Abstract
Protein arginine methyltransferases (PRMTs) catalyze the post-translational methylation of arginine residues. PRMT1 is the predominant mammalian isozyme, and is responsible for generating the majority of the asymmetrically dimethylated arginine found in vivo. The dysregulation of this enzyme has been implicated in heart disease and cancer; thus, its inhibition would be useful in the treatment of these diseases. Herein, we describe the most potent PRMT1 inhibitor described to date. This compound, denoted C21, is a chloroacetamidine-containing peptide that is able to irreversibly bind and inactivate the enzyme selectively. We have also shown that the coactivator activity of PRMT1 is selectively inhibited by the compound in cellulo.
Keywords: Protein Arginine Methyltransferase, Citrulline, Cl-amidine, transcription, inactivator
Over the last decade, arginine methylation has emerged as an important post-translational modification (PTM), and dysregulation of this process appears to contribute to the onset and/or progression of multiple human pathologies, including heart disease and cancer.[1–3] The Protein Arginine Methyltransferases (PRMTs) catalyze this modification, using S-adenosyl methionine as the methyl donor. To date, ≥10 putative and confirmed PRMT isozymes have been described in humans, and these enzymes have been shown to contribute to the regulation of numerous physiological processes, including RNA splicing, nuclear-cytoplasmic shuttling, and transcriptional regulation.[1–3]
Among the PRMTs, PRMT1 is arguably the best characterized enzyme at both the molecular and physiological levels. For example, its structure has been determined at atomic resolution,[4] and the enzyme has been shown to utilize a rapid equilibrium random mechanism, with the formation of dead-end EAP and EBQ complexes, to catalyze the formation of mono- and asymmetrically dimethylated arginine (ADMA) in a partially processive fashion.[5, 6] In vivo, this enzyme has been shown to catalyze the methylation of histone H4 at arginine 3, a modification that is associated with the increased transcription of various genes under the control of several nuclear receptors (e.g. the estrogen receptor (ER)), p53, and NF-κB.[2, 3] Additionally, PRMT1 methylates ERα at R260, and this modification results in the formation of a cytoplasmic complex that activates signal transduction cascades mediated by MAPK, Akt, and PKC.[7]
