Abstract
The genomic architecture of the 10q22q23 region is characterised by two low-copy repeats (LCRs3 and 4), and deletions in this region appear to be rare. We report the clinical and molecular characterisation of eight novel deletions and six duplications within the 10q22.3q23.3 region. Five deletions and three duplications occur between LCRs3 and 4, whereas three deletions and three duplications have unique breakpoints. Most of the individuals with the LCR3–4 deletion had developmental delay, mainly affecting speech. In addition, macrocephaly, mild facial dysmorphisms, cerebellar anomalies, cardiac defects and congenital breast aplasia were observed. For congenital breast aplasia, the NRG3 gene, known to be involved in early mammary gland development in mice, is a putative candidate gene. For cardiac defects, BMPR1A and GRID1 are putative candidate genes because of their association with cardiac structure and function. Duplications between LCRs3 and 4 are associated with variable phenotypic penetrance. Probands had speech and/or motor delays and dysmorphisms including a broad forehead, deep-set eyes, upslanting palpebral fissures, a smooth philtrum and a thin upper lip. In conclusion, duplications between LCRs3 and 4 on 10q22.3q23.2 may lead to a distinct facial appearance and delays in speech and motor development. However, the phenotypic spectrum is broad, and duplications have also been found in healthy family members of a proband. Reciprocal deletions lead to speech and language delay, mild facial dysmorphisms and, in some individuals, to cerebellar, breast developmental and cardiac defects.
Keywords: 10q22.3q23.2, NRG3, BMPR1A, PTEN, GRID1, breast development
Introduction
Patients with submicroscopic aberration syndromes have traditionally been recognised by a specific combination of clinical features.1 When chromosomal banding techniques became available in the 1970s, the cytogenetic basis of many of these syndromes was revealed.2, 3, 4, 5 The development of subtelomeric fluorescent in situ hybridisation (FISH), targeting all telomeres in a single assay, led to a shift from the original ‘phenotype-first' approach to a ‘genotype-first' approach. In the absence of a recognisable phenotype, individuals were screened by FISH for novel submicroscopic chromosomal abnormalities. On the basis of similar genomic aberrations in various patients the clinical presentation was delineated, by a process designated ‘reverse phenotypics'.6
Using new molecular karyotyping techniques such as subtelomeric MLPA and microarray analysis, reverse phenotypics has proven to be successful, by the constantly increasing list of microdeletion/microduplication syndromes.7 Some of these have a recognisable phenotype, such as the 17q21.31 microdeletion syndrome.8, 9, 10 Others, such as deletions of chromosome band 1q21.1, 15q13.3 or 16p13.11, give rise to less consistent phenotypes.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 These aberrations show incomplete penetrance, as demonstrated by their presence in clinically unaffected relatives. Several of these aberrations are associated with an increased risk of neuropsychiatric disorders, such as schizophrenia and autism.15, 16, 22 Chromosomal anomalies, with an unpredictable and inconsistent phenotypic outcome, represent a difficult counselling situation for clinicians. Microarray analysis during pregnancy is now offered in several countries, and prenatal detection of these submicroscopic changes is a particularly challenging counselling situation.23 Therefore, the need for extensive information about phenotypic outcomes of such recurrent aberrations is essential.
Recurrent deletions of 10q22.3q23.2 have been characterised by cognitive and behavioural abnormalities.24 The 10q22.3q23.2 region is characterised by a complex set of low-copy repeats (LCRs), which can give rise to various genomic changes mediated by non-allelic homologous recombination (NAHR).25 Balciuniene et al24 described three probands with interstitial deletions in this region. The breakpoints in two of these mapped within these LCRs (LCR3 and LCR4) and inheritance was reported in one. This deletion segregated with a wide range of cognitive and behavioural phenotypes within a large family. The third proband had a complex non-contiguous rearrangement consisting of two deletions with breakpoints in a unique genomic sequence, with the most proximal breakpoint between LCR3 and LCR4 and a telomeric breakpoint ∼630 kb distal of LCR4. A considerable overlap of cognitive and behavioural phenotypes was noted among probands and affected family members. The presence of LCRs in this region suggests that this locus has an increased susceptibility to chromosomal rearrangements.24 In contrast to expectations based on the chromosomal architecture defined by LCR3 and LCR4, only six novel deletions and no duplications comprising this region have been reported.26 Of these, four were clinically assessed.
In this report, we describe the clinical and molecular characterisation of eight novel deletions and six duplications within the 10q22.3q23.3 region. Five deletions and three duplications occur between LCRs3 and 4, whereas three deletions and three duplications have unique breakpoints.
Methods
Patients and DNA samples
In this study, 14 individuals with an aberration in 10q22q23 were studied. DNA was isolated according to standard procedures. Except for individual 4, who had a 47,XYY karyotype, all others had normal karyotypes by G-banded chromosome studies. Clinical information was obtained from the respective physicians.
The submicroscopic aberrations were detected by array analysis using different array platforms.
