Abstract
The spread of parasites is inherently a spatial process often embedded in physically complex landscapes. It is therefore not surprising that infectious disease researchers are increasingly taking a landscape genetics perspective to elucidate mechanisms underlying basic ecological processes driving infectious disease dynamics and to understand the linkage between spatially-dependent population processes and the geographic distribution of genetic variation within both hosts and parasites. The increasing availability of genetic information on hosts and parasites when coupled to their ecological interactions can lead to insights for predicting patterns of disease emergence, spread, and control. Here, we review research progress in this area based on four different motivations for the application of landscape genetics approaches: (1) assessing the spatial organization of genetic variation in parasites as a function of environmental variability, (2) using host population genetic structure as a means to parameterize ecological dynamics that indirectly influence parasite populations, e.g. gene flow and movement pathways across heterogeneous landscapes and the concurrent transport of infectious agents, (3) elucidating the temporal and spatial scales of disease processes, and (4) reconstructing and understanding infectious disease invasion. Throughout this review, we emphasise that landscape genetic principles are relevant to infection dynamics across a range of scales from within host dynamics to global geographic patterns and that they can also be applied to unconventional “landscapes” such as heterogeneous contact networks underlying the spread of human and livestock diseases. We conclude by discussing some general considerations and problems for inferring epidemiological processes from genetic data and try to identify possible future directions and applications for this rapidly expanding field.
Introduction
Population genetics can be a powerful tool for quantifying disease dynamics from within individuals to across large-scale geographic regions. Interactions among hosts and pathogens at the landscape scale are of particular interest since heterogeneous features of habitat and environment associated with different spatial scales affect many infectious disease patterns (Archie et al. 2009). While an increasing trend in addressing landscape level phenomena in epidemiology is apparent it is also important to recognize historical precedents for attending to environmental spatial heterogeneity in understanding disease dynamics. Researchers have long been interested in how environmental variables contribute to the emergence and distribution of parasites and infectious diseases (Ostfeld et al. 2005) and in a number of cases this research has also harnessed the power of molecular markers to assess spatial and temporal differentiation and change (Barrett et al. 2008; Nadler 1995). The aim of the current review is to illustrate how modern genetic analyses can contribute to our understanding of infectious disease processes in spatially complex environments and to review both novel applications and developments at the interface of landscape genetics and epidemiology. At the same time, we highlight previous work that predates the formal introduction of “landscape genetics” as a separate research area.
For some authors, a landscape genetics approach requires a spatially explicit statistical analysis of genetic data often restricted to a local to regional spatial scale (Manel et al. 2003). However, for the purpose of this review, we take a broader view, that includes spatially implicit studies as well phylogeographic approaches. In a sense, we are using ‘landscape’ as shorthand for heterogeneous space that can influence microevolutionary processes in parasite populations across scales, from within individual hosts to global species distributions. In essence, we are suggesting that “landscape genetics” be defined by a conceptual approach rather than a set of analytic tools. An approach that highlights the inevitable linkage between the spatial context of ecological dynamics and evolutionary process, which is revealed in genetic signatures that reflect the underlying forces shaping evolutionary and ecological trajectories. We believe this wider definition is useful because it allows us to consider a broader range of studies and analytical approaches and to also draw from the instructive examples coming from human and livestock diseases. While the spread of some these agents may not take place in natural landscapes, environmental heterogeneity often affects their genetic structure nonetheless, so that these cases can be conceptually informative and often provide superlative illustrations of the fundamental linkages between landscape ecology and population genetics. In addition, some of these definitional boundaries can become rather blurred depending on the infectious organisms in question. In the context of rapidly evolving parasites such as RNA viruses or bacteria for example, phylogeographic analyses can consider the same small spatial and temporal scales that are the focus of most landscape genetic studies (Holmes 2004; Real et al. 2005).
Apart from redefining what constitutes a ‘landscape’, there are a number of other considerations that are particularly or uniquely relevant when trying to apply landscape genetics principles to infectious organisms:
Because parasites ultimately rely on host resources, landscapes exert an organising influence on their populations in large part by affecting the distribution, accessibility and abundance of susceptible hosts. Environmental factors may often be the ultimate drivers of these patterns, in which case parallels can be drawn to non-parasitic species dependent on patchily distributed resources. However, environmental heterogeneity is not a necessary requirement for genetic discontinuities to emerge, as seen in sexually transmitted diseases where structure may result exclusively from varying host contact patterns.
As a consequence of the previous, infectious organisms (especially microparasites) have the potential to modify the landscape they are embedded in, for example by reducing the density of susceptible hosts through death or acquired immunity. Genetic signatures resulting from the interactions between epidemiological dynamics and physical environment can thus vary markedly and over short time periods, such as before and after an outbreak. Important from an analytical point of view, such rapid fluctuations can render the use of population genetic models that rely on stability assumptions (e.g. migration-drift equilibrium) inappropriate.
Landscape genetics focuses on contemporary microevolutionary processes but distinguishing these from historical effects can be difficult. As a general rule, parasites are characterised by relatively high rates of mutation, population growth and population turnover, traits that facilitate the emergence of novel population genetic structure (that may in fact often override any historic signatures). However, for studies focussing on more slowly evolving parasites, host or vector species, the question how population genetic patterns relate to contemporary epidemiological dynamics still arises commonly. Similarly, for parasites maintaining a high effective population size, genetic discontinuities may take considerably longer to become apparent due to the reduced effect of drift.
Many parasites have multiple means of dispersal, in particular if they have environmental reservoirs, broad host ranges or infect both wild and human-associated species. Due to multiple transmission processes potentially acting simultaneously in those cases, complex spatial genetic patterns may arise that cannot easily be related to environmental predictors and thus present considerable analytical challenges. At the same time, landscape genetic approaches may play a useful role in these situations by helping to discriminate between alternative epidemiological hypotheses or by quantifying the relative contribution of different ecological processes to overall pattern, effectively partitioning the contribution of different forces associated with landscape features or population processes.
These points will be reiterated and further illustrated throughout this review, which is loosely based on major research themes in landscape genetics identified by Storfer, et al. (2007), who also emphasise spatially-dependent ecological processes without reference to any particular scale.
We start by discussing the role of environmental factors in structuring the geographic distribution of genetic variation within parasite populations. We contrast this with studies that have tried to infer disease dynamics indirectly based on genetic population structure in the host. The focus on dynamical processes, as opposed to stable population genetic patterns, is developed further in the two subsequent sections, where we demonstrate how genetics can provide information about (1) the temporal and spatial scales of epidemiological processes and (2) how diseases invade and occupy new spatial locations both within and among hosts. We conclude by discussing some general considerations and current limitations of landscape genetic approaches with respect to infectious diseases and by pointing to new lines of research for which cross-fertilisation between the landscape ecology and population genetics of disease could be particularly stimulating and productive.
Environmental determinants of parasite genetic structure
Parasites differ greatly in the degree to which their populations are genetically partitioned, as might be expected given their wide range of life history patterns and modes of dispersal (Barrett et al. 2008; Criscione et al. 2005; Nadler 1995). Extremely fine-grained structure can be found if transmission takes place exclusively through local contact processes. Such patterns of local differentiation have been observed in, for example, soil-born pathogens (Campbell & Madden 1990) or hosts characteristically living within tight social groups (Nadler 1995) where local contact processes dominate transmission. At the other end of the extreme, wind-dispersed plant pathogens may undergo gene flow on continental and global scales (Brown & Hovmoller 2002). Regardless of the specific dispersal mechanism, spatial dissemination will often be modulated by local environmental conditions (Barrett et al. 2008), which can include a wide range of factors from particular landscape components and overall landscape structure to environmental gradients or physical patterns associated with wind and water flow.
For the large group of parasites that depend on their hosts or vectors for dispersal, genetic discontinuities should correlate with environmental features that have a strong influence on host or vector movement and population structure. Such a structuring influence may be overridden however, if alternative means of long-distance dispersal exist for example due to a more mobile intermediate or definitive host or human involvement (Criscione & Blouin 2004; Jancovich et al. 2005; Keeney et al. 2009). Mountain ranges and rivers are important landscape elements that have not only been implicated in slowing the spread of many wildlife diseases (see following sections) but also in limiting the geographic distribution of particular genotypes or strains.