Because our understanding of the role that PRMT1 plays in human cell signaling is still limited, and because of its relevance to human disease, our efforts have been focused on characterizing the molecular mechanism of PRMT1 catalysis and developing inhibitors targeting this enzyme.[5, 6, 8] Our efforts have also been focused on developing inhibitors for the Protein Arginine Deiminases (PADs), a separate family of arginine modifying enzymes whose activity is also dysregulated in a variety of human diseases.[9] These latter efforts have been quite successful and led to the development of F- and Cl-amidine, which are highly potent fluoro- and chloroacetamidine-based PAD inactivators (Figure 1).[10, 11]
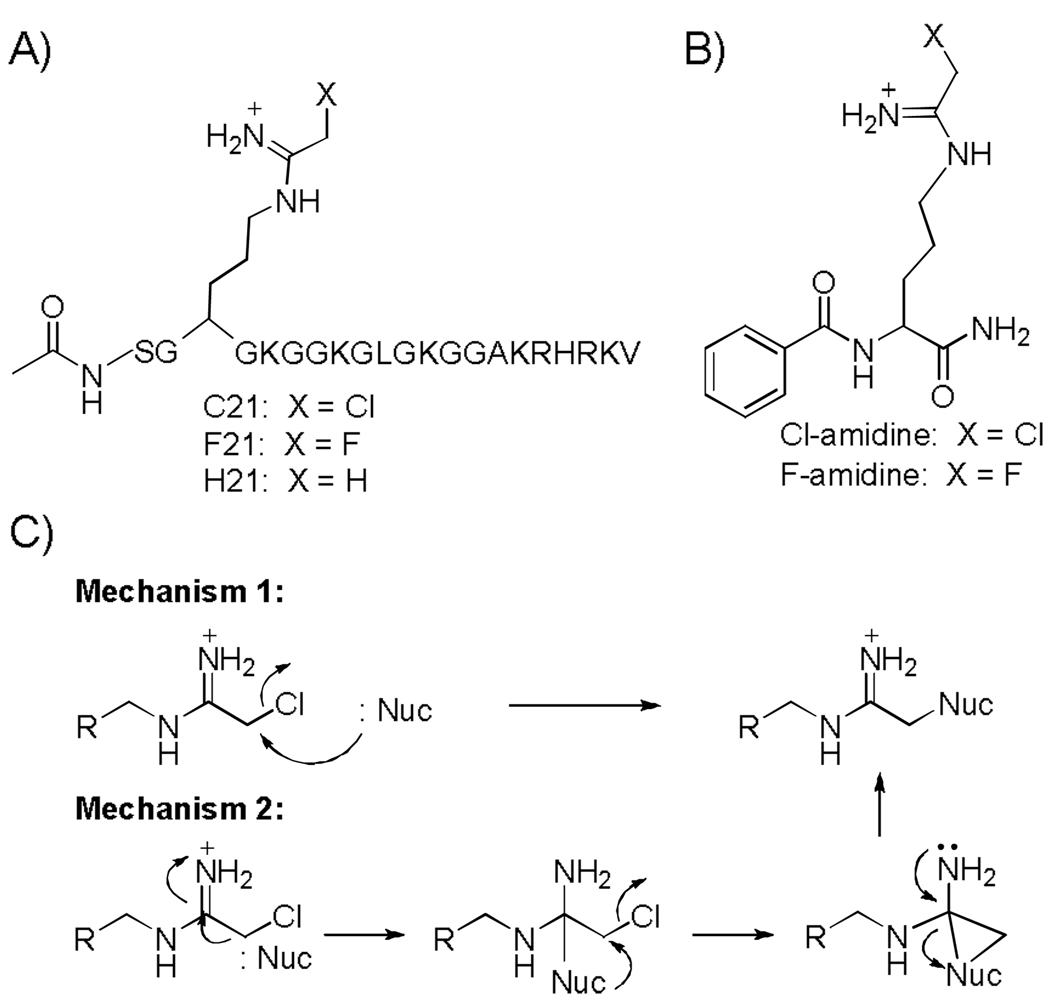
Figure 1.
Structures of haloacetamidine-based PRMT and PAD inactivators. A) Structures of C21 and F21. B) Structures of the PAD inactivators F- and Cl-amidine. C) Possible mechanisms of PRMT inhibition.
Because PADs and PRMTs both modify guanidinium groups, we reasoned that one or both of these compounds might also inactivate the PRMTs through one of two mechanisms (Figure 1). Although inhibition studies with F-[11] and Cl-amidine demonstrated that these compounds were rather weak PRMT1 inhibitors (Table 1; Figure S1), we were intrigued by the fact that the IC50 value for Cl-amidine was significantly lower than that obtained for F-amidine, suggesting that the greater reactivity of the chloroacetamidine moiety might facilitate reaction with a nucleophile in the active site of PRMT1.
Table 1.
IC50 values for PRMT and PAD inhibitors and inactivators
| Compound | IC50 for PRMT1 (µM) |
IC50 for PRMT3 (µM) |
IC50 for PRMT4 (µM) |
IC50 for PRMT6 (µM) |
IC50 for PAD4 (µM) |
|---|---|---|---|---|---|
| C21 | 1.8 ± 0.1 | > 500 | > 500 | 8.8 ± 0.5 | 145 ± 20 |
| Cl-amidine | 225 ± 10 | > 500 | > 500 | > 500 | 5.9 ± 0.3 |
| F21 | 94 ± 17 | 109 ± 28 | > 500 | > 250 | 117 ± 14 |
| F-amidine | > 500 | > 500 | > 500 | > 500 | 21.6 ± 2.1 |
If this hypothesis were correct, we reasoned that the poor potency of F- and Cl-amidine may well be due to a lack of recognition elements specific for PRMT1. Consistent with this possibility is the fact that shorter peptides lacking distal positively charged residues are relatively poor PRMT1 substrates.[5] To test our hypothesis, we synthesized two 21 residue peptides, denoted F21 and C21, incorporating fluoro- and chloroacetamidine warheads (Figure 1). We chose this peptide sequence because it is based on the N-terminus of histone H4, a PRMT1 substrate, and the parent peptide, i.e. AcH4-21, is an excellent substrate for this enzyme, and it is regiospecifically methylated at arginine 3.[5] The syntheses of F21 and C21 are described in detail below.