All deletions were fine-mapped using high-resolution, custom-tiled oligonucleotide CGH arrays. These were designed, manufactured, processed and analysed by Roche NimbleGen Systems Inc. (Madison, WI, USA). Except for patient 3, all patients were analysed using arrays (design ID 2794) that contained 385 000 isothermal oligonucleotide probes finely tiled across the human chromosome 10 region from 44 to 92 Mb (human genome reference sequence version hg18). Data analysis included 371 000 ‘unique' probes providing an average coverage of one probe every 130 nucleotides. A probe was considered ‘unique' if it differed by at least five nucleotides from homologous hits retrieved by a Blastn search against the entire human genome. Less stringent criteria for ‘uniqueness' were applied for probes in LCR sequences, and probes showing only one nucleotide mismatch to paralogous sequences were included for the analysis. Patient 3 was analysed using a different design array (design ID 5621). The chromosome 10 region from 77 to 92 Mb (human genome reference sequence version hg18) was finely tiled with 68 291 probes, which yielded a mean coverage of one probe per 210 nucleotides. All probes on this array were ‘unique', on the basis of the aforementioned criteria. Each sample was subjected to one hybridisation experiment, except for patient 2 whose DNA was subjected to two independent hybridisation arrays. Test DNA samples were labelled with Cy3 and paired with a reference DNA sample labelled with Cy5. We used a human DNA reference sample (Promega) provided by Roche NimbleGen Systems, which is a pool of genomic DNA from six anonymous males. Probe hybridisation signals were expressed as the log2 ratio of signal intensities of a test sample versus signal intensities of a reference sample. Array data were analysed with the segMNT or DNACopy algorithms. More detailed description of probe design, array construction, DNA labelling, hybridisation and data analysis can be found on the Roche Nimblegen Systems webpage (http://www.nimblegen.com/products/lit/index.html).
In the case of copy number variations (CNVs) with breakpoints mapping within LCRs, the automated segmentation algorithm performed poorly. The breakpoints assigned by analyses of different averaging windows were inconsistent and fell within large genomic intervals up to 5 Mb in length. Instead, we visualised the data using our previously published colour binning method and compared the data of our patients with those from three previously obtained control samples.24 For patient 2, log2 ratios from two independent experiments were averaged, and the averaged values were used for colour binning analysis. Breakpoint genomic coordinates provided in the result section represent our best estimate, and are in reference to the hg18 version of the Human Genome assembly (http://genome.ucsc.edu/).
Results
Molecular findings
The breakpoints of the deletions and duplications are shown in Figures 1 and 2, respectively. In addition to the aberration in the 10q22q23 region, patient 3 had a de novo 722 kb gain in 2q36.3q36.3 (167.2–167.7 Mb), patient 4 a 47,XYY karyotype, patient 12 a de novo 1.5 Mb deletion at 16p13.11 (14.8–16.3 Mb) and patient 14 an inherited 2.2 Mb gain in 6q25.1 (149.4–151.6 Mb). All 10q22q23 and additional aberrations are summarised in supplementary Table A.
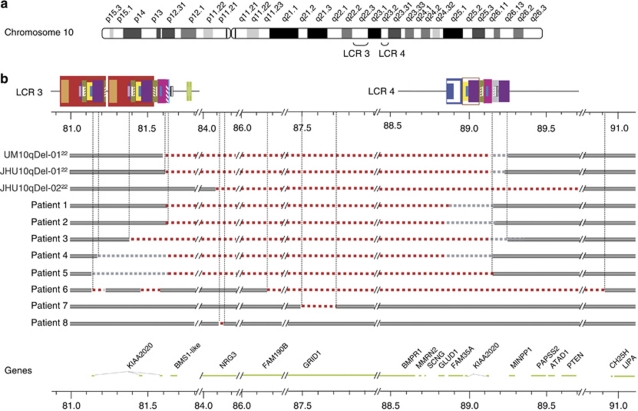
Figure 1.
Schematic representation of 10q genomic rearrangements identified in the probands. (a) Structure of LCRs located in 10q22.3-q23.2 is shown.24 Blocks of the same colour and/or pattern denote paralogous sequences. Degree of sequence identity between paralogous sequences ranges from 90.8 to 99.8%. Genomic position (in Mb) is shown on a scale below the LCRs, and corresponds to human reference human genome reference sequence version hg18. (b) Map of deletions predicted by oligonucleotide CGH arrays in patients 1–8 and of previously studied individuals JHU10qDel-01, JHU10qDel-02, UM10qDel-01.24 Grey bar indicates intact DNA sequence. Red dashed line indicates hemizygous deletion. Grey dashed line denotes the genomic area that contains a breakpoint. Genes located in this genomic region are shown below the diagram of the rearrangements.
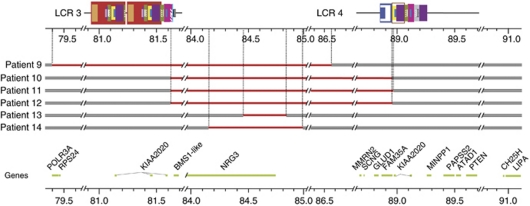
Figure 2.
Map of duplications predicted by oligonucleotide CGH arrays in patients 9–14. Grey bar indicates intact DNA sequence. Red solid line indicates duplication. Genes located in this genomic region are shown below the diagram of the rearrangements. Genomic position (in Mb) is shown on a scale below the LCRs, and corresponds to human reference human genome reference sequence version hg18.
Clinical data
The clinical findings of the patients are described in detail in Supplementary information. The clinical features of probands 1–5 with an LCR3–4 deletion are summarised in Table 1 and of probands 10–12 with an LCR3–4 duplication in Table 2. The latter table also includes clinical features of patient 9, who had a largely overlapping duplication with breakpoints located in unique sequences outside LCRs3 and 4. The clinical features of probands 6–8 and of probands 13–14 are displayed in Tables 3 and 4, respectively. Photographs of six patients (cases 1, 3, 4, 9, 10 and 11), whose parents consented for publication, are shown in Figure 3.