Simian immunodefiency viruses (SIV) provide a good example of the effects of physical barriers in shaping the interaction between host movement and pathogen genetic structure. Major rivers appear to be strong dispersal barriers for many African primates including mandrill, gorillas and chimpanzees (Anthony et al. 2007; Gagneux et al. 2001; Telfer et al. 2003), species that have also been identified as natural hosts of specific strains of SIV either throughout or within part of their range. Consistent with host ecology, large rivers also correlate well with presence or absence of SIV within the same host species or with boundaries among distributions of different SIV sub-strains (Takehisa et al. 2009; Tsujimoto et al. 1988; Van Heuverswyn et al. 2007). These landscape genetic patterns thus help to better understand the complex history of host-parasite co-evolution in the case of SIV. Furthermore, they have implications for understanding zoonotic disease emergence by indicating where and when two chimpanzee-associated strains of SIV, one of which ultimately gave rise to the global HIV-1 group M pandemic, could have entered the human population (Keele et al. 2006). The fact that each of the two types is only found in chimpanzee communities inhabiting a relatively small catchment area pinpoints the geographic context for these host jumps with some confidence.
For the Zaire strain of Ebolavirus, another zoonotic pathogen circulating in Central Africa, the linkage between landscape barriers, host movement, and pathogen genetic structure may be useful in identifying candidate animal reservoirs of the virus. Both wildlife mortality patterns and genetic data suggest that for several years virus outbreaks were restricted to the west side of a major river, the Oguee, before flaring up just east of it (Lahm et al. 2007; Walsh et al. 2005). While such a barrier effect is consistent with transmission among terrestrial hosts, such as primates, it would be unexpected for volant species, such as fruit bats, which so far are considered the most likely natural reservoir (Leroy et al. 2005). Limited host dispersal ability also represents an important consideration when interpreting genetic data for pathogens that have emerged in amphibians in recent decades. Low variation and genetic homogeneity on regional and continental scales among isolates of iridovirus (Jancovich et al. 2005) and chythrid fungus (Goka et al. 2009; James et al. 2009) suggest high landscape permeability to pathogen transmission that is inconsistent with amphibian ecology but instead indicative of recent human-mediated spread.
The structure of a landscape, such as the size range and spatial distribution of habitat patches supporting host populations, can be another critical environmental determinant of parasite genetic structure. Again, the genetic pattern emerging for a particular parasite species will largely depend on how dispersal and overall population dynamics are affected by this type of spatial heterogeneity. A well-studied system in this regard is the anther smut, Microbotryum violaceum - a fungal plant pathogen infecting host populations with typically a patchy distribution. Fine-scale genetic analysis of the pathogen indicates that spatial aggregation within the host population structure can strongly affect the movement of the insects that vector fungal spores. Transmission by the vector occurs predominantly among neighbouring plants and appears to be very rare among different host populations irrespective of distance to the nearest patch (Giraud 2004).
Although air-borne pathogens are expected to show much less genetic structure than pathogens restricted by local contact or limited dispersal, geographic and temporal variation in prevailing wind patterns can have a profound effect on the distribution of these air-borne pathogens and the genetic composition of their populations. The first outbreak of bluetongue virus (BTV) in the United Kingdom in 2007, for example, is thought to be the consequence of highly unusual wind conditions promoting the transport of infected midge vectors across the North Sea from continental Europe. Meteorological records show that these conditions were, in fact, limited to a particular day, yielding a highly precise estimate for the virus' introduction (Gloster et al. 2008). The evidence for such a scenario was strengthened by the simultaneous detection of two virus genotypes, previously documented only from Holland and Denmark. The introduction of several plant pathogens into novel geographic regions has also often been associated with unique weather events that have promoted long-distance establishment followed by sustained propagation (Campbell & Madden). Examples include the transport of coffee rust (Hemileia vastatrix) from Angola to Brazil (Bowden et al. 1971), the movement of sugarcane rust (Puccinia melanocephela) into the Americas from Africa (Purdy et al. 1985), and the dispersal of aphids carrying maize dwarf mosaic virus pathogens of sweet corn into Minnesota from its origin in the southern Great Plains (Berger et al. 1987). As these and similar examples (Sellers & Maarouf 1991) illustrate, it is usually the deviation from normal weather conditions that has revealed their effect. Future work should examine whether meteorological data could be used more generally to produce predictive models for wind-dispersed pathogens, models that could then be validated using spatial-genetic data.
Many of the concepts and considerations we've discussed regarding spatial structure and its impact on parasite population genetics are equally relevant to infectious diseases maintained in humans or domestic species. Host population structure in these cases may be best described in terms of spatial networks with varying degrees of contact rather than physical landscapes. Yet by focusing on the identification of genetic discontinuity or connectivity and their underlying mechanisms, landscape genetics and molecular epidemiology clearly share conceptual common ground in many ways. Relevant research examples of human diseases in this context include efforts to genetically track the spread of viruses within and among local networks of high-risk groups (e.g. intravenous drug users) in order to identify heterogeneity in host contact structure (Lewis et al. 2008; Wylie et al. 2005). Similarly, transmission of certain pathogen lineages or strains has been compared to patterns of livestock trade and movement (Cottam et al. 2008; Smith et al. 2006).
Inferring disease dynamics from host genetic structure
In cases where disease transmission can unequivocally be attributed to a particular wild host or vector species, landscape genetic analyses are sometimes directed at these species rather than the disease agent itself. Such an approach can offer distinct advantages, for example, for infectious organisms exhibiting insufficient genetic variation. Prion diseases, such as chronic wasting disease (CWD), are illustrative of this scenario. Blanchong et al., (2008) tested whether potential dispersal barriers had affected gene flow and spread of CWD in white-tailed deer in Wisconsin and found increased genetic differentiation among populations separated from the core CWD-affected area by two types of landscape barrier, highways and rivers (Fig. 1). This type of information is critical from a disease management perspective because it can be used as the basis for developing local strategies for CWD surveillance and control (Blanchong et al. 2008).
Figure 1.
Chronic wasting disease (CWD) in white-tailed deer as an example of using host population genetics to identify landscape determinants of disease spread. (a) Prevalence of CWD in 15 study areas in Wisconsin and (b) genetic differentiation (FST) of deer host populations in study areas relative to core area of CWD infection, with study areas grouped based on the type of landscape feature separating them from the core-area. Reprinted from Blanchong et al. (2008) with permission.
Importantly, Blanchong et al. (2008) also found that higher genetic differentiation was associated with lower relative disease prevalence (Fig. 1), which establishes a link between deer dispersal and gene flow and the level of CWD spread. While such a linkage is commonly assumed, its strength should in fact be highly dependent on the manner in which parasite life history and host ecology interact. Contact patterns inferred from host genetics may overestimate the opportunities for spatial spread if the infectious period is short relative to the time scale of dispersal (Cross et al. 2005) or if infected dispersers bear a high fitness cost. Also, individuals may be most likely to become infected after dispersing because the pathogen is sexually transmitted and thus skewed towards reproductive individuals. Conversely, measures of host gene flow may underestimate disease spread if the probability of long-distance transmission is high relative to the chance of successfully immigrating and reproducing. For example, a narrow hybrid zone separating two rabbit subspecies on the Iberian peninsula that represents a substantial barrier to interspecific gene flow (Geraldes et al. 2006) appeared to have had no limiting effect on the rate of spatial spread of rabbit hemorrhagic disease virus (Muller et al. 2009).
Because of the above limitations, host genetic structure is generally most useful as a qualitative, rather than quantitative, measure of potential disease spread by revealing the most likely invasion pathway over a specific landscape. This approach was illustrated in the analysis of raccoon rabies spread and raccoon gene flow across two major rivers at the boundary between the US and Canada (Cullingham et al. 2009). One of these rivers appeared to have halted the spread of raccoon rabies for over a decade with no detection of this rabies virus strain on the Canadian side, whereas the other had experienced multiple documented invasions over a similar time frame. Since the disease control efforts in the two regions were equivalent, the authors hypothesised that the spatial heterogeneity in disease incidence may be driven by different racoon dispersal patterns. Consistent with the absence of rabies, they found permeability of the first river to raccoon gene flow to be low with one distinct genetic cluster identified on either side and a low percentage of potential immigrants assigned across the barrier. In contrast, the genetic structure of raccoons in the second case showed no association with the river, in accord with the observed pattern of repeated raccoon rabies invasion. The puzzling finding that the same type of environmental feature can have such different effects on host movement and disease spread certainly highlights the implication that landscape complexity and its population genetic consequences cannot be reduced to a simple search for dispersal barriers in an otherwise homogeneous matrix. Cunningham propose that differences in resource distribution (as the driver for raccoon density and dispersal distance) as well as landscape configuration and shape may have contributed to the observed pattern. Clearly, more studies of this type are needed to better understand how different environmental factors in natural landscapes interact to drive host population genetic, demographic, and epidemiological processes.