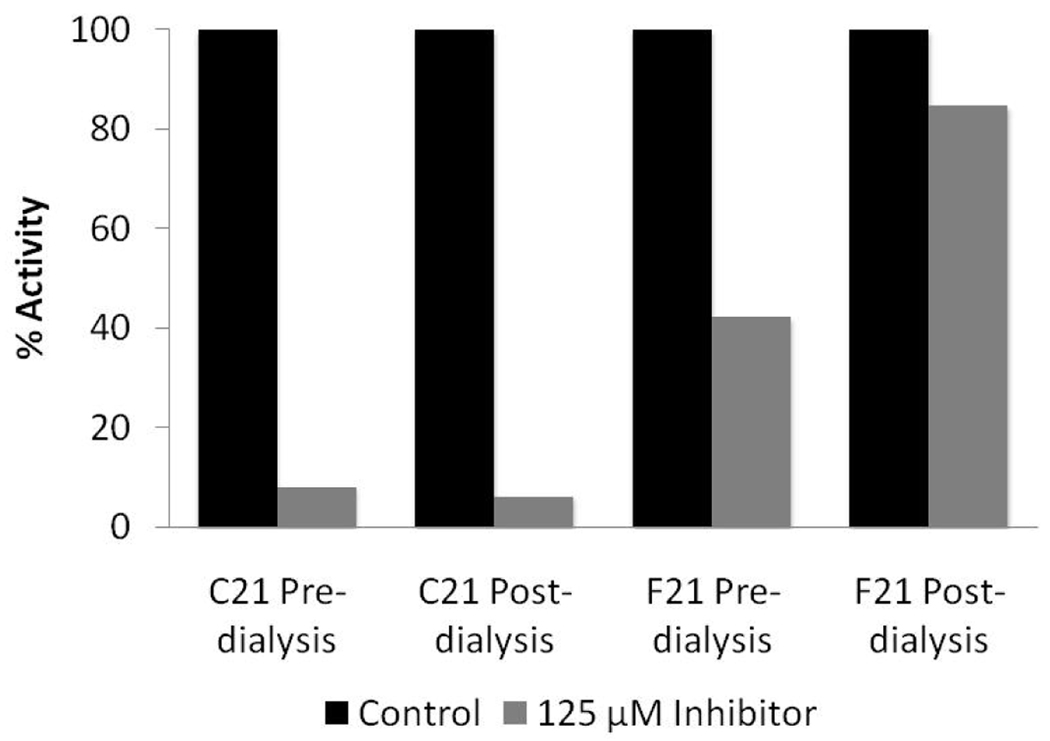
To begin to evaluate their ability to inhibit PRMT1, IC50 values were determined and compared to those obtained for F- and Cl-amidine (Table 1). The results indicate that F21 and C21 are at least 125-fold more potent than either F- or Cl- amidine. Significantly, C21 is ~52-fold more potent than F21. Dialysis experiments were then performed to differentiate between reversible and irreversible inhibition. For these experiments, F21 and C21 (125 µM final) were incubated with PRMT1 for 1 h to allow for enzyme inactivation. After this incubation, the enzyme inhibitor complex, as well as controls incubated in the absence of inhibitor, was dialyzed for 24 h, at which point the residual enzyme activity was measured using our gel-based assay system. As depicted in Figure 2, activity was only observed for the enzyme incubated with F21, indicating that C21, but not F21, irreversibly inactivates PRMT1. Consistent with this conclusion is the fact that a fluorescein-tagged version of C21, but not F21, covalently labels PRMT1 (manuscript in preparation). Further inhibition studies revealed that F21 is a competitive inhibitor (Kis = 1.8 ± 0.9 µM).
Figure 2.
C21, but not F21, irreversibly inactivates PRMT1. For these experiments, 125 µM F21 or C21 were incubated with PRMT1 for 1 h to allow for enzyme inactivation. After this incubation, the enzyme inhibitor complex, as well as controls incubated in the absence of inhibitor, were dialyzed for 24 h. Residual activity measurements were made prior to and after dialysis.