Table 1. Clinical features in literature cases and probands with a LCRs3–4 10q22.3q23.3 deletion.
| UM10qDel-0122 | JHU10qDel-0122 | Case 126 | Case 226 | Case 326 | Case 426 | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Deletion position (Mb) | 81.6–89.1/89.2 | 81.6–89.1/89.3 | 81.6–88.9 | 81.6–88.9 | 81.6–88.9 | 81.6–88.9 | 81.6–88.9/89.1 | 81.6–88.9/89.1 | 81.4–89.1/89.3 | 81.6–88.6/89.1 | 81.6−89.1 |
| LCR involvement | LCR3–LCR4 | LCR3–LCR4 | LCR3–LCR4 | LCR3–LCR4 | LCR3–LCR4 | LCR3–LCR4 | LCR3–LCR4 | LCR3–LCR4 | LCR3–LCR4 | LCR3–LCR4 | LCR3–LCR4 |
| Inheritance | Inherited mat | u | de novo | de novo | de novo | de novo | de novo | U | de novo | de novo | Inherited mat |
| Additional aberrations | Gain 2q36.3q36.3 | 47,XYY | |||||||||
| Developmental delay | + | + | + | Borderline | + | + | + | + | + | + | |
| Autism | + | − | + | − | − | − | − | ||||
| Speech delay | + | NA | + | + | + | − | + | + | + | + | |
| Hyperactivity | + | − | − | + | − | ||||||
| Seizures | − | − | − | − | + | − | − | ||||
| OFC | P97 | P97 at birth | >P97 | P10 | P3 | P10 | >P97 | P84 | |||
| Height | u | P10-P90 | P50 | P10 | <P3 | >P97 | P30 | ||||
| Cardiac evaluation | u | u | PDA | Normal | AVSD | Normal | Tricuspid regurgitation | Normal | |||
| Pulmonic regurgitation | |||||||||||
| Brain anomalies | u | Retrocerebellar cyst | u | − | u | u | Chiari I malformation | − | u | ||
| Small cerebellum | |||||||||||
| Wide ventricles | |||||||||||
| Dysmorphisms and congenital anomalies | Minor features | White forelock | Micrognathia Thin upper lip Mild hypertelorism Upsl palp fissures Earlobe creases Overfolding lateral pinna Arachnodactyly Joint hyperextensibility | Mild facial asymmetry Prognathia Downsl palp fissures Mild arachnodactyly Joint hyperextensibility | Hypertelorism Mild epicanthal folds Low-set ears Narrow ear canals Bilateral club feet Bilateral hearing loss | Frontal bossing Narrow philtrum Retrognathia Small mouth Downsl palp fissures Hypertelorism Small ears | Ptosis Low set small ears Hypotelorism Upsl palp fissures Broad thumbs Broad halluxes Breast aplasia | Telecanthus Low set ears Hypertelorism Anteverted nares Flat nasal bridge Large mouth | Dolichocephaly Low-set prominent ears Hypertelorism Epicanthal folds Flat nasal bridge Midface retraction High arched palate Maligned teeth Retroplaced thumbs | Long face Hypertelorism Radioulnar synostosis Scoliosis Kyphosis Pectus excavatum Café-au-lait spots | Low set ears Hypertelorism Broad base of nose Flat nasal bridge Shawl scrotum Club feet |
Abbreviations: AVSD, atrial ventricular septal defect; LCR, low-copy repeat; PDA, persistent ductus arteriosus; u, unknown; upsl/downsl palp fissures, upward/downslanting slanting palpebral fissures.
aBrain anomalies in the same person.
Table 2. Clinical features of four patients with a duplication of 10q22.3q23.2.
| Patient 9 | Sibpair patients 10–11 | Patient 12 | |
|---|---|---|---|
| Duplication position (Mb) | 79.4–86.6 | 81.6–89.0 | 81.6–89.0 |
| LCR involvement | No | LCR3–CR4 | LCR3–LCR4 |
| Inheritance | Unknown | Inherited mat | de novo |
| Additional aberrations | − | − | 16p13.1 del |
| Developmental delay | + | + | + |
| Speech delay | + | + | − |
| Seizures | − | − | |
| Birth weight | P50 | P20 | |
| Recurrent ear infections infancy | + | ||
| Current weight | P30 | P25 | |
| Current height | P50 | >P90 | |
| Current OFC | P50 | P25 | |
| Behavioural problems | Impaired social interaction | Impaired social interaction | |
| Deep-set eyes | + | + | + |
| Upslanting palpebral fissures | + | + | − |
| Hypotelorism | − | + | + |
| Low and prominent ears | + | − | − |
| Smooth philtrum | + | + | − |
| Thin upper lip | + | + | + |
| Full lower lip | + | − | + |
| Strabismus | − | + | − |
| Micrognathia | − | + | − |
| Full cheeks | + | − | + |
| Large wide spaced teeth | + | + | |
| Thin built | − | + | − |
Abbreviation: LCR, low-copy repeat.
Three patients with an LCRs3–4 duplication and one patient with a duplication in unique sequences, but sharing 5 Mb of overlap with other three duplications.
Table 3. Clinical features in probands with a 10q22.3q23.3 deletion, with breakpoints in unique sequences.
| Patient 6 | Patient 7 | Patient 8 | |
|---|---|---|---|
| Deletion position (Mb) | 86.2–90.9 | 87.5–87.7 | 84 118–84 148 |
| LCR involvement | None | None | None |
| Inheritance | de novo | u | Inherited pat |
| Additional aberrations | − | − | − |
| Developmental delay | − | − | + |
| Autism | − | − | + |
| Hyperactivity | + | − | − |
| Seizures | − | − | + |
| OFC | >P97 | P95 | P10 |
| Height | P70 | <P3 | P50 |
| Cardiac evaluation | u | AVSD | u |
| Brain anomalies | u | NA | u |
| Dysmorphisms and congenital anomalies | Upturned nose GI polyps Hypertelorism | Prominent forehead Downsl palp fissures Hypertelorism Anteverted nares Small low set ears Diaphragmatic eventration Undescended testes | − |
Abbreviations: downsl palp fissures, downslanting palpebral fissures; GI, gastrointestinal; LCR, low-copy repeat; u, unknown.