One aspect of assessing landscape permeability from the host point of view is that it can allow us to assess prospectively, even in the absence of the parasite, which areas would be predicted to be most prone to disease invasion in the future. Bataille et al., (2009) addressed this type of question regarding the potential spread of West Nile Virus (WNV) into the Galapagos archipelago, an event that would be expected to have catastrophic consequences for the endemic bird and reptile fauna. Because WNV is transmitted by mosquito vectors, including a species that recently established populations on the Galapagos, patterns of contemporary gene flow in the vector could provide a basis for assessing the risk of disease invasion. Genetic results for the mosquito clearly show that inadvertent introductions from the mainland to some islands via aircraft are common and ongoing. Furthermore, these introductions are often followed by further admixture among different island populations (Bataille et al. 2009). Consequently, human transportation networks appear to have all but removed the natural impediments to vector movement associated with impermeable oceanic barriers. Although WNV has not yet reached mainland Ecuador, it is expected to do so within the next few years. Recently implemented control programs against mosquito transfer (Bataille et al. 2009) will have to be strictly adhered to if the virus' introduction to the Galapagos is to be prevented.
These types of prospective genetic approaches are likely to become increasingly common, given that predicting and quantifying future infectious disease threats remains one of the major research challenges in disease ecology and epidemiology (Jones et al. 2008). However, this also raises the need for more studies trying to verify these genetically-derived predictions and to test their accuracy at least on a qualitative, if not on a quantitative, level. Ideally, this would involve a comprehensive collection of comparative genetic data for both parasite and host. One system that has received considerable interest in this regard is avian influenza and the looming risk of the highly pathogenic H5N1 strain being introduced from Eurasia to North America through wild bird species. To examine this issue from the host perspective, Winker et al., (2007) conducted genetic analyses for several dominant waterfowl species. Results indicated that gene flow across continental boundaries likely occurs at a small but epidemiologically relevant level in several species (Winker et al. 2007). This is generally consistent with rarely observed intercontinental transmission of individual avian influenza virus segments (Makarova et al. 1999) even though migration of complete virus has not yet been documented (Dugan et al. 2008). More recent work has corroborated that Eurasian virus segments have been repeatedly introduced into North America over the last few decades but also found evidence that the range of host species capable of facilitating such an exchange may be even greater than previously thought (Bahl et al. 2009). The large number of wild species potentially involved in avian influenza transmission represents one of the major problems for monitoring the virus and for developing risk models of intercontinental spread based on genetic data.
Temporal and spatial scales of epidemiological processes
Epidemiological processes can be highly variable over space and time. Being able to put results of spatial genetic analyses of infectious diseases into a temporal context is therefore often critical for their correct interpretation. Estimating divergence times from genetic data requires knowledge about evolutionary rates, which when combined with coalescent theory can give detailed insights not only about the dates associated with emergence and spread of parasites but also their population dynamics (Kingman 1982; Pybus & Rambaut 2009). Among parasites, coalescent approaches are most commonly applied to RNA viruses, for which epidemiological and evolutionary dynamics fall onto similar time scales due to their rapid rates of evolution (Holmes 2004; Pybus & Rambaut 2009).
Influenza A virus, more specifically the H3N2 strain predominantly responsible for human seasonal flu, provides a striking example of the complex spatio-temporal dynamics that can be inferred from viral sequence data (Rambaut et al. 2008; Russell et al. 2008). Analyses of several thousand H3N2 sequences collected worldwide have revealed that the virus is not able to sustain itself in temperate regions, where it causes seasonal epidemics during the winter but disappears over the summer months. Furthermore, viruses circulating in the same temperate location are more closely related to those from other temperate regions than to those that had been present during the previous season, suggesting annual seeding from a common source (Rambaut et al. 2008). This source is thought to be southeast Asia where, possibly due to particular environmental conditions, the virus persists throughout the year and where new antigenic variants arise capable of escaping recognition by the human immune system (Russell et al. 2008). From this tropical reservoir, viruses are able to spread each year along human transportation networks around the world to start new seasonal epidemics in temperate regions (Fig. 2). Molecular evidence for such a scenario comes from new genetic and antigenic variants appearing earlier in southeast Asia and observing that variants from this area tend to branch off directly from the trunk of the phylogenetic tree (Russell et al. 2008). In further support of this model, it has been shown that new drug resistant viruses arose and circulated in southeast Asia before reaching a global distribution (Nelson et al. 2009). Influenza A thus represents a clear example of source-sink dynamics of an infectious disease agent playing out on a global scale, an observation that has obvious implications for vaccine design and other control measures.
Figure 2.
Schematic depiction of source-sink dynamics characterising the seasonal spread of influenza A from tropical, permanent source populations (probably located in southeast Asia, see Russel et al., 2008) to temperate regions on both hemispheres where the virus is introduced and subsequently goes extinct each season. Reprinted from Rambaut et al. (2008) with permission from Macmillan Publishers Ltd: Nature.
The critical role of endemic core areas relative to peripheral populations that only support infections temporally is particularly well documented for measles (Grenfell et al. 2001) but probably characterises the dynamics of many diseases across different spatial scales. For individual hosts, for example, the issue of certain organs acting as spatial refugia from treatment or immune attack commonly arises (Sobesky et al. 2007; Wong et al. 1997). Within natural landscapes, persistence of diseases maintained in wild host reservoirs are often thought to depend on particular habitat types or landscape features, usually identified using GIS-based methods (Glass et al. 2007; Ostfeld et al. 2005; Wimberly et al. 2008). As shown for influenza, genetic inference could play a major part in validating and quantifying the relative contribution of these source populations to long-term disease dynamics and infection risk across the landscape.
A common and somewhat puzzling result of coalescent analysis of RNA viruses is that their population history can often only be traced back to a most recent common ancestor a few hundred years ago. This includes viral pathogens which are thought to have been around for several millennia including measles and mumps (Holmes 2003; Pomeroy et al. 2008). Common ancestry in the recent past therefore implies fairly regular and complete population turnover that has effectively removed all historic genetic diversity in these viruses. This lack of ancestral diversity is noteworthy from a landscape genetics point of view because it implies complete permeability of host populations to viral spread over a time scale of a few centuries. This may be quite plausible for many of the human viruses for which movement and contact patterns over such time scales, even on a global scale, may not be limited by physical boundaries. Older ancestors can be seen in viruses of domesticated species, such as dog rabies, presumably because of reduced connectivity among populations compared to human hosts (Bourhy et al. 2008). However, significantly more spatial structure and even deeper divergence times would be expected for viruses associated with wildlife reservoirs, especially in terrestrial species for which populations are significantly structured by environmental features.
Rodents and rodent-borne viruses may be good test case for this. Sin Nombre virus (Hantaviridae), for example, is a zoonotic pathogen maintained through asymptomatic infection of mice in the genus Peromyscus. Intriguingly, coalescent analysis of Sin Nombre virus sampled across western North America suggests an ancestor within the last 100 years (Black et al. 2009). Based on the spatial distances among sampling locations, this estimate implies a spreading rates of >5km per year at a minimum. Such a high rate would be surprising for a directly transmitted parasite considering small host body size and the complexity of the largely mountainous landscape, which appears to affect the genetic structure of Peromyscus populations (Dragoo et al. 2006; Root et al. 2003). Even if this spreading rate may be an overestimate, given that the underlying evolutionary rates are fast compared to those derived from larger data sets of related viruses (Ramsden et al. 2008), the result raises the general question of whether the resistance of natural landscapes to disease spread may be significantly lower than one might suspect based on host ecology and dispersal ability. So far, few comparative studies have examined how these host attributes affect spatial transmission processes and parasite gene flow (Blouin et al. 1995; Brown & Hovmoller 2002; Criscione & Blouin 2004) and whether general predictions in this regard are possible.
Spatial and temporal analysis of pathogen genetic data is widely applied to viral diseases but rarely to other parasites including some with potentially high rates of evolution, such as bacteria. Bacterial populations are frequently characterised by rapid clonal expansion (Keim & Wagner 2009; Smith et al. 2006) making it difficult to establish sufficiently resolved phylogenetic relationships. One noticeable exception is work by Girard et al. (2004) who examined the population genetic structure of Yersinia pestis, the bacterium causing plague, at two different landscape scales during an extensive prairie dog die-off in Arizona in 2001. In addition to field data, the authors used a passage experiment with field-derived plague isolates to empirically determine the pathogen's mutation rate, allowing them to infer time scales of epidemiological processes from genetic distances. On a regional scale, several distinct genetic groups could be detected, indicative of almost simultaneous but independent introduction of plague into prairie dog populations from an unknown reservoir. These introductions were likely triggered by environmental conditions. In addition, isolates belonging to the same grouping tended to cluster in space, especially among populations connected by open grassland, which represents principal prairie dog habitat. In contrast, populations separated by forest rarely shared the same genotype of plague suggesting that these landscape characteristics affected pathogen dissemination possibly by influencing prairie dog movement.