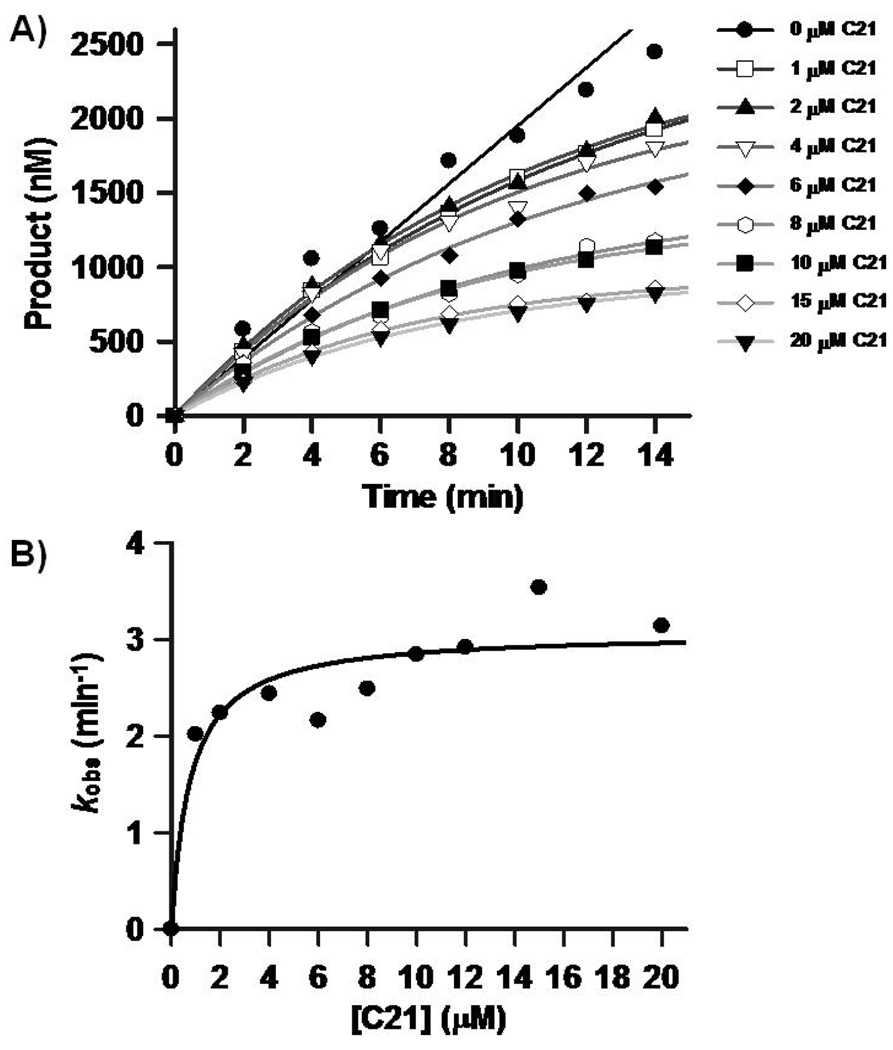
Further analyses with C21, revealed that this compound inactivates PRMT1 in a time- and concentration-dependent fashion (Figure 3). Plots of the pseudo first order rate constants of inactivation versus C21 concentration are saturatable, consistent with a 2 step mechanism of inactivation. The KI, kinact, and kinact/KI values are ≤0.8 ± 0.4 µM, 3.1 ± 0.2 min−1, and 4.6 × 106 min−1M−1. Although the specific residue modified by this compound is not known, the fact that higher substrate concentrations can protect against inactivation (Figure S2), strongly suggests that enzyme inactivation is due to the modification of an active site residue.
Figure 3.
In vitro evaluation of a PRMT1-selective inactivator. A) Progress curves depicting product formation versus time at increasing concentrations of C21. B) Pseudo first order rates of inactivation, i.e. kobs, versus C21 concentration.
To probe the selectivity of this compound, IC50 values were determined for CARM1 (coactivator associated arginine methyltransferase 1 aka PRMT4), a PRMT that is also known to play key roles in mammalian transcriptional regulation,[1–3] as well as PRMTs 3 and 6, the latter of which is known to possess transcriptional corepressor activity.[12, 13] The results indicate that C21 is specific for PRMT1, as demonstrated by the fact that the IC50 values for PRMT3 and CARM1 are >500 µM (Table 1). This high selectivity is consistent with the fact that the substrate specificities of PRMTs 1 and 3 differ substantially,[14] and CARM1 preferentially methylates the N-terminus of histone H3[1–3], whereas PRMT1 preferentially methylates histone H4. Although C21 also inhibits PRMT6 with micromolar potency, this compound retains a distinct ~5-fold preference for PRMT1 (Table 2).
Table 2.