Table 4. Clinical features of probands with small sized 10q22.3q23.3 duplications with breakpoints in unique sequences.
| Patient 13 | Patient 14 | |
|---|---|---|
| Duplication position (Mb) | 84.5–84.8 | 84.2–85 |
| LCR involvement | No | No |
| Inheritance | Inherited pat | Inherited pat |
| Additional aberrations | − | Inh 6q25.1 dup |
| Developmental delay | − | − |
| Speech delay | + (Blindness related) | − |
| Seizures | − | − |
| Birth weight | P50 | <P3 |
| Current height | P10 | P75 |
| Current weight | P3 | >P97 |
| Deep-set eyes | + | − |
| Full lower lip | + | − |
| Strabismus | + | − |
| Anteverted nares | + | − |
| Full cheeks | + | − |
| Urogenital anomalies | − | + |
| Cardiac defect | − | + |
| Fusion of sacral vertebrae | − | + |
| Long slender ribs and clavicles | − | + |
Abbreviation: LCR, low-copy repeat.
On the basis of phenotype and inheritance pattern, these aberrations most likely represent non-pathogenic variants.
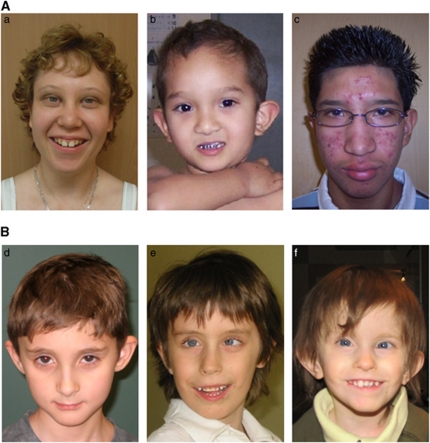
Figure 3.
(A) Photographs of probands 1, 3 and 4 with deletions between LCR3 and LCR4. Proband 1 (a) at 13 years of age; she has upward-slanting palpebral fissures, hypotelorism and low-set ears. Proband 3 (b) at 3 years and 7 months of age; he has dolichocephaly, low-set and prominent ears, hypertelorism, epicanthal folds and a flat midface. Proband 4 (c) at 12 years of age; he has hypertelorism, almond-shaped eyes, low-set ears and full lips. (B) Photographs of patients 9, 10 and 11 with duplications of 10q22.3q23.2. Patient 9 (d) at 9 years of age; he has a triangular face, a broad forehead, upslanting palpebral fissures, slightly deep-set eyes, lateral flaring of eyebrows, prominent ears with thickened horizontal superior helices, a thin upper lip and a smooth philtrum. Sibling patients 10 (e) and 11 (f) show upslanting palpebral fissures, strabismus, hypotelorism, a smooth philtrum and anteverted nares.
Discussion
In recent years, many recurrent rearrangements have been added to the increasing list of genomic disorders,27 including deletions in the 10q22.3q23.2 region.24 The breakpoints of this deletion are flanked by LCRs. LCR3 flanks the proximal breakpoint and harbours two large (>300 kb) highly homologous (99.8% identity) segmental duplications. They are composed of smaller modules with different orientations that are dispersed elsewhere on chromosome 10 and on other chromosomes. LCR4 flanks the distal breakpoint and contains ∼170 kb of sequence homologous to LCR3, as well as >100 kb of sequence homologous to LCRs located near the chromosome 10 centromere.24 On the basis of this genomic architecture, an increased susceptibility of chromosomal rearrangements between LCRs3 and 4 can be expected in this region. However, since the description of two deletions by Balciuniene et al,24 only six additional cases have been reported.26 The additional five new cases with the LCR3–4 deletion in the current study were detected within a cohort of 30 991 individuals with MR and/or multiple congenital anomalies, leading to a frequency of 0.016%. This is a much lower frequency than reported for other genomic disorders, such as Williams syndrome (0.31%),28 proximal 16p11.2 deletions (0.6%),29 17q21.31 deletions (0.64%)9 and 15q13.3 deletions (0.24%).21
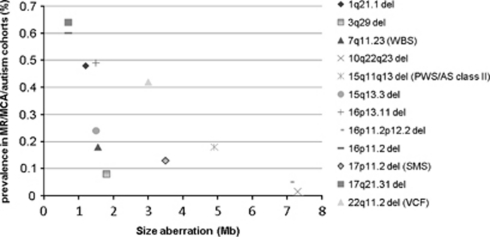
On the basis of genomic architecture, the lower frequency might be explained by the ∼7 Mb unique sequence between the LCRs in the 10q22.3q23.2 region, which is much larger than the distance between segmental duplications in other LCR-mediated recurrent rearrangements reported thus far. The distance between two LCRs is known to be one of the genomic architectural features that influences the efficiency of NAHR.30 In fact, on the basis of the 18 new genomic disorders described since 2005, recently reviewed by Mefford and Eichler,31 the distance between LCRs3 and 4 on 10q22.3q23.2 is the largest in size (Figure 4). Except for the 16p11.2p12.2 deletion, having a size of 7 Mb, all aberrations are <2.5 Mb. However, the latter study did not include previously described LCR-mediated syndromes delineated through a ‘phenotype-first approach'. Some of these also include intermediate-sized deletions, such as the Williams (MIM no. 19450), velocardiofacial (MIM no. 192430), Smith-Magenis (MIM no. 182290), Prader-Willi (MIM no. 176270) and Angelman (MIM no. 105830) syndromes (Figure 4).31, 32, 33, 34, 35, 36 In addition, it has been observed that larger-sized genomic rearrangements, using LCRs positioned farther apart, are regularly associated with larger LCRs.30 Although the aberration on 10q22.3q23.2 is largest in size, its LCRs are intermediate in size compared with LCRs in other genomic rearrangements.
