RW, Oh WK, Xie W, et al. Inherited variation in the androgen pathway is associated with the efficacy of androgen-deprivation therapy in men with prostate cancer. J Clin Oncol. 2008;26:842–7. doi: 10.1200/JCO.2007.13.6804. [DOI] [PubMed] [Google Scholar]
- 17.Kader AK, Sun J, Isaacs SD, et al. Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5,895 prostate cancer patients. Prostate. 2009;69:1195–205. doi: 10.1002/pros.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh WK, Hayes J, Evan C, et al. Development of an integrated prostate cancer research information system. Clin Genitourin Cancer. 2006;5:61–6. doi: 10.3816/CGC.2006.n.019. [DOI] [PubMed] [Google Scholar]
- 19.Sun T, Lee GS, Werner L, et al. Inherited Variations in AR, ESR1 and ESR2 Genes Are Not Associated With Prostate Cancer Aggressiveness or With Efficacy of Androgen Deprivation Therapy. Cancer Epidemiol Biomarkers Prev. 2010;19:1871–8. doi: 10.1158/1055-9965.EPI-10-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–74. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 22.Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71:1242–7. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Mizutani K, Sud S, McGregor NA, et al. The chemokine CCL2 increases prostate tumor growth and bone metastasis through macrophage and osteoclast recruitment. Neoplasia. 2009;11:1235–42. doi: 10.1593/neo.09988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loberg RD, Ying C, Craig M, Yan L, Snyder LA, Pienta KJ. CCL2 as an important mediator of prostate cancer growth in vivo through the regulation of macrophage infiltration. Neoplasia. 2007;9:556–62. doi: 10.1593/neo.07307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loberg RD, Ying C, Craig M, et al. Targeting CCL2 with systemic delivery of neutralizing antibodies induces prostate cancer tumor regression in vivo. Cancer Res. 2007;67:9417–24. doi: 10.1158/0008-5472.CAN-07-1286. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y. DNA variations in human and medical genetics: 25 years of my experience. J Hum Genet. 2009;54:1–8. doi: 10.1038/jhg.2008.6. [DOI] [PubMed] [Google Scholar]
- 28.Joven J, Coll B, Tous M, et al. The influence of HIV infection on the correlation between plasma concentrations of monocyte chemoattractant protein-1 and carotid atherosclerosis. Clin Chim Acta. 2006;368:114–9. doi: 10.1016/j.cca.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 29.Mühlbauer M, Bosserhoff AK, Hartmann A, et al. A novel MCP-1 gene polymorphism is associated with hepatic MCP-1 expression and severity of HCV-related liver disease. Gastroenterology. 2003;125:1085–93. doi: 10.1016/s0016-5085(03)01213-7. [DOI] [PubMed] [Google Scholar]
- 30.Brown KS, Nackos E, Morthala S, Jensen LE, Whitehead AS, Von Feldt JM. Monocyte chemoattractant protein-1: plasma concentrations and A(−2518)G promoter polymorphism of its gene in systemic lupus erythematosus. J Rheumatol. 2007;34:740–6. [PubMed] [Google Scholar]
- 31.Wright EK, Jr, Page SH, Barber SA, Clements JE. Prep1/Pbx2 complexes regulate CCL2 expression through the −2578 guanine polymorphism. Genes Immun. 2008;9:419–30. doi: 10.1038/gene.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thye T, Nejentsev S, Intemann CD, et al. MCP-1 promoter variant −362C associated with protection from pulmonary tuberculosis in Ghana, West Africa. Hum Mol Genet. 2009;18:381–8. doi: 10.1093/hmg/ddn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nyquist P, Zhang J, De Graba TJ. The −928 G/C and −362 G/C single-nucleotide polymorphisms in the promoter of MCP-1: Increased transcriptional activity and novel binding sites. Cerebrovasc Dis. 2010;29:242–7. doi: 10.1159/000267849. [DOI] [PubMed] [Google Scholar]
- 34.Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Münch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–8. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 36.Helfand BT, Loeb S, Meeks JJ, Fought AJ, Kan D, Catalona WJ. Pathological outcomes associated with the 17q prostate cancer risk variants. J Urol. 2009;181:2502–7. doi: 10.1016/j.juro.2009.01.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levin AM, Machiela MJ, Zuhlke KA, Ray AM, Cooney KA, Douglas JA. Chromosome 17q12 variants contribute to risk of early-onset prostate cancer. Cancer Res. 2008;68:6492–5. doi: 10.1158/0008-5472.CAN-08-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao BY, Pao JB, Lin VC, et al. Individual and cumulative association of prostate cancer susceptibility variants with clinicopathologic characteristics of the disease. Clin Chim Acta. 2010;411:1232–1237. doi: 10.1016/j.cca.2010.04.028. [DOI] [PubMed] [Google Scholar]