Fold selectivity of C21
| Enzyme | Fold Selectivity of C21 |
|---|---|
| PRMT1 | 1 |
| PRMT3 | > 250 |
| PRMT4/CARM1 | > 250 |
| PRMT6 | 4.9 |
| PAD4 | 80.6 |
Given that the PADs are also inactivated by haloacetamidine-containing compounds, the kinact/KI value for PAD4 was determined to assess inter-enzyme selectivity trends. The results indicate that C21 also inactivates PAD4 via a two-step mechanism. However, the kinact/KI value for this compound is only 300 M−1min−1, indicating that, with respect to PAD4, C21 inactivates PRMT1 with >15,000-fold selectivity. These results are quite significant because they indicate that some measure of inter-enzyme selectivity can be achieved by varying either the identity of the warhead or its context.
We additionally demonstrated that C21 can be used to selectively inhibit PRMT1 in cellulo (Figure 4). For these experiments, 293T cells were transfected with a luciferase reporter gene controlled by GAL4 response elements, an expression vector encoding the GAL4-DNA binding domain fused to the C-terminal end of the steroid receptor coactivator GRIP1 (GRIP1C), and vectors encoding either, PRMT1, a catalytically deficient PRMT1 mutant (PRMT1mut), CARM1, or a catalytically deficient CARM1 mutant (CARM1mut). Cells were then treated with 1 to 10 µM of C21 that was transported into cells using a peptide transfection reagent. The results reveal that the coactivator activity of PRMT1 is almost completely inhibited even at concentrations as low as 1 µM C21. In contrast, no inhibition of CARM1 activity was observed. The fact that no coactivator activity was observed with the catalytically deficient CARM1 mutant indicates that, in this assay, CARM1 coactivator activity requires its methyltransferase activity; thus, the lack of an effect of C21 on the coactivator activity of CARM1 is not due to an artifact of the assay. Additionally, no inhibition was observed in these assays with H21, a control peptide that lacks the chloro group; the IC50 for this compound with PRMT1 is 23 ± 2 µM. The lack of an effect with the control peptide, coupled with the lack of an effect in the CARM1 assays, demonstrates that the effect of C21 on PRMT1 activity in cellulo is not due to toxicity or a non-specific effect on the assay.
Figure 4.
In vivo evaluation of a PRMT1-selective inactivator. C21 selectively inhibits PRMT1, but not CARM1 activity in cellulo. 293T cells in 12-well plates were transfected with 0.1 µg of GK-1 luciferase reporter plasmid (which has GAL4 response elements) and 0.01 µg of a plasmid encoding GAL4 DBD fused to GRIP1C, and 0.3 µg of each of the plasmids encoding PRMT1, PRMT1 E153Q methyltransferase deficient mutant, or CARM1. To test the bioavailability of C21, cells were treated with 1 µM to 10 µM C21 that was transported into cells using the Chariot Protein Delivery Reagent (Active Motif). Luciferase activity was measured after 40 h.
In summary, the studies described herein highlight the versatility of the haloacetamidine warhead as a tool for targeting arginine modifying enzymes. Furthermore, the development of C21 is significant because in comparison to other known PRMT1 inhibitors, for example, arginine methyltransferase inhibitor 1 (IC50 = 350 ± 36 µM)[8, 15] and an in situ generated bisubstrate analogue (IC50 = 18.5 ± 4.2 µM),[8] C21 is the most potent and selective PRMT1 inhibitor described to date. The fact that C21 can selectively inhibit PRMT1 activity in cellulo suggests that this compound will be useful for discerning the role of PRMT1 in transcriptional control, nuclear-cytoplasmic transport, and RNA splicing. Additionally, the covalent nature of the interaction between C21 and PRMT1 indicates that it will be possible to develop activity based proteomic probes that selectively modify PRMT1, similarly to those that have been developed for PAD4 and other enzymes involved in human cell signaling.[16, 17] Such compounds will be useful for identifying the factors that regulate in vivo PRMT1 activity. Finally, by incorporating the chloroacetamidine warhead into libraries, it will be possible to characterize both the substrate specificity of the enzyme and identify inhibitors with even greater potencies and selectivities.