Figure 4.
Frequency of LCR-mediated microdeletions in cohorts of individuals with MR, multiple congenital anomalies and/or autism (y axis) 9, 12, 20, 21, 29, 32, 74, 75 compared with the size of the unique sequence between LCRs (x axis).31, 32, 33, 34, 35, 36 WBS, Williams–Beuren Syndrome; PWS, Prader Willi Syndrome; AS, Angelman syndrome; SMS, Smith-Magenis syndrome; VCF, velocardiofacial syndrome. Frequencies of PW/AS and VCF syndromes were corrected for the most common deletion causing these syndromes.
The low frequency of 10q22.3q23.2 deletions among MR/MCA individuals may reflect lack of ascertainment because of the mild phenotype in some individuals with this deletion. Balciuniene et al24 reported multiple family members having this deletion, which segregated with a wide range of cognitive and behavioural phenotypes, including learning difficulties, speech and language delay, ADHD and autism. Individuals with a similar mild phenotype will often not be tested for submicroscopic aberrations. Similar phenotypic variability has been described for other rearrangements, such as on chromosomes 1q21.1, 15q13.3 and 16p13.11. All aberrations are enriched in affected persons compared with control individuals.12, 13, 14, 15, 16, 18, 19, 20, 21 A less likely explanation for the low frequency might be that 10q22.3q23.2 deletions in foetuses often lead to miscarriages. Although miscarriages were not reported in our families, the family reported by Balciuniene et al24 showed spontaneous abortions and a stillbirth.
In addition to the six clinically reported 10q22.3q23.2 cases, we present the clinical features of five novel patients with this deletion. Of these 11 deletions, seven had arisen de novo, two were inherited and for two inheritance remained unknown. Two patients had additional chromosome abnormalities. The phenotype of patient 2 might have been influenced by the additional de novo duplication of chromosome 2q36.3. However, this aberration has also been reported in a healthy father of an affected individual (DECIPHER database ID 248304), and several partially overlapping duplications in this region have been reported in the Database of Genomic Variants. The phenotype of patient 4 might have been influenced by the 47,XYY karyotype. In general, individuals with an XYY karyotype have lower IQ scores, mainly verbally, as expected for their social background, but not impaired in relation to general population norms. In addition, problems with anger control and also with attention have been described.37
Cognitive development was impaired in all 11 probands, varying from mild to moderate impairment. All patients had apparent speech and language problems. Three patients also had motor developmental delay, but their speech was more severely affected than their motor development.
Patients were mildly dysmorphic, and low-set ears, hypertelorism and a flat nasal bridge were frequently noted. Head circumference ranged from the 3rd centile to above the 97th centile, but the majority of patients were macrocephalic.
Behavioural problems were present in some, but were non-overlapping in type; two patients had autism,24, 26 two showed hyperactivity26 and patient 4 demonstrated aggressive behaviour. In the latter, this may have been due to his 47,XYY karyotype.
Two patients had cerebellar anomalies; low-lying cerebellar tonsils in patient 3 and a retrocerebellar cyst with a small cerebellum in patient JHU10qDel-01 were reported by Balciuniene et al.24
Patient 1 had unilateral congenital breast aplasia. Except for two patients, all other LCR3–LCR4 deletion probands were male. Patients 2 and 4 and case 1 of Alliman et al had a cardiac defect (AVSD, tricuspid and pulmonic regurgitation and persistent ductus arteriosus, respectively) and patient 3 had epilepsy. An AVSD was also noted in patient 7 and epilepsy in patient 8. However, these two latter patients had smaller intragenic deletions, located in unique sequences.
The breakpoints of the deletion in proband 6 were also located in unique sequences, including LCR4 and the PTEN gene. This patient had macrocephaly and juvenile colonic polyps, common features of contiguous gene deletions including tumour suppression genes BMPR1A and PTEN.38 Germline loss-of-function heterozygous mutations in PTEN have been described, causing an overlapping phenotype including macrocephaly and gastrointestinal polyps in patients with Cowden and Banayan–Riley–Ruvalcaba syndrome.39 Furthermore, heterozygous germline loss-of-function mutations in BMPR1A cause juvenile polyposis.40
Three currently reported patients (2, 4 and 7) had a cardiac defect, indicating that a dosage-sensitive gene or genes involved in the pathogenesis of cardiac defects may be located in the 10q22.3q23.2 region. There are two interesting candidate genes located in this region, BMPR1A and GRID1. BMPR1A is deleted in patients 3 and 4 and in all individuals who have been reported with overlapping deletions and cardiac septal defects.41, 42, 43, 44, 45 Of these deletions, at least one did not include the GRID1 gene.42 Cardiac-specific deletion of BMPR1A disrupts cardiac morphogenesis in mice, showing ventricular septum, trabeculae, compact myocardium and endocardial cushion defects.46
BMPR1A was not deleted in patient 7 with a cardiac defect, who had an intragenic deletion of GRID1, which is located only about 500 kb upstream of BMPR1A. The inheritance of this deletion was unknown, hampering conclusions on pathogenicity. Similar deletions have not been reported in the Database of Genomic Variants, nor in the chromosomal databases DECIPHER and ECARUCA. GRID1 encodes for a subunit of glutamate receptor channels, which have a role in mediating excitatory synaptic transmission in the central nervous system.47 The deletion in patient 7 removes exons 5–8 of the GRID1 gene. The removal of these exons would lead to an in-frame protein that would partly lack the extracellular ligand-binding domain, and therefore may represent a dominant-negative mutation. A recent meta-analysis of genome-wide association data, aimed to identify common genetic variants associated with cardiac structure and function, proposed GRID1 as a candidate gene for left ventricle wall thickness.48
Remarkably, this glutamatergic gene has also been associated with an entirely different pathology, namely schizophrenia.49, 50, 51, 52 The glutamatergic system is the major excitatory neurotransmitter system in the central nervous system,53 and changes in this system are the basis of the ‘glutamate hypothesis' aiming to explain symptoms of schizophrenia.54, 55 After finding linkage evidence for schizophrenia in 10q22 in different populations,56, 57, 58, 59GRID1 was implicated as being the causative gene in several association studies.49, 50, 51, 52
Another gene in the 10q22 region, associated with schizophrenia risk and presentation, is neuregulin 3 (NRG3).56 This gene is located 2.6 Mb proximal of GRID1 and affected in copy number variation in 12 patients in this study. Neuregulins are signalling proteins that mediate cell–cell interactions in the nervous system, heart, breast and other organ systems.60 Although several family members of proband 8 had a psychiatric disorder, the intragenic deletion found in the proband and his father did not segregate in the rest of the family. In addition, this intragenic deletion minimally overlaps a CNV (loss) described in two healthy individuals61 and in our patient 7; however, it extends beyond the CNV telomeric boundary. Therefore, the pathogenic significance of this CNV remains uncertain.