On a more local scale, genetic differentiation was even detectable among plague isolates within a single outbreak location so that the molecular data could be used to track spatial progression of disease across a prairie dog town (Fig. 3; Girard et al. 2004). Comparing the observed genetic pattern to the experimentally determined mutation rate revealed that the outbreak must have taken place in two distinct phases: the high frequency and wide distribution of a single genotype implied that initial progression was extremely rapid, leaving insufficient time for mutational events to occur. Only during a second slower phase, probably characterised by local burrow-to-burrow transmission, was the emergence of new, spatially restricted variants apparent. Interestingly, this two-stage invasion process does not seem to be limited to the local scale but is also characteristic for global patterns of plague colonisation (Keim & Wagner 2009). While population genetic studies of this kind are still rare for bacterial diseases, this situation is likely to change as rapid advances in sequencing technology makes whole bacterial genomes, and thus spatially informative polymorphisms, increasingly available (Harris et al. 2010).
Figure 3.

Phylogenetic and spatial analyses of a plague outbreak within a single prairie dog town in Arizona, 2001. (A) Unrooted phylogeny of plague isolates based on maximum-parsimony. (B) Individual genotypes are represented by colored symbols and spatially mapped by using ARCVIEW. Genotypes observed only once are represented by squares and are numbered. More common genotypes are represented by colored circles and defined in the legend. Reprinted from Girard et al. (2004) with permission based on Copyright (2004) National Academy of Sciences, U.S.A.
The general lack of genetic variation has also so far limited molecular investigations of chytrid fungus, an important emerging pathogen in amphibian populations. While genetic monomorphism is generally consistent with the idea that the fungus became globally distributed rather recently, more diverse markers have been identified and are permitting the test of epidemiological scenarios on a more local scale (Goka et al. 2009; James et al. 2009).
Genetic variation in parasite and host and its relative distribution across space and time is of course not only of interest where it reflects neutral markers but also as the basis for adaptive change. It has long been recognised that spatial population structure can strongly influence the process of co-adaptation between parasite and host and the evolution of virulence. We do not discuss these issues here but point to the rich body of literature examining these issues in detail (Dybdahl & Storfer 2003). Instead, we highlight some recent examples of environmental variation influencing the emergence and spatial spread of resistance in both host and parasite.
Avian malaria is a major threat to the endemic avifauna of Hawaii but resistance has recently been documented in one of the affected native bird species (Eggert et al. 2008). Disease pressure on these birds varies along an environmental gradient because cooler, high elevation habitats that don't support the mosquito vector are essentially disease free. Whether resistance evolved in the populations at high elevation, which are large but under little selective pressure, or in small surviving remnant populations at low elevation remains an open question. Molecular studies supported the latter scenario in that resistant lowland and susceptible highland populations are genetically distinct (Eggert et al. 2008). Interestingly, two related species that don't show the same genetic segregation with altitude have so far failed to evolve resistance and remain rare at low elevation.
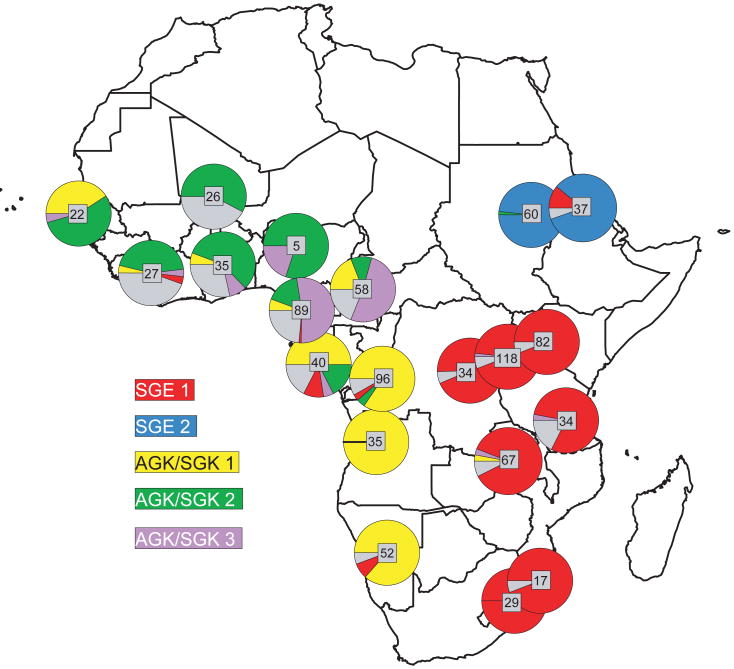
Pearce et al. (2009) suggest that the evolution and geographic extent of drug resistance in the malaria parasite Plasmodium falciparum to commonly used drugs arose independently in at least five different areas in Africa. Furthermore, the current spatial distribution of different resistance alleles falls into geographical clusters with little overlap (Fig 4). One intriguing possibility is that dispersal barriers or other changes in environmental conditions are preventing alleles from increasing their current distribution (Anderson 2009). However, before researchers can focus on understanding the basis for these genetic boundaries, alternative explanations need to be ruled out. Because the current distribution map is essentially a snapshot it is possible that these alleles, which all appeared within the last few decades, are continuing to spread and simply haven't had enough time yet to spatially admix (Pearce et al. 2009). The latter scenario best explains a very similar pattern of spatial clustering found in a cougar virus, where it is thought to be the result of a recent population bottleneck followed by spatial expansion that is still ongoing (Biek et al. 2006).
Figure 4.
Distribution and local frequency of five resistance alleles lineages against malarial drugs that arose independently within Plasmodium falciparum populations in sub-Saharan Africa. Resistance alleles whose flanking microsatellite haplotypes did not conform to a defined major lineage are shown in grey. Reprinted from Pearce et al. (2009).
The question whether genetic discontinuities are really explained by environmental variables rather than random or temporary processes arises as a fundamental problem in landscape genetics (Cushman & Landguth, this issue). Spatial genetic boundaries in parasite populations can arise spontaneously and without any landscape influence, especially if they are inferred from a single locus (Real & Biek 2007). Data from additional unlinked loci and from host or vector genetics are therefore often needed to identify genuine structure. Even for a well-studied system like malaria these types of data are currently not available at the appropriate scale and resolution (Pearce et al. 2009).
Invasion dynamics
The emergence of new infectious diseases or their spread to novel areas are often dramatic events with significant health and economic consequences as illustrated by the recent examples of SARS, West Nile virus, foot-and-mouth disease virus, or the H5N1 and H1N1 strains of influenza A. Because of their impact, these diseases are usually recognised early and their progression documented with samples taken over the course of invasion. The substantial record of samples and analysis associated with important emergence events has often enabled researchers to study the temporal and spatial process of invasion in great detail (CSMEC 2004; Smith et al. 2009; Wallace et al. 2007). This has been especially the case for viral diseases, which, as also seen from the list above, are responsible for the vast majority of emerging microbial threats (Cleaveland et al. 2001). In addition, the characteristic of RNA viruses to accumulate mutations frequently enough to track the underlying ecological process of viral spread means that a wealth of epidemiological information can commonly be gleaned from molecular data when analysed in a temporal coalescent framework (Pybus & Rambaut 2009).
A key epidemiological question for any new disease outbreak is: from where did this disease originate and how has spatial diffusion proceeded from there? Even if viral samples are not available until the disease has already spread to additional locations, the resulting sequence data will frequently contain information about the temporal order in which populations were seeded and the frequency of viral exchange among them. Different methods have been applied to infer this but all have in common that they use the spatial phylogenetic information associated with the samples (i.e. the tree tips) to estimate ancestral spatial states for all internal nodes of the phylogeny including the root. If the spatial states are discrete (i.e. countries, habitat types, etc) a parsimony principle can be used to infer the minimum number of spatial state changes required throughout the phylogeny to explain the data represented by the tree tip. It is worth noting that while this is the same parsimony principle as the one used to find phylogenetic trees, the spatial reconstruction is independent of the tree search and in fact assumes that the phylogeny is already known. Using this method, Wallace et al (2007) deduced the origin and phylogeographic spread of H5N1 influenza virus in Eurasia based on close to 200 sequences from two viral genes. They identified Guandong province in China as the likely source of the epidemic and quantified the amount of viral movement between locations at both local and regional scales. Statistical support for the inferred migration rates was assessed by comparing them to a distribution generated from repeatedly randomising the spatial states at the tree tips while keeping the phylogeny constant (Wallace et al. 2007).