Experimental Section
Chemicals
N-α-Fmoc-amino acids, HOBt, HBTU, dithiothreitol (DTT), N-(2-hydroxyethyl)piperazine-N’-2-ethane-sulfonic acid (HEPES), and ethylene-dinitrilo-tetra-acetate (EDTA) were obtained from VWR. 14C-methyl-SAM and 14C-labeled bovine serum albumin (BSA) were obtained from Sigma and Perkin Elmer, respectively. PRMT4 (CARM1) was purchased from Millipore (Upstate). The syntheses of F- and Cl-amidine has been described previously.[10, 11]
Synthesis of C21
The AcH4-21(R3Orn(Dde)) peptide was synthesized using standard Fmoc solid phase peptide chemistry on Wang resin using a Rainin PS3 automated peptide synthesizer. Removal of the Dde-protecting group was accomplished by two 1 h incubations of the resin with 2% hydrazine in DMF. The resin containing free Orn was treated with ethylchloroacetimidate hydrochloride (4 eq.) and triethylamine (8 eq.) in DMF twice for 8 h. The resin was washed three times each with DMF, ethanol, and DCM. Cleavage and complete deprotection of C21 was accomplished by treatment with TFA/TIS/H2O (95/2.5/2.5) for 1 h. After the volatiles were removed, crude C21 was precipitated with ether, isolated by centrifugation, and purified by RP-HPLC using a H2O(0.05% TFA)/ACN(0.05% TFA) linear gradient. The identity of C21 was verified by MALDI-MS. Expected [C90H162ClN36O24]+ 2166.23; found 2166.66.
Synthesis of F21
The AcH4-21(R3Orn(Dde)) peptide, synthesized as described above, was treated twice with 2% hydrazine in DMF for 1 h to afford removal of the Dde-protecting group. The resin containing free Orn was treated with ethylfluoroacetimidate hydrochloride (4 eq.) and triethylamine (8 eq.) in DMF twice for 8 h. The methodology outlined above for C21 was then used to isolate pure F21. The identity of F21 was verified by MALDI-MS. Expected [C90H162FN36O24]+ 2150.25; found 2150.46.
Synthesis of H21
The AcH4-21(R3Orn(Dde)) peptide, synthesized as described above, was treated twice with 2% hydrazine in DMF for 1 h to afford removal of the Dde-protecting group. The resin was then treated with ethylacetimidate hydrochloride (4 eq.) and triethylamine (8 eq.) in DMF twice for 8 h. The methodology outlined above for C21 was then used to isolate pure F21. The identity of F21 was verified by MALDI-MS. Expected [C90H163N36O24]+ 2132.26; found 2132.62.
Protein Purification
The purification of recombinant human PRMT1 has been previously described.[5] His-tagged PRMTs 3[4] and 6[13] were purified analogously. Briefly, expression constructs, encoding N-terminal hexa-histidine tagged PRMTs were expressed in E. coli BL21(DE3) cells and purified by immobilized metal ion affinity chromatography on a nickel(II) chelating sepharose column. Anion exchange chromatography was used to further purify the enzyme.
IC50 Assays
IC50 values for PRMT1 were determined as previously described.[5, 8] Briefly, various inhibitor concentrations were incubated with 200 nM PRMT1 and 15 µM 14C-methyl-SAM in Assay Buffer (50 mM HEPES pH 8.0, 50 mM NaCl, 1 mM EDTA and 0.5 mM dithiothreitol) at 37 °C for 10 min. The reaction was initiated by the addition of peptide substrate (i.e., 25 µM AcH4-21) and quenched with tris-tricine gel loading dye after 15 min. Samples were run on 16.5% tris-tricine polyacrylamide gels and incorporated radioactivity was quantified by phosphorimage analysis (Molecular Dynamics). IC50 values were determined by fitting the data thus obtained to equation 1,
| (eq 1) |
using the GraFit version 5.0.11 software package,[18] where [I] is the concentration of inhibitor and IC50 is the concentration of inhibitor that yields half-maximal activity.
IC50 values for PRMTs 3, 4, and 6 were determined in an analogous fashion. Briefly, for PRMT3, various inhibitor concentrations were incubated with PRMT3 (500 nM) and 14C-methyl-SAM (15 µM) in Assay Buffer at 37 °C for 10 min. The reaction was initiated by the addition of peptide substrate (i.e., 250 µM RGG[4]) and quenched with tris-tricine gel loading dye after 30 min. For CARM1/PRMT4, various inhibitor concentrations were incubated with 150 nM CARM1/PRMT4 and 15 µM 14C-methyl-SAM in Assay Buffer at 30 °C for 10 min. The reaction was initiated by the addition of 10 µM histone H3 and quenched with tris-tricine gel loading dye after 30 min. For PRMT6, various inhibitor concentrations were incubated with 500 nM PRMT6 and 15 µM 14C-methyl-SAM in Assay Buffer at 37 °C for 10 min. The reaction was initiated by the addition of substrate (i.e., 30 µM histone H3) and quenched with tris-tricine gel loading dye after 30 min.