Taking into account that most probands in this study are still young, definitive evidence linking haploinsufficiency of NRG3 or GRID1 to psychiatric disease could not be established.
NRG3 is also a candidate gene for congenital breast aplasia, which was present in patient 1. NRG3 has been implicated in early mammary gland development, and Nrg3 signalling in mice promotes differentiation of mammary epithelial cells at the initial stages of organ formation. Mice harbouring a hypomorphic allele of Nrg3 exhibit defects in mammary gland formation, such as hypoplastic and supernumerary mammary gland formation.62, 63
NRG3 was also involved in two patients with an intragenic NRG3 duplication and congenital anomalies. Patient 13 had vision problems, most likely because of prematurity, whereas patient 14 had tetralogy of fallot, hypospadias and ambiguous genitalia. Both patients inherited the duplication from an unaffected parent. Although a possible role in organ development cannot be excluded, these duplications did not influence neurocognitive functioning and are most likely non-pathogenic variants.
Three patients (10–12) had a duplication between LCR3 and LCR4. Patient 9 had a duplication that did not involve LCR4, but shared a 5 Mb overlap with the three LCR3–4 duplications. Sibpair patients 10 and 11 inherited this duplication from their healthy mother. The duplication in patient 12 occurred de novo. This patient also had a de novo 16p13.11 deletion, which has been reported as a risk factor, strongly predisposing for, but not sufficient to cause, autism, epilepsy and mental retardation.13, 20 Duplications involving the 10q22-q23 region are rare. To date, only four cases overlapping this region have been reported.64, 65, 66, 67 Two of these patients had microcephaly and three had cardiac defects. Precise genotype–phenotype correlations are hampered because these aberrations have often been detected by conventional cytogenetic techniques. In addition, except for one case reported by Dufke et al,65 these duplications were much larger in size. Patients 9 and 12 displayed speech delay and impaired social interaction. Sibpair patients 10 and 11 were mentally retarded. Probands 9–11 and the patient described by Dufke et al had a strikingly similar facial appearance including a broad forehead, deep-set eyes, upslanting palpebral fissures, a smooth philtrum and a thin upper lip.
As could be noted in the healthy mother of sibpair patients 10 and 11, duplication of the LCR3–4 region does not always lead to an abnormal phenotype. Incomplete penetrance of chromosomal aberrations is a well-known phenomenon, which has been described for several abnormalities such as 1q21.1, 15q13.3, 16p13.11 and 22q11.2 aberrations,11, 12, 13, 14, 20, 21, 68, 69, 70, 71 with reciprocal duplications of the same interval often less penetrant.72, 73 To determine the full spectrum and penetrance of the LCR3–4 duplication, more information about probands carrying this duplication, and especially information about other carriers in their families, is needed.
In conclusion, duplications of the LCR3–LCR4 region on 10q22.3q23.2 are associated with variable phenotypic penetrance. In affected individuals, these aberrations may lead to a distinct facial appearance and developmental speech and/or motor function delays. Deletions between LCRs3 and 4 are clinically penetrant in all cases reported thus far. These deletions are associated with cognitive impairment, mainly affecting speech development, and cardiac defects. In addition, these deletions may also be associated with cerebellar and breast developmental defects.