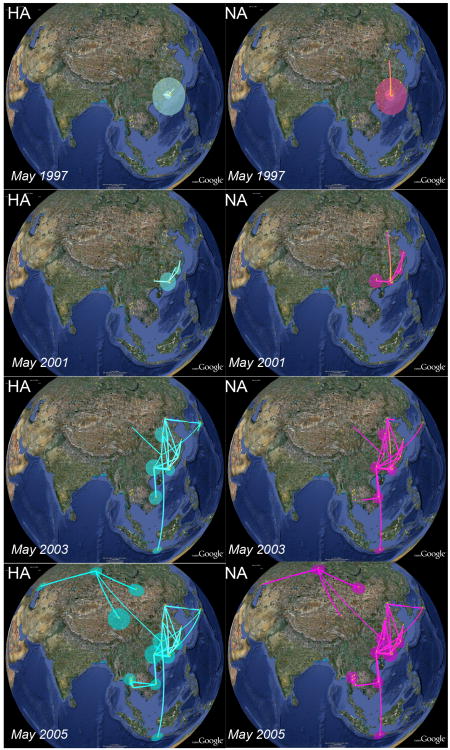
Although the parsimony method is intuitive and easy to implement, it ignores two important sources of uncertainty: the underlying tree and the estimation of the spatial state itself. A Bayesian inference approach that addresses both these issues has been developed recently and was used to re-analyse the same H5N1 data set (Lemey et al. 2009). This study confirmed Guandong province as the most likely origin but showed that some of the earlier conclusions were no longer statistically supported once additional sources of uncertainty had been taken into account. Dates associated with all samples were used to estimate evolutionary rates and to calibrate the phylogeny, which provided a means to visualise the spatial diffusion process through temporal snapshots (Fig. 5). The study also introduced novel ways of using the spatial information more explicitly through the formulation of distance–informed priors on migration rates. Future extensions, already explored in biogeography (Sanmartìn et al. 2008), may also be able to scale migration rates with local host population size akin to the use of gravity models in epidemiology (Xia et al. 2004). One major limitation of the current approach is that the inferred migration rates are constrained to be symmetrical with viral movement from A to B being equivalent to movement from B to A. However, a revised version has already been developed that is able to also accommodate the kind of directional imbalances one would expect during an invasion (P. Lemey, personal communication).
Figure 5.
Spread of avian influenza A H5N1 across Asia, May 1997 – May 2005, as inferred from phylogeographic anayses of two viral genes, hemaglutinin (HA) and neuraminidase (NA). Lines between locations represent branches in the Maximum Clade Credibility (MCC) tree along which the relevant location transition occurs. Location circle diameters are proportional to square root of the number of MCC branches maintaining a particular location state at each time-point. The white-green and yellow-magenta color gradients inform the relative age of the transitions for HA and NA respectively (older-recent). Reprinted from Lemey et al. (2009).
While the two molecular H5N1 studies above were successful at quantifying viral movement they provide little information about the specific factors that drive geographical variation in the invasion process. Gilbert (2008) investigated outbreaks in several southeast Asian countries using GIS and remote sensing data to identify environmental predictors of H5N1 occurrence. Human population size, duck numbers and rice cropping intensity were associated with increased disease risk in their model. Other work has examined the importance of additional factors, particularly wild bird migration and poultry trade, in the global spread of avian influenza virus (Kilpatrick et al. 2006). Spatial risk maps from these types of approaches could thus be used in future analyses to test whether the identified environmental variables are able to explain the observed phylogeographic patterns of H5N1 spread at various scales.
The large number of potential host species, both wild and domestic, and their complex movement patterns clearly makes spatial inference particularly challenging in the case of avian influenza. The same complexities do not apply to rabies virus, where several large-scale invasions have occurred over the last century, usually in a characteristic wave-like fashion and driven by one or few terrestrial wild carnivore species (Biek et al. 2007; Bourhy et al. 1999; Real et al. 2005). As a consequence, wildlife rabies represents a particularly informative and well-studied model for the landscape genetics of infectious disease invasion. In Europe, an epizootic in red foxes that started in northeast Europe during the mid 20th century continuously spread westwards to reach France in 1986. Molecular analysis of this outbreak revealed a distinct ladder-like pattern within the phylogenetic tree, consistent with spatial progression: viruses from the outbreak origin in northeastern Europe branched off close to the tree root whereas those from the areas in western Europe invaded last, branched off near the tips (Bourhy et al. 1999; Holmes 2004). The same study was also the first to show that spatial genetic structure of rabies virus may be organised along major rivers and mountain ranges.
Two other major rabies invasions occurred during the last century, both in North America. The first originated in arctic foxes through which it entered into northern Canada before spreading southwards and becoming associated with red foxes the current principal host species. A fine-scale genetic analysis of the virus in southern Ontario, one of the southernmost areas reached by this invasion, revealed high viral diversity and strong spatial clustering of the same genotypes within a relatively small area (Nadin-Davis et al. 1994). While this pattern was originally interpreted as being driven by environmental variables, later analysis by Real et al. (2005) showed that it is in fact fully consistent with an isolation-by-distance process. These analyses further revealed that invasion of southern Ontario took place in the form of two distinct waves, representing two arms of the invasion front that had entered the area from two different directions. Curiously, no sign of spatial admixture among the two wave fronts was detected. Furthermore, the strength of the isolation-by-distance pattern differed markedly between the two lineages suggesting that both were characterised by very different epidemiological dynamics.
The second North American rabies epizootic took place among raccoon populations along the Atlantic coast from 1977 onwards. A unique feature of this outbreak is that its starting point and spatial progression have been documented at a relatively fine resolution from a very early stage. In all likelihood, this invasion was initiated by the translocation of raccoons from Florida, where raccoon rabies had been endemic for at least several decades, to West Virginia where raccoon populations were completely naïve to the virus. From this single point of introduction, the infection wave radiated out into all directions across a major portion of the eastern US and could only be prevented from northward expansion into Canada and from further spread to the west through the massive and ongoing deployment of an oral rabies vaccine (ORV) (Rupprecht & Smith 1994).
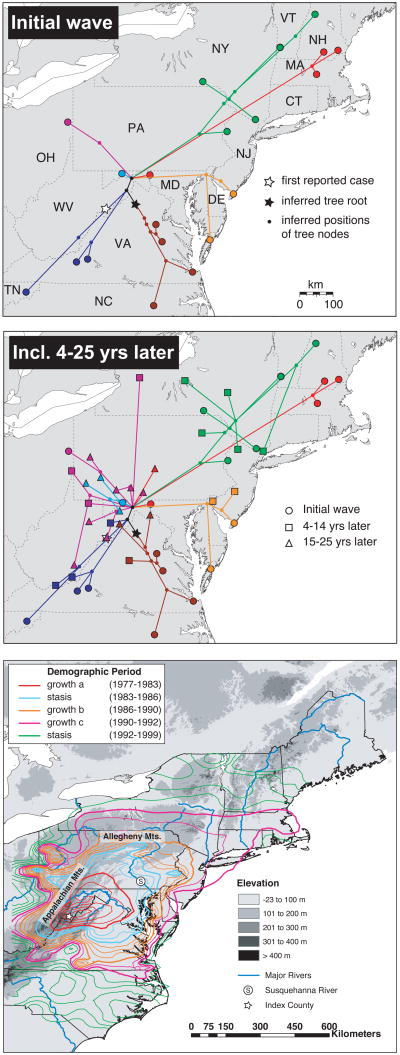
Genetic analysis showed that the pattern of spatial radiation in this outbreak finds a clear reflection in its phylogenetic structure, with different viral lineages representing different general directions of spread (Fig. 6; Biek et al. 2007). This orderly pattern permitted the estimation of ancestral spatial states, here in the form of continuous geographic coordinates (i.e. latitude/ longitude) rather than discrete states, from the phylogeny. While the uncertainty associated with this method is high, the point estimate for the epizootic origin fell reassuringly close to the documented first case (Fig. 6). Results further indicated that rabies lineages had encountered very different levels of landscape resistance to viral spread. For example, spatial progression of genetic lineages that entered into the Appalachians, supporting lower density of raccoons and likely restricting their movements, more or less stalled in most cases. In contrast, lineages that spread into urban areas along the coast, which support particularly high raccoon densities, had been able to traverse these areas very rapidly (Fig. 6). Demographic reconstruction based on temporal coalescent methods further showed that these landscape heterogeneities had repeatedly caused notable changes in the overall number of infections over the course of the invasion.
Figure 6.
Phylogenetic relationships among virus samples collected during a raccoon rabies virus (RRV) epizootic in eastern North America. (A) Portion of Bayesian phylogeny that corresponds to initial infection wave. Estimated locations of internal nodes and the tree root are as shown as black symbols. White star marks the epizootic's documented origin in 1977. (B) Full MCC tree projected onto landscape, including samples collected 4 –14 yrs (squares) and 15 –25 yrs (triangles) after the first case within a county. (C) Annual rate of RRV spread along the US mid- Atlantic relative to elevation and major rivers, 1977 -1999. See Biek et al. (2007) for further details. Reprinted with permission based on Copyright (2007) National Academy of Sciences, U.S.A.
