1. DISEASE CHARACTERISTICS
1.1 Name of the disease (synonyms)
Wolf–Hirschhorn syndrome (4p- syndrome, monosomy 4p. Includes Pitt–Rogers–Danks syndrome).
1.2 OMIM# of the disease
194190.
1.3 Name of the analysed genes or DNA/chromosome segments
4p16.3.
1.4 OMIM# of the gene(s)
WHSC1 602952.
LETM1 604407.
1.5 Mutational spectrum
Not applicable.
1.6 Analytical methods
Not applicable.
1.7 Analytical validation
Not applicable.
1.8 Estimated frequency of the disease (incidence at birth (‘birth prevalence') or population prevalence)
1:50 000 births with a 2:1 female/male ratio.
1.9 If applicable, prevalence in the ethnic group of investigated person
Not applicable.
1.10 Diagnostic setting

Comment:
2. TEST CHARACTERISTICS
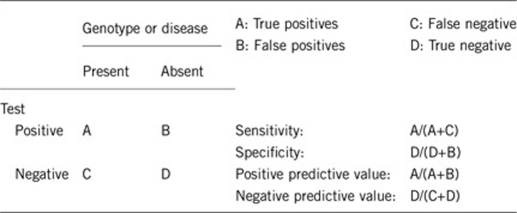
2.1 Analytical sensitivity (proportion of positive tests if the genotype is present)
The technology utilized for testing should be designed to detect deletion of the critical region for WHS, which includes at least portions of the LETM1 and WHSC1 genes. Therefore, either fluorescence in situ hybridization (FISH) with a probe covering this region or genomic microarray with coverage of this region should yield greater than 99% clinical sensitivity. Standard chromosome studies may not identify microdeletions of this region and would be predicated to have only a 50–60% clinical and analytical sensitivity. Also, as unbalanced translocations are frequently identified in this syndrome, genomic microarray or FISH with the subtelomeric probes should be considered to identify any concurrent duplications with the 4p deletion.
As the phenotype and genotype are interrelated for the diagnosis, then the analytical and clinical sensitivity and specificity should all be greater than 99% if the appropriate testing technology is utilized.
2.2 Analytical specificity (proportion of negative tests if the genotype is not present)
>99%.
2.3 Clinical sensitivity (proportion of positive tests if the disease is present)
The clinical sensitivity can be dependent on variable factors such as age or family history. In such cases a general statement should be given, even if a quantification can only be made case by case.
>99%.
2.4 Clinical specificity (proportion of negative tests if the disease is not present)
The clinical specificity can be dependent on variable factors such as age or family history. In such cases a general statement should be given, even if a quantification can only be made case by case.
>99%.
2.5 Positive clinical predictive value (lifetime risk to develop the disease if the test is positive)
Not applicable.
2.6 Negative clinical predictive value (probability of not developing the disease if the test is negative)
Assume an increased risk based on family history for a non-affected person. Allelic and locus heterogeneity may need to be considered.
Index case in that family had been tested:
Not applicable.
Index case in that family had not been tested:
Not applicable.
3. CLINICAL UTILITY
3.1 (Differential) diagnosis: The tested person is clinically ffected
(To be answered if in 1.10 ‘A' was marked)
3.1.1 Can a diagnosis be made other than through a genetic test?

3.1.2 Describe the burden of alternative diagnostic methods to the patient
Not applicable.
3.1.3 How is the cost effectiveness of alternative diagnostic methods to be judged?
Not applicable.
3.1.4 Will disease management be influenced by the result of a genetic test?