Acknowledgments
We thank all the parents and children who participated in this study. This work was supported by a grant from the European commission: AnEUploidy project (LSHG-CT-2006–037627) under FP6 (BvB, HB, BBAdV), supplemental grants from The Netherlands Organisation for Health Research and Development (ZonMW 917–86–319 to BBAdV), Hersenstichting Nederland (BBAdV), Cure Autism Now (SBS), the Harrison Endowment and Autism Initiative Funds (JB, SBS).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Web Resources
Human Genome Browser: http://genome.ucsc.edu/cgi-bin/hgGateway (for March 2006 assembly)
DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources: https://decipher.sanger.ac.uk
Database of Genomic Variants: http://projects.tcag.ca/variation/?source=hg18
European Cytogeneticists Association Registered Unbalanced Chromosome Aberrations: www.ecaruca.net
Supplementary Material
References
- Schmickel RD. Contiguous gene syndromes: a component of recognizable syndromes. J Pediatr. 1986;109:231–241. doi: 10.1016/s0022-3476(86)80377-8. [DOI] [PubMed] [Google Scholar]
- Ewart AK, Morris CA, Atkinson D, et al. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- Driscoll DA, Budarf ML, Emanuel BS. A genetic etiology for DiGeorge syndrome: consistent deletions and microdeletions of 22q11. Am J Hum Genet. 1992;50:924–933. [PMC free article] [PubMed] [Google Scholar]
- Bowen P, Biederman B, Hoo JJ. The critical segment for the Langer-Giedion syndrome: 8q24.11----q24.12. Ann Genet. 1985;28:224–227. [PubMed] [Google Scholar]
- Ledbetter DH, Riccardi VM, Airhart SD, et al. Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N Engl J Med. 1981;304:325–329. doi: 10.1056/NEJM198102053040604. [DOI] [PubMed] [Google Scholar]
- de Vries BB, Winter R, Schinzel A, et al. Telomeres: a diagnosis at the end of the chromosomes. J Med Genet. 2003;40:385–398. doi: 10.1136/jmg.40.6.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek AM. Novel microdeletion syndromes detected by chromosome microarrays. Hum Genet. 2008;124:1–17. doi: 10.1007/s00439-008-0513-9. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C, Pittman AM, Willatt L, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Sharp AJ, Hurst JA, et al. Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet. 2008;45:710–720. doi: 10.1136/jmg.2008.058701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JL, O'Donovan MC, Gurling H, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, Lanpher B, German JR, et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46:382–388. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, Mullen S, Helbig I, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Hum Mol Genet. 2009;18:3625–3631. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannes FD, Sharp AJ, Mefford HC, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 2009;46:223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bon BW, Mefford HC, Menten B, et al. Further delineation of the 15q13 microdeletion and duplication syndromes: a clinical spectrum varying from non-pathogenic to a severe outcome. J Med Genet. 2009;8:511–523. doi: 10.1136/jmg.2008.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnamenta AT, Wing K, Akha ES, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009;5:687–692. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Veyver I, Patel A, Shaw CA, et al. Clinical use of array comparative genomic hybridization (aCGH) for prenatal diagnosis in 300 cases. Prenat Diagn. 2009;29:29–39. doi: 10.1002/pd.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciuniene J, Feng N, Iyadurai K, et al. Recurrent 10q22-q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am J Hum Genet. 2007;80:938–947. doi: 10.1086/513607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliman S, Coppinger J, Marcadier J, et al. Clinical and molecular characterization of individuals with recurrent genomic disorder at 10q22.3q23.2. Clin Genet. 2010;78:162–168. doi: 10.1111/j.1399-0004.2010.01373.x. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders ten years on. Genome Med. 2009;1:42. doi: 10.1186/gm42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson RE, Procopio-Allen AM, Schroer RJ, et al. Genetic syndromes among individuals with mental retardation. Am J Med Genet A. 2003;123A:29–32. doi: 10.1002/ajmg.a.20492. [DOI] [PubMed] [Google Scholar]
- Walters RG, Jacquemont S, Valsesia A, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Bejjani BA, Torchia B, et al. The identification of microdeletion syndromes and other chromosome abnormalities: cytogenetic methods of the past, new technologies for the future. Am J Med Genet C Semin Med Genet. 2007;145C:335–345. doi: 10.1002/ajmg.c.30152. [DOI] [PubMed] [Google Scholar]
- Bayes M, Magano LF, Rivera N, et al. Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Bi W, Lupski JR. Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet. 2002;71:1072–1081. doi: 10.1086/344346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makoff AJ, Flomen RH. Detailed analysis of 15q11-q14 sequence corrects errors and gaps in the public access sequence to fully reveal large segmental duplications at breakpoints for Prader-Willi, Angelman, and inv dup(15) syndromes. Genome Biol. 2007;8:R114. doi: 10.1186/gb-2007-8-6-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett V, Jacobs P, Nation K, et al. Neurocognitive outcomes of individuals with a sex chromosome trisomy: XXX, XYY, or XXY: a systematic review. Dev Med Child Neurol. 2010;52:119–129. doi: 10.1111/j.1469-8749.2009.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delnatte C, Sanlaville D, Mougenot JF, et al. Contiguous gene deletion within chromosome arm 10q is associated with juvenile polyposis of infancy, reflecting cooperation between the BMPR1A and PTEN tumor-suppressor genes. Am J Hum Genet. 2006;78:1066–1074. doi: 10.1086/504301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C. PTEN: one gene, many syndromes. Hum Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- Howe JR, Bair JL, Sayed MG, et al. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- Shapiro SD, Hansen KL, Pasztor LM, et al. Deletions of the long arm of chromosome 10. Am J Med Genet. 1985;20:181–196. doi: 10.1002/ajmg.1320200122. [DOI] [PubMed] [Google Scholar]
- Menko FH, Kneepkens CM, de LN, et al. Variable phenotypes associated with 10q23 microdeletions involving the PTEN and BMPR1A genes. Clin Genet. 2008;74:145–154. doi: 10.1111/j.1399-0004.2008.01026.x. [DOI] [PubMed] [Google Scholar]