Another important insight from the raccoon rabies study is that the speed at which viruses spread during wave front expansion was markedly higher than after local establishment (Biek et al. 2007). Combined with the rapid divergence into separate lineages along different diffusion axes, this means that spatial genetic structure is largely determined by the initial invasion process but remains relatively stable once the virus has become enzootic (Fig. 6). This fits into a more general recognition of the importance of stochastic processes along wave fronts during colonisation processes and their long lasting impact on the colonisers' population genetic structure (Excoffier & Ray 2008). In the case of rabies, this phenomenon relates to colonisation reducing the density of susceptible hosts available for infection. As a consequence, viruses may have a relatively low probability of successfully dispersing into areas already colonised by another virus (of equal fitness). Landscape elements limiting host movement are likely to further accentuate this pattern by forming natural boundaries among different genetic groups. However, even a complete absence of detectable viral gene flow across a barrier in an endemic situation would not imply a lack of movement of infected hosts. This has important implication for designing control strategies based on spatial genetic structure in the virus because the same landscape barrier may pose few limits to viral spread when separating infected and susceptible populations, greatly reducing its epidemiological importance.
Conclusions: future directions for studying landscapes and infection dynamics
The interface of landscape genetics and epidemiology is a rapidly expanding research area that is poised to make relevant contributions to our ecological and evolutionary understanding of host-parasite interactions and disease. Such a contribution is also timely given the large impact infectious diseases are having not only on human populations and their economies but also wild species and ecosystems. Many of our examples focused on microparasites, especially viruses, reflecting their prominent role as emerging pathogens (Woolhouse 2002). The rapid genetic changes typically seen in these organisms further results in genetic patterns that can be matched to environmental heterogeneities at precise and well-defined temporal and spatial scales, making these systems well-suited for microevolutionary studies. The biological characteristics of many viruses and bacteria (e.g. haploid genomes, insufficient recombination resulting in essentially one locus, rapid spread over continental or global scales), however, means that the analytical approaches applied to them tend to be different from those most commonly seen in the landscape genetics literature. Phylogenetic and coalescent-based methods for example are much more prominent and have only recently made more explicit use of spatial information within a statistical framework (e.g. Lemey et al. 2009; Lemey et al. 2010). There is therefore good reason to believe that current conceptual and methodological advances in landscape genetics as a whole, including some of those covered in this issue, could significantly enhance the analytical toolbox available to infectious disease researchers.
GIS are a well-established tool in spatial studies of infectious disease (Clements & Pfeiffer 2009; Waller & Gotway 2004) but remain strangely underused in molecular epidemiology. Emerging pathogens such as avian influenza and Sin Nombre virus, which have been well studied from both perspectives, could be obvious test cases for a better integration of pathogen genetic data into GIS. Possible applications range from the identification of spatial genetic patterns (e.g. interpolation and clustering methods) to the specification of geographical models and hypotheses, which then could be tested against molecular data (Kidd & Ritchie 2006). Techniques focused on differential landscape permeability, such as least cost path analysis, non-Euclidian distance matrices or isolation by resistance (Cushman et al. 2006; Kidd & Ritchie 2006; McRae 2006), should be widely applicable, especially in the context of pathogen invasion and persistence. GIS are a particularly powerful analytical platform for this because they can accommodate such a wide variety of data which, depending on the pathogen systems, may range from habitat or climatic variables to patterns of human transportation and livestock movement. Ecological niche modelling is another spatial tool that is already being applied to understand past, current and future patterns of infectious disease distribution (Peterson 2006). Similar to those mentioned above, this technique has considerable potential to generate testable predictions about the genetic structure of parasite populations and how they may respond to climatic and other environmental changes. Finally, our understanding of how landscapes simultaneously affect epidemiological and genetic processes could be greatly enhanced by a broader application of simulation studies, which can yield important insights regarding the reliability and power of different analytical techniques and which have an increasingly important role to play in generating meaningful null hypotheses (Frantz et al. 2009; Epperson et al., this issue).
A common tactic for examining the potential or realised spread of infectious disease through a landscape is by investigating the genetic structure of the host, especially if the parasite is closely associated with a particular host species (Frantz et al. 2009). However, as some of our examples as well as experiences from co-evolution and biogeography show (Nieberding & Olivieri 2007), even a very strong association of host and parasite may not necessarily lead to genetically congruent patterns. Concordance between gene flow of parasite and host can thus rarely be assumed a priori but should ideally be confirmed through comparative genetic analysis of both. There is a clear need for more combined micro-evolutionary studies to determine how parasite and host populations experience and genetically respond to the same physical landscape. Such studies need to be carefully designed because issues related to the appropriate temporal and spatial scales of inference (Anderson et al., this issue; Balkenhol et al. 2009) may be exacerbated when examining host-parasite pairs due to differences in evolutionary rates, effective population sizes or demographic variability (Bruyndonckx et al. 2009). One critical research question is how spatial variation in host density affects the dynamics of both transmission and host dispersal and how these factors interact in driving the local and long-distance spread of disease. Intensifying research in this area would thus help to understand how host-parasite interactions at the micro-evolutionary level may scale up to create larger patterns but also offer interesting applications. For example, it may ultimately be possible to develop predictive models of disease spread in novel environments by combining information about host ecology and genetics, geography, and parasite life history.
While epidemiological studies should benefit greatly from engaging more broadly with the tools and concepts available from landscape genetics, they also have much to offer in return, as increased knowledge about the processes governing host-parasite co-evolution in heterogeneous environments should open up exciting research avenues in multiple directions. The high temporal and spatial resolution of genetic structure afforded in many parasites offers unique opportunities to quantify temporal and spatial variation in host contact processes. This ‘parasite as host marker’ approach has considerable potential for applied landscape ecology and conservation (Whiteman & Parker 2005) but is still rarely used for this purpose. The potential for rapid genetic change also makes infectious organisms a promising target to examine the geographical context of adaptive evolution in continuous populations, one of the current frontiers in landscape genetics (Manel et al., this issue). Further, some of the selective forces acting on parasite populations, such as drugs or host immunity, are well defined and even amendable to experimental manipulation, a rare constellation that could present novel research opportunities for examining how landscape structure affects adaptive dynamics.
In conclusion, there are many reasons why a larger and multidirectional exchange of data, methods and ideas among epidemiologists, landscape ecologists and population geneticists would be desirable and should lead to new and innovative lines of research. It is becoming increasingly recognized that landscape genetics has a much broader scientific remit and relevance as a discipline than initial work in this field had suggested (Balkenhol et al. 2009). The study of infectious disease dynamics and genetics in heterogeneous environments should become an integral part of such a broadened agenda for landscape genetic research.
Acknowledgments
This research was supported by a University of Glasgow Kelvin-Smith Fellowship to RB, National Institutes of Health grant RO1 AI047498 to LAR and by the RAPIDD Program of the Science and Technology Directorate, Department of Homeland Security and the Fogarty International Center, National Institutes of Health. We thank several reviewers for their helpful comments on an earlier draft of this manuscript and some authors whose work we reproduced for sharing their graphic material.