3.2 Predictive Setting: The tested person is clinically unaffected but carries an increased risk based on family history
(To be answered if in 1.10 ‘B' was marked)
Not applicable.
3.2.1 Will the result of a genetic test influence lifestyle and prevention?
If the test result is positive (please describe):
Not applicable.
If the test result is negative (please describe):
Not applicable.
3.2.2 Which options in view of lifestyle and prevention does a person at-risk have if no genetic test has been done (please describe)?
Not applicable.
3.3 Genetic risk assessment in family members of a diseased person
(To be answered if in 1.10 ‘C' was marked)
Risk to family members depends on the mechanism of origin of the deletion. If the deletion is a simple deletion and the parents are phenotypically normal, they are very unlikely to carry a deletion. If the deletion is part of an unbalanced translocation, then there is a significant risk (greater than 50%) that either parent would carry a balanced version of the translocation.
3.3.1 Does the result of a genetic test resolve the genetic situation in that family?
Yes, as it will likely determine the aetiology of the disease and the recurrence risk.
3.3.2 Can a genetic test in the index patient save genetic or other tests in family members?
It will likely result in more genetic testing within other family members if an unbalanced translocation is identified.
3.3.3 Does a positive genetic test result in the index patient enable a predictive test in a family member?
For certain types of genetic results (eg, unbalanced translocation) this would increase the likelihood of an abnormal genetic test in either parent
3.4 Prenatal diagnosis
(To be answered if in 1.10 ‘D' was marked)
Prenatal testing is available to families in which one parent is known to be a carrier of a chromosome rearrangement.
3.4.1 Does a positive genetic test result in the index patient enable a prenatal diagnostic?
Not necessarily, parental testing should be done first to determine recurrence risk.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21
4. IF APPLICABLE, FURTHER CONSEQUENCES OF TESTING
Please assume that the result of a genetic test has no immediate medical consequences. Is there any evidence that a genetic test is nevertheless useful for the patient or his/her relatives? (Please describe)
Acknowledgments
This work was supported by EuroGentest, an EU-FP6-supported NoE, contract number 512148 (EuroGentest Unit 3: ‘Clinical genetics, community genetics and public health', Workpackage 3.2).
References
- Battaglia A.Deletion 4p: Wolf-Hirschhorn syndromein Cassidy SB, Allanson JE (eds): Management of Genetic Syndromes Hoboken, NJ: Wiley-Liss and Sons Inc.2010249–261. [Google Scholar]
- Battaglia A, Carey JC. Update on the clinical features and natural history of Wolf Hirschhorn syndrome (WHS): experience with 48 cases. Am J Hum Genet. 2000;67:127. [Google Scholar]
- Battaglia A, Carey JC. Seizure and EEG patterns in Wolf-Hirschhorn (4p-) syndrome. Brain Dev. 2005;27:362–364. doi: 10.1016/j.braindev.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Carey JC, Cederholm P, Viskochil DH, Brothman AR, Galasso C. Natural history of Wolf-Hirschhorn syndrome: experience with 15 cases. Pediatrics. 1999;103:830–836. doi: 10.1542/peds.103.4.830. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Carey JC, Cederholm P, Viskochil DH, Brothman AR, Galasso C. Storia naturale della sindrome di Wolf-Hirschhorn: esperienza con 15 casi. Pediatrics. 1999;11:236–242. doi: 10.1542/peds.103.4.830. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Carey JC, Thompson JA, Filloux FM. EEG studies in the Wolf-Hirschhorn (4p-) syndrome. EEG Clin Neurophysiol. 1996;99:324. [Google Scholar]
- Battaglia A, Carey JC, Viskochil DH, Cederholm P, Opitz JM. Wolf-Hirschhorn syndrome (WHS): a history in pictures. Clin Dysmorphol. 2000;9:25–30. doi: 10.1097/00019605-200009010-00005. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Carey JC, Wright TJ. Wolf-Hirschhorn (4p-) syndrome. Adv Pediatr. 2001;48:75–113. [PubMed] [Google Scholar]
- Battaglia A, Filippi T, Carey JC. Update on the clinical features and natural history of Wolf-Hirschhorn (4p-) syndrome: experience with 87 patients and recommendations for routine health supervision. Am J Med Genet Part C Semin Med Genet. 2008;148C:246–251. doi: 10.1002/ajmg.c.30187. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Filippi T, South ST, Carey JC. Spectrum of epilepsy and EEG patterns in Wolf-Hirschhorn syndrome: experience with 87 patients. Dev Med Child Neurol. 2009;51:373–380. doi: 10.1111/j.1469-8749.2008.03233.x. [DOI] [PubMed] [Google Scholar]
- Fisch GS, Battaglia A, Parrini B, Youngblom J, Simensen R. Cognitive-behavioral features of children with Wolf-Hirschhorn syndrome: preliminary report of 12 cases. Am J Med Genet. 2008;148C:252–256. doi: 10.1002/ajmg.c.30185. [DOI] [PubMed] [Google Scholar]
- Giglio S, Calvari V, Gregato G, et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas NM, Van Buggenhout G, Hannes F, et al. Genotype-phenotype correlation in 21 patients with Wolf-Hirschhorn syndrome using high resolution array comparative genome hybridisation (CGH) J Med Genet. 2008;45:71–80. doi: 10.1136/jmg.2007.052910. [DOI] [PubMed] [Google Scholar]
- South ST, Bleyl SB, Carey JC. Two unique patients with novel microdeletions in 4p16.3 that exclude the WHS critical regions: implications for critical region designation. Am J Med Genet A. 2007;143A:2137–2142. doi: 10.1002/ajmg.a.31900. [DOI] [PubMed] [Google Scholar]
- South ST, Whitby H, Battaglia A, Carey JC, Brothman AR. Comprehensive analysis of Wolf-Hirschhorn syndrome using array CGH indicates a high prevalence of translocations. Eur J Hum Genet. 2008;16:45–52. doi: 10.1038/sj.ejhg.5201915. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Krause M, Majewski F, et al. Effect of the size of the deletion and clinical manifestation in Wolf-Hirschhorn syndrome: analysis of 13 patients with a de novo deletion. Eur J Hum Genet. 2000;8:519–526. doi: 10.1038/sj.ejhg.5200498. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Krause M, Majewski F, et al. Unexpected high frequency of de novo unbalanced translocations in patients with Wolf-Hirschhorn syndrome (WHS) J Med Genet. 2000;37:798–804. doi: 10.1136/jmg.37.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TJ, Ricke DO, Denison K, et al. A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum Mol Genet. 1997;6:317–324. doi: 10.1093/hmg/6.2.317. [DOI] [PubMed] [Google Scholar]
- Zollino M, Lecce R, Fischetto R, et al. Mapping the Wolf-Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am J Hum Genet. 2003;72:590–597. doi: 10.1086/367925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollino M, Lecce R, Selicorni A, et al. A double cryptic chromosome imbalance is an important factor to explain phenotypic variability in Wolf–Hirschhorn syndrome. Eur J Hum Genet. 2004;12:797–804. doi: 10.1038/sj.ejhg.5201203. [DOI] [PubMed] [Google Scholar]
- Zollino M, Murdolo M, Marangi G, et al. On the nosology and pathogenesis of Wolf-Hirschhorn syndrome: genotype-phenotype correlation analysis of 80 patients and literature review. Am J Med Genet Part C Semin Med Genet. 2008;148C:257–269. doi: 10.1002/ajmg.c.30190. [DOI] [PubMed] [Google Scholar]