The ability of F21 and C21 to inhibit PAD4 was determined as previously described.[10, 11] Briefly, PAD4 (0.2 µM) was preincubated in a buffer containing 100 mM HEPES (pH 7.6), 50 mM NaCl, 10 mM CaCl2, and 0.25 mM TCEP with different amounts of either F21 or C21 (0–200 µM) for 15 min before initiating the reaction with Benzoyl Arginine Ethyl Ester (BAEE; 10 mM final). The reactions were quenched after 15 min of incubation and Cit production was measured using a previously established method.[19, 20] Initial rates were fit to equation 1 using GraFit (version 5.0.11).[18]
Note that in all cases, experiments were performed at least in duplicate and activity generally agreed within 20% standard deviation.
Time Course Inactivation Assays
Progress curves were generated by incubating 25 µM AcH4-21, 15 µM 14C-SAM, and the specified concentration of inhibitor in Assay Buffer at 37 °C for 10 min. The reaction was initiated by the addition of 200 nM PRMT1 and quenched with tris-tricine loading dye at the appropriate time. Samples were processed as described above. The data obtained for F21 was fit by linear regression. As the data obtained with C21 were nonlinear, the progress curves were generated by fitting the data to equation 2,
| (eq 2) |
using the GraFit version 5.0.11 software package,[18] where vi is the initial velocity, kobs.app is the apparent pseudo first order rate constant for inactivation, and t is time. Equation 3,
| (eq 3) |
was used to correct the apparent pseudo-first-order inactivation rate constants, obtained from this analysis, for substrate concentration and the pseudo-first-order inactivation rate constants, i.e. kobs, thus obtained, were plotted against the tested inhibitor concentrations. As the data are consistent with a two step mechanism of inactivation, they were fit to equation 4,
| (eq 4) |
using the GraFit version 5.0.11 software package,[18] where KI is the concentration of inactivator that yields half-maximal inactivation, kinact is the maximal rate of inactivation, and [I] is the concentration of inactivator.
Progress curves for PAD4 were determined at various concentrations of C21 by pre-incubating BAEE (10 mM) in the same buffer as described above. The reactions were initiated by the addition of PAD4 (0.2 µM). Aliquots (60 µL) of the reactions were removed and flash frozen in liquid nitrogen at various time points. The amount of Cit was quantified using the same procedure described above. The inactivation constants were determined using the methodology outlined above for PRMT1.
Substrate Protection Assays
To assess the ability of substrate to protect against inhibition/inactivation, 20 µM C21 or 50 µM F21, and no inhibitor controls, were incubated in Assay Buffer in the presence of 14C-SAM (15 µM final), and either 25 µM or 100 µM of the AcH4-21 peptide, for 10 min at 37 °C. The reaction was initiated by the addition of 200 nM PRMT1 and quenched with tris-tricine loading dye at the indicated times. The samples were the processed using the methodology outlined above, and the data acquired were fit to either a straight line or to equation 2, using GraFit version 5.0.11. [18]
Dialysis Experiments
For dialysis experiments, 10 µM PRMT1 was incubated for 1 h at 37 °C with 125 µM C21 or F21 in Assay Buffer for a total reaction volume of 150 µL. Control reactions were only incubated with reaction buffer. The samples were then dialyzed for 24 h at 4 °C against 2 L of PRMT1 long term storage buffer (100 mM HEPES pH 8.0, 200 mM NaCl, 1 mM DTT, 2 mM EDTA, 10% glycerol). Residual activity assays were performed on pre- and post dialysis samples. Activity was measured in the presence of 25 µM AcH4-21, 15 µM 14C-SAM, and Assay Buffer at 37 °C for 15 min. The reactions were quenched with tris-tricine loading dye and processed as described above. The amount of product formed in the control reaction was determined to be 100% and used to determine the percent activity remaining in the inhibited reactions.