References
- Anderson T. Mapping the spread of malaria drug resistance. PLoS Medicine. 2009;6:e1000054. doi: 10.1371/journal.pmed.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony NM, Johnson-Bawe M, Jeffery K, et al. The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proceedings of the National Academy of Sciences of the USA. 2007;104:20432–20436. doi: 10.1073/pnas.0704816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archie EA, Luikart G, Ezenwa VO. Infecting epidemiology with genetics: a new frontier in disease ecology. Trends in Ecology & Evolution. 2009;24:21–30. doi: 10.1016/j.tree.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Bahl J, Vijaykrishna D, Holmes EC, Smith GJ, Guan Y. Gene flow and competitive exclusion of avian influenza A virus in natural reservoir hosts. Virology. 2009;390:289–297. doi: 10.1016/j.virol.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkenhol N, Gugerli F, Cushman SA, et al. Identifying future research needs in landscape genetics: where to from here? Landscape Ecology. 2009;24:455–463. [Google Scholar]
- Barrett LG, Thrall PH, Burdon JJ, Linde CC. Life history determines genetic structure and evolutionary potential of host-parasite interactions. Trends in Ecology & Evolution. 2008;23:678–685. doi: 10.1016/j.tree.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille A, Cunningham AA, Cedeno V, et al. Evidence for regular ongoing introductions of mosquito disease vectors into the Galapagos Islands. Proceedings of the Royal Society B-Biological Sciences. 2009;276:3769–3775. doi: 10.1098/rspb.2009.0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger PH, Zeyen RTJ, Groth JV. Aphid retention of maize dwarf mosaic virus (potyvirus): epidemiological implications. Annals of Applied Biology. 1987;111:337–344. [Google Scholar]
- Biek R, Drummond AJ, Poss M. A virus reveals population structure and recent demographic history of its carnivore host. Science. 2006;311:538–541. doi: 10.1126/science.1121360. [DOI] [PubMed] [Google Scholar]
- Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proceedings of the National Academy of Sciences of the USA. 2007;104:7993–7998. doi: 10.1073/pnas.0700741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black WCt, Doty JB, Hughes MT, Beaty BJ, Calisher CH. Temporal and geographic evidence for evolution of Sin Nombre virus using molecular analyses of viral RNA from Colorado, New Mexico and Montana. Virology Journal. 2009;6:102. doi: 10.1186/1743-422X-6-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchong JA, Samuel MD, Scribner KT, et al. Landscape genetics and the spatial distribution of chronic wasting disease. Biology Letters. 2008;4:130–133. doi: 10.1098/rsbl.2007.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin MS, Yowell CA, Courtney CH, Dame JB. Host movement and the genetic structure of populations of parasitic nematodes. Genetics. 1995;141:1007–1014. doi: 10.1093/genetics/141.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy H, Kissi B, Audry L, et al. Ecology and evolution of rabies virus in Europe. Journal of General Virology. 1999;80:2545–2557. doi: 10.1099/0022-1317-80-10-2545. [DOI] [PubMed] [Google Scholar]
- Bourhy H, Reynes JM, Dunham EJ, et al. The origin and phylogeography of dog rabies virus. Journal of General Virology. 2008;89:2673–2681. doi: 10.1099/vir.0.2008/003913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J, Gregory PH, Johnson CG. Possible wind transport of coffee leaf rust across the Atlantic Ocean. Nature. 1971;229:500–501. doi: 10.1038/229500b0. [DOI] [PubMed] [Google Scholar]
- Brown JK, Hovmoller MS. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- Bruyndonckx N, Henry I, Christe P, Kerth G. Spatio-temporal population genetic structure of the parasitic mite Spinturnix bechsteini is shaped by its own demography and the social system of its bat host. Molecular Ecology. 2009;18:3581–3592. doi: 10.1111/j.1365-294X.2009.04299.x. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Madden LV. Introduction to plant disease epidemiology. John Wiley & Sons; New York: 1990. [Google Scholar]
- Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements AC, Pfeiffer DU. Emerging viral zoonoses: frameworks for spatial and spatiotemporal risk assessment and resource planning. Veterinary Journal. 2009;182:21–30. doi: 10.1016/j.tvjl.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam EM, Thebaud G, Wadsworth J, et al. Integrating genetic and epidemiological data to determine transmission pathways of foot-and-mouth disease virus. Proceedings of the Royal Society B-Biological Sciences. 2008;275:887–895. doi: 10.1098/rspb.2007.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscione CD, Blouin MS. Life cycles shape parasite evolution: comparative population genetics of salmon trematodes. Evolution. 2004;58:198–202. doi: 10.1111/j.0014-3820.2004.tb01587.x. [DOI] [PubMed] [Google Scholar]
- Criscione CD, Poulin R, Blouin MS. Molecular ecology of parasites: elucidating ecological and microevolutionary processes. Molecular Ecology. 2005;14:2247–2257. doi: 10.1111/j.1365-294X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Cross PC, Lloyd-Smith JO, Johnson PLF, Getz WM. Duelling timescales of host movement and disease recovery determine invasion of disease in structured populations. Ecology Letters. 2005;8:587–595. [Google Scholar]
- CSMEC Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Cullingham CI, Kyle CJ, Pond BA, Rees EE, White BN. Differential permeability of rivers to raccoon gene flow corresponds to rabies incidence in Ontario, Canada. Molecular Ecology. 2009;18:43–53. doi: 10.1111/j.1365-294X.2008.03989.x. [DOI] [PubMed] [Google Scholar]
- Cushman SA, McKelvey KS, Hayden J, Schwartz MK. Gene flow in complex landscapes: testing multiple hypotheses with causal modeling. American Naturalist. 2006;168:486–499. doi: 10.1086/506976. [DOI] [PubMed] [Google Scholar]
- Dragoo JW, Lackey JA, Moore KE, et al. Phylogeography of the deer mouse (Peromyscus maniculatus) provides a predictive framework for research on hantaviruses. Journal of General Virology. 2006;87:1997–2003. doi: 10.1099/vir.0.81576-0. [DOI] [PubMed] [Google Scholar]
- Dugan VG, Chen R, Spiro DJ, et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathogens. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybdahl MF, Storfer A. Parasite local adaptation: Red Queen versus Suicide King. Trends in Ecology & Evolution. 2003;18:523–530. [Google Scholar]
- Eggert LS, Terwilliger LA, Woodworth BL, et al. Genetic structure along an elevational gradient in Hawaiian honeycreepers reveals contrasting evolutionary responses to avian malaria. BMC Evolutionary Biology. 2008;8:315. doi: 10.1186/1471-2148-8-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends in Ecology & Evolution. 2008;23:347–351. doi: 10.1016/j.tree.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Frantz AC, Cellina S, Krier A, Schley L, Burke T. Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? Journal of Applied Ecology. 2009;46:493–505. [Google Scholar]
- Gagneux P, Gonder MK, Goldberg TL, Morin PA. Gene flow in wild chimpanzee populations: what genetic data tell us about chimpanzee movement over space and time. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2001;356:889–897. doi: 10.1098/rstb.2001.0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Ferrand N, Nachman MW. Contrasting patterns of introgression at X-linked loci across the hybrid zone between subspecies of the European rabbit (Oryctolagus cuniculus) Genetics. 2006;173:919–933. doi: 10.1534/genetics.105.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Xiao X, Pfeiffer DU, et al. Mapping H5N1 highly pathogenic avian influenza risk in Southeast Asia. Proceedings of the National Academy of Sciences of the USA. 2008;105:4769–4774. doi: 10.1073/pnas.0710581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard JM, Wagner DM, Vogler AJ, et al. Differential plague-transmission dynamics determine Yersinia pestis population genetic structure on local, regional, and global scales. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8408–8413. doi: 10.1073/pnas.0401561101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T. Patterns of within population dispersal and mating of the fungus Microbotryum violaceum parasitising the plant Silene latifolia. Heredity. 2004;93:559–565. doi: 10.1038/sj.hdy.6800554. [DOI] [PubMed] [Google Scholar]
- Glass GE, Shields T, Cai B, Yates TL, Parmenter R. Persistently highest risk areas for hantavirus pulmonary syndrome: potential sites for refugia. Ecological Applications. 2007;17:129–139. doi: 10.1890/1051-0761(2007)017[0129:phrafh]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Gloster J, Burgin L, Witham C, Athanassiadou M, Mellor PS. Bluetongue in the United Kingdom and northern Europe in 2007 and key issues for 2008. Veterinary Record. 2008;162:298–302. doi: 10.1136/vr.162.10.298. [DOI] [PubMed] [Google Scholar]
- Goka K, Yokoyama J, Une Y, et al. Amphibian chytridiomycosis in Japan: distribution, haplotypes and possible route of entry into Japan. Molecular Ecology. 2009 doi: 10.1111/j.1365-294X.2009.04384.x. [DOI] [PubMed] [Google Scholar]
- Grenfell BT, Bjornstad ON, Kappey J. Travelling waves and spatial hierarchies in measles epidemics. Nature. 2001;414:716–723. doi: 10.1038/414716a. [DOI] [PubMed] [Google Scholar]
- Harris SR, Feil EJ, Holden MT, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. Molecular clocks and the puzzle of RNA virus origins. Journal of Virology. 2003;77:3893–3897. doi: 10.1128/JVI.77.7.3893-3897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. The phylogeography of human viruses. Molecular Ecology. 2004;13:745–756. doi: 10.1046/j.1365-294x.2003.02051.x. [DOI] [PubMed] [Google Scholar]