- Salviati L, Patricelli M, Guariso G, et al. Deletion of PTEN and BMPR1A on chromosome 10q23 is not always associated with juvenile polyposis of infancy. Am J Hum Genet. 2006;79:593–596. doi: 10.1086/507151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigman AF, Lavine JE, Jones MC, et al. Localization of the Bannayan-Riley-Ruvalcaba syndrome gene to chromosome 10q23. Gastroenterology. 1997;113:1433–1437. doi: 10.1053/gast.1997.v113.pm9352843. [DOI] [PubMed] [Google Scholar]
- Sweet K, Willis J, Zhou XP, et al. Molecular classification of patients with unexplained hamartomatous and hyperplastic polyposis. JAMA. 2005;294:2465–2473. doi: 10.1001/jama.294.19.2465. [DOI] [PubMed] [Google Scholar]
- Gaussin V, Van de PT, Mishina Y, et al. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc Natl Acad Sci USA. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Araki K, Shibata A, et al. Molecular cloning of a cDNA encoding a novel member of the mouse glutamate receptor channel family. Biochem Biophys Res Commun. 1992;183:886–892. doi: 10.1016/0006-291x(92)90566-4. [DOI] [PubMed] [Google Scholar]
- Vasan RS, Glazer NL, Felix JF, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Muhleisen TW, Frank J, et al. Dissection of phenotype reveals possible association between schizophrenia and glutamate receptor delta 1 (GRID1) gene promoter. Schizophr Res. 2009;111:123–130. doi: 10.1016/j.schres.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Guo SZ, Huang K, Shi YY, et al. A case-control association study between the GRID1 gene and schizophrenia in the Chinese Northern Han population. Schizophr Res. 2007;93:385–390. doi: 10.1016/j.schres.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Kalbfleisch T, Brennan MD, et al. A microRNA gene is hosted in an intron of a schizophrenia-susceptibility gene. Schizophr Res. 2009;109:86–89. doi: 10.1016/j.schres.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Avramopoulos D, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan DT, Bridges RJ, Cotman CW. The excitatory amino acid receptors: their classes, pharmacology, and distinct properties in the function of the central nervous system. Annu Rev Pharmacol Toxicol. 1989;29:365–402. doi: 10.1146/annurev.pa.29.040189.002053. [DOI] [PubMed] [Google Scholar]
- Kim JS, Kornhuber HH, Schmid-Burgk W, et al. Low cerebrospinal fluid glutamate in schizophrenic patients and a new hypothesis on schizophrenia. Neurosci Lett. 1980;20:379–382. doi: 10.1016/0304-3940(80)90178-0. [DOI] [PubMed] [Google Scholar]
- Coyle JT. Glutamate and schizophrenia: beyond the dopamine hypothesis. Cell Mol Neurobiol. 2006;26:365–384. doi: 10.1007/s10571-006-9062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Avramopoulos D, Lasseter VK, et al. Fine mapping on chromosome 10q22-q23 implicates neuregulin 3 in schizophrenia. Am J Hum Genet. 2009;84:21–34. doi: 10.1016/j.ajhg.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Wolyniec PS, et al. Genomewide linkage scan for schizophrenia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 10q22. Am J Hum Genet. 2003;73:601–611. doi: 10.1086/378158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Hwu HG, Liu CM, et al. Genome scan of Han Chinese schizophrenia families from Taiwan: confirmation of linkage to 10q22.3. Am J Psychiatry. 2006;163:1760–1766. doi: 10.1176/ajp.2006.163.10.1760. [DOI] [PubMed] [Google Scholar]
- Liu J, Juo SH, Dewan A, et al. Evidence for a putative bipolar disorder locus on 2p13-16 and other potential loci on 4q31, 7q34, 8q13, 9q31, 10q21-24, 13q32, 14q21 and 17q11-12. Mol Psychiatry. 2003;8:333–342. doi: 10.1038/sj.mp.4001254. [DOI] [PubMed] [Google Scholar]
- Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- Shaikh TH, Gai X, Perin JC, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B, Panchal H, McCarthy A, et al. Identification of the scaramanga gene implicates neuregulin3 in mammary gland specification. Genes Dev. 2005;19:2078–2090. doi: 10.1101/gad.338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard BA. The role of NRG3 in mammary development. J Mammary Gland Biol Neoplasia. 2008;13:195–203. doi: 10.1007/s10911-008-9082-8. [DOI] [PubMed] [Google Scholar]
- Erdogan F, Belloso JM, Gabau E, et al. Fine mapping of a de novo interstitial 10q22-q23 duplication in a patient with congenital heart disease and microcephaly. Eur J Med Genet. 2008;51:81–86. doi: 10.1016/j.ejmg.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Dufke A, Singer S, Borell-Kost S, et al. De novo structural chromosomal imbalances: molecular cytogenetic characterization of partial trisomies. Cytogenet Genome Res. 2006;114:342–350. doi: 10.1159/000094224. [DOI] [PubMed] [Google Scholar]
- Han JY, Kim KH, Jun HJ, et al. Partial trisomy of chromosome 10(q22-q24) due to maternal insertional translocation (15;10) Am J Med Genet A. 2004;131:190–193. doi: 10.1002/ajmg.a.30307. [DOI] [PubMed] [Google Scholar]
- Goss PW, Voullaire L, Gardner RJ. Duplication 10q22.1-q25.1 due to intrachromosomal insertion: a second case. Ann Genet. 1998;41:161–163. [PubMed] [Google Scholar]
- Donald-McGinn DM, Kirschner R, Goldmuntz E, et al. The Philadelphia story: the 22q11.2 deletion: report on 250 patients. Genet Couns. 1999;10:11–24. [PubMed] [Google Scholar]
- Digilio MC, Marino B, Giannotti A, et al. Familial deletions of chromosome 22q11. Am J Med Genet. 1997;73:95–96. [PubMed] [Google Scholar]
- Ben-Shachar S, Ou Z, Shaw CA, et al. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008;82:214–221. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodningen OK, Prescott T, Eriksson AS, et al. 1.4Mb recurrent 22q11.2 distal deletion syndrome, two new cases expand the phenotype. Eur J Med Genet. 2008;51:646–650. doi: 10.1016/j.ejmg.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Berg JS, Brunetti-Pierri N, Peters SU, et al. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genet Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- Shinawi M, Liu P, Kang SH, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioral problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2009;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Robinson WP, Huang B, et al. Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients. Am J Hum Genet. 1995;57:40–48. [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Hornor SA, Jenkins E, et al. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.