F21 Inhibition Assays
To determine the inhibition constants and the mechanism by which F21 inhibits PRMT1, the steady state kinetic parameters for the methylation of the AcH4-21 peptide were determined in the absence or presence of increasing concentrations of F21. For these assays, the AcH4-21 peptide was the varied substrate and the concentration of SAM was fixed (15 µM final). SAM, F21, and the AcH4-21 peptide were preincubated for 10 min at 37 °C, prior to the addition of PRMT1 (200 nM final) to initiate the reaction. The reactions were allowed to proceed for 15 min, at which point the assays were quenched with tris-tricine loading dye and processed as described above. The acquired data were fit to equations representative of linear competitive inhibition (eq 5), linear noncompetitive inhibition (eq 6), linear mixed inhibition (eq 7), and linear uncompetitive inhibition (eq 8), using a nonlinear least fit squares approach by GraFit version 5.0.11.
| (eq 5) |
| (eq 6) |
| (eq 7) |
| (eq 8) |
In the equations, Kii is the intercept Ki, and Kis is the slope Ki. The nature of the inhibition induced by F21 on PRMT1 was determined by the best fit of the data to equations 5 – 8. Visual inspection of the fits, and a comparison of the standard errors, was used to confirm these assignments.
Transient Transfection Assay
The transfection procedure, using Targefect F1 reagent (Targeting Systems), has been described previously.[21] The plasmids used in the transfection assay have been previously described: (i) Reporter plasmid: GK1-Luciferase;[21] (ii) PRMT1 protein expression plasmids: pSG5.HA-PRMT1 wild type and methyltransferase deficient E153Q mutant;[22] (iii) CARM1 protein expression plasmid: wild type and methyltransferase deficient E267Q mutant;[23] and (iv) Gal4DBD-GRIP1C, encoding the Gal4 DNA binding domain fused to the C-terminal domain of GRIP1.[23]
Briefly, 293T cells in 12-well plates were transfected with 0.1 µg of GK-1 luciferase reporter plasmid (which has GAL4 response elements) and 0.01 µg of a plasmid encoding GAL4 DBD fused to GRIP1C, and 0.3 µg of each of the plasmids encoding PRMT1, PRMT1 E153Q methyltransferase deficient mutant, or CARM1 E267Q methyltransferase deficient mutant. To test the bioavailability of C21, cells were treated with 1 µM to 10 µM C21 that was transported into cells using the Chariot Protein Delivery Reagent (Active Motif)a Luciferase activity was measured after 40 h. The experiments were each performed in duplicate and representative data from one of 3 separate experiments are depicted in Figure 4. The error bars depict the average deviation.
Supplementary Material
Footnotes
This work was supported by funds provided by the University of South Carolina and in part by NIH grant GM079357 to PRT and DK055274 to MRS
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.Copeland RA, Solomon ME, Richon VM. Nat Rev Drug Discov. 2009;8:724. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 2.Bedford MT, Richard S. Mol Cell. 2005;18:263. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Bedford MT, Clarke SG. Mol Cell. 2009;33:1. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Cheng X. Structure. 2003;11:509. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Biochemistry. 2007;46:13370. doi: 10.1021/bi701558t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obianyo O, Osborne TC, Thompson PR. Biochemistry. 2008;47:10420. doi: 10.1021/bi800904m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L. Mol Cell. 2008;31:212. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 8.Osborne T, Roska RL, Rajski SR, Thompson PR. J Am Chem Soc. 2008;130:4574. doi: 10.1021/ja077104v. [DOI] [PubMed] [Google Scholar]
- 9.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Curr Opin Drug Discov Devel. 2009;12:616. [PMC free article] [PubMed] [Google Scholar]
- 10.Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Thompson PR. Biochemistry. 2006;45:11727. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Y, Knuckley B, Lee YH, Stallcup MR, Thompson PR. J Am Chem Soc. 2006;128:1092. doi: 10.1021/ja0576233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Nature. 2007;449:933. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 13.Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. J Biol Chem. 2007 doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 14.Tang J, Gary JD, Clarke S, Herschman HR. J Biol Chem. 1998;273:16935. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
- 15.Cheng D, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT. J Biol Chem. 2004;279:23892. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 16.Jessani N, Cravatt BF. Curr Opin Chem Biol. 2004;8:54. doi: 10.1016/j.cbpa.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Luo Y, Knuckley B, Bhatia M, Thompson PR. J Am Chem Soc. 2006;128:14468. doi: 10.1021/ja0656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leatherbarrow RJ. Erathicus Software. UK: Staines; 2004. [Google Scholar]
- 19.Knipp M, Vasak M. Anal Biochem. 2000;286:257. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- 20.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Biochemistry. 2005;44:10570. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 21.Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Mol Cell Biol. 2002;22:3621. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Genes Dev. 2005;19:1466. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Science. 1999;284:2174. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.