- James TY, Litvintseva AP, Vilgalys R, et al. Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathogens. 2009;5:e1000458. doi: 10.1371/journal.ppat.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancovich JK, Davidson EW, Parameswaran N, et al. Evidence for emergence of an amphibian iridoviral disease because of human-enhanced spread. Molecular Ecology. 2005;14:213–224. doi: 10.1111/j.1365-294X.2004.02387.x. [DOI] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney DB, King TM, Rowe DL, Poulin R. Contrasting mtDNA diversity and population structure in a direct-developing marine gastropod and its trematode parasites. Molecular Ecology. 2009;18:4591–4603. doi: 10.1111/j.1365-294X.2009.04388.x. [DOI] [PubMed] [Google Scholar]
- Keim PS, Wagner DM. Humans and evolutionary and ecological forces shaped the phylogeography of recently emerged diseases. Nature Reviews Microbiology. 2009;7:813–821. doi: 10.1038/nrmicro2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd DM, Ritchie MG. Phylogeographic information systems: putting the geography into phylogeography. Journal of Biogeography. 2006;33:1851–1865. [Google Scholar]
- Kilpatrick AM, Chmura AA, Gibbons DW, et al. Predicting the global spread of H5N1 avian influenza. Proceedings of the National Academy of Sciences of the USA. 2006;103:19368–19373. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman JFC. On the genealogy of large populations. Journal of Applied Probability. 1982;19A:27–43. [Google Scholar]
- Lahm SA, Kombila M, Swanepoel R, Barnes RF. Morbidity and mortality of wild animals in relation to outbreaks of Ebola haemorrhagic fever in Gabon, 1994-2003. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2007;101:64–78. doi: 10.1016/j.trstmh.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Computational Biology. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Welch JJ, Suchard MA. Phylogeography takes a relaxed random walk in continuous space and time. Molecular Biology and Evolution. 2010 doi: 10.1093/molbev/msq067. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy EM, Kumulungui B, Pourrut X, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Medicine. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova NV, Kaverin NV, Krauss S, Senne D, Webster RG. Transmission of Eurasian avian H2 influenza virus to shorebirds in North America. Journal of General Virology. 1999;80(Pt 12):3167–3171. doi: 10.1099/0022-1317-80-12-3167. [DOI] [PubMed] [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology & Evolution. 2003;18:189–197. [Google Scholar]
- McRae BH. Isolation by resistance. Evolution. 2006;60:1551–1561. [PubMed] [Google Scholar]
- Muller A, Freitas J, Silva E, et al. Evolution of rabbit haemorrhagic disease virus (RHDV) in the European rabbit (Oryctolagus cuniculus) from the Iberian Peninsula. Veterinary Microbiology. 2009;135:368–373. doi: 10.1016/j.vetmic.2008.09.057. [DOI] [PubMed] [Google Scholar]
- Nadin-Davis SA, Casey GA, Wandeler AI. A molecular epidemiological study of rabies virus in central Ontario and western Quebec. Journal of General Virology. 1994;75:2575–2583. doi: 10.1099/0022-1317-75-10-2575. [DOI] [PubMed] [Google Scholar]
- Nadler SA. Microevolution and the genetic structure of parasite populations. Journal of Parasitology. 1995;81:395–403. [PubMed] [Google Scholar]
- Nelson MI, Simonsen L, Viboud C, Miller MA, Holmes EC. The origin and global emergence of adamantane resistant A/H3N2 influenza viruses. Virology. 2009;388:270–278. doi: 10.1016/j.virol.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieberding CM, Olivieri I. Parasites: proxies for host genealogy and ecology? Trends in Ecology & Evolution. 2007;22:156–165. doi: 10.1016/j.tree.2006.11.012. [DOI] [PubMed] [Google Scholar]
- Ostfeld RS, Glass GE, Keesing F. Spatial epidemiology: an emerging (or re-emerging) discipline. Trends in Ecology & Evolution. 2005;20:328–336. doi: 10.1016/j.tree.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Pearce RJ, Pota H, Evehe MS, et al. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Medicine. 2009;6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AT. Ecologic niche modeling and spatial patterns of disease transmission. Emerging Infectious Diseases. 2006;12:1822–1826. doi: 10.3201/eid1212.060373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy LW, Bjornstad ON, Holmes EC. The evolutionary and epidemiological dynamics of the paramyxoviridae. Journal of Molecular Evolution. 2008;66:98–106. doi: 10.1007/s00239-007-9040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy LH, Krupa SV, Dean JL. Introduction of sugar cane rust into the Americas and its spread in Florida. Plant Diseases. 1985;69:469–474. [Google Scholar]
- Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nature Reviews Genetics. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Pybus OG, Nelson MI, et al. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden C, Melo FL, Figueiredo LM, Holmes EC, Zanotto PM. High rates of molecular evolution in hantaviruses. Molecular Biology and Evolution. 2008;25:1488–1492. doi: 10.1093/molbev/msn093. [DOI] [PubMed] [Google Scholar]
- Real LA, Biek R. Spatial dynamics and genetics of infectious diseases on heterogeneous landscapes. Journal of the Royal Society Interface. 2007;4:935–948. doi: 10.1098/rsif.2007.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real LA, Henderson JC, Biek R, et al. Unifying the spatial population dynamics and molecular evolution of epidemic rabies virus. Proceedings of the National Academy of Sciences of the USA. 2005;102:12107–12111. doi: 10.1073/pnas.0500057102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root JJ, Black WCt, Calisher CH, et al. Analyses of gene flow among populations of deer mice (Peromyscus maniculatus) at sites near hantavirus pulmonary syndrome case-patient residences. Journal of Wildlife Diseases. 2003;39:287–298. doi: 10.7589/0090-3558-39.2.287. [DOI] [PubMed] [Google Scholar]
- Rupprecht CE, Smith JS. Raccoon rabies: the re-emergence of an epizootic in a densely populated area. Seminars in Virology. 1994;5:155–164. [Google Scholar]
- Russell CA, Jones TC, Barr IG, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- Sanmartìn I, Mark Pvd, Ronquist F. Inferring dispersal: a Bayesian approach to phylogeny-based island biogeography, with special reference to the Canary Islands. Journal of Biogeography. 2008;35:428–449. [Google Scholar]
- Sellers RF, Maarouf AR. Possible introduction of epizootic hemorrhagic disease of deer virus (serotype 2) and bluetongue virus (serotype 11) into British Columbia in 1987 and 1988 by infected Culicoides carried on the wind. Canadian Journal of Veterinary Research. 1991;55:367–370. [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Smith NH, Gordon SV, de la Rua-Domenech R, Clifton-Hadley RS, Hewinson RG. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nature Reviews Microbiology. 2006;4:670–681. doi: 10.1038/nrmicro1472. [DOI] [PubMed] [Google Scholar]
- Sobesky R, Feray C, Rimlinger F, et al. Distinct hepatitis C virus core and F protein quasispecies in tumoral and nontumoral hepatocytes isolated via microdissection. Hepatology. 2007;46:1704–1712. doi: 10.1002/hep.21898. [DOI] [PubMed] [Google Scholar]
- Storfer A, Murphy MA, Evans JS, et al. Putting the “landscape” in landscape genetics. Heredity. 2007;98:128–142. doi: 10.1038/sj.hdy.6800917. [DOI] [PubMed] [Google Scholar]
- Takehisa J, Kraus MH, Ayouba A, et al. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. Journal of Virology. 2009;83:1635–1648. doi: 10.1128/JVI.02311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer PT, Souquiere S, Clifford SL, et al. Molecular evidence for deep phylogenetic divergence in Mandrillus sphinx. Molecular Ecology. 2003;12:2019–2024. doi: 10.1046/j.1365-294x.2003.01877.x. [DOI] [PubMed] [Google Scholar]
- Tsujimoto H, Cooper RW, Kodama T, et al. Isolation and characterization of simian immunodeficiency virus from mandrills in Africa and its relationship to other human and simian immunodeficiency viruses. Journal of Virology. 1988;62:4044–4050. doi: 10.1128/jvi.62.11.4044-4050.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn F, Li Y, Bailes E, et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368:155–171. doi: 10.1016/j.virol.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Wallace RG, Hodac H, Lathrop RH, Fitch WM. A statistical phylogeography of influenza A H5N1. Proceedings of the National Academy of Sciences of the USA. 2007;104:4473–4478. doi: 10.1073/pnas.0700435104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller LA, Gotway CA. Applied Spatial Statistics for Public Health Data. Wiley; New York: 2004. [Google Scholar]
- Walsh PD, Biek R, Real LA. Wave-like spread of Ebola Zaire. PLoS Biology. 2005;3:e371. doi: 10.1371/journal.pbio.0030371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman NK, Parker PG. Using parasites to infer population history: a new rationale for parasite conservation. Animal Conservation. 2005;8:175–181. [Google Scholar]
- Wimberly MC, Baer AD, Yabsley MJ. Enhanced spatial models for predicting the geographic distributions of tick-borne pathogens. International Journal of Health Geographics. 2008;7:15. doi: 10.1186/1476-072X-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winker K, McCracken KG, Gibson DD, et al. Movements of birds and avian influenza from Asia into Alaska. Emerging Infectious Diseases. 2007;13:547–552. doi: 10.3201/eid1304.061072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JK, Ignacio CC, Torriani F, et al. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. Journal of Virology. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse ME. Population biology of emerging and re-emerging pathogens. Trends in Microbiology. 2002;10:S3–7. doi: 10.1016/s0966-842x(02)02428-9. [DOI] [PubMed] [Google Scholar]
- Wylie JL, Cabral T, Jolly AM. Identification of networks of sexually transmitted infection: a molecular, geographic, and social network analysis. Journal of Infectious Diseases. 2005;191:899–906. doi: 10.1086/427661. [DOI] [PubMed] [Google Scholar]
- Xia Y, Bjornstad ON, Grenfell BT. Measles metapopulation dynamics: a gravity model for epidemiological coupling and dynamics. American Naturalist. 2004;164:267–281. doi: 10.1086/422341. [DOI] [PubMed] [Google Scholar]