Abstract
Background. Volunteer challenge studies have provided detailed data on viral shedding from the respiratory tract before and through the course of experimental influenza virus infection. There are no comparable quantitative data to our knowledge on naturally acquired infections.
Methods. In a community-based study in Hong Kong in 2008, we followed up initially healthy individuals to quantify trends in viral shedding on the basis of cultures and reverse-transcription polymerase chain reaction (RT-PCR) through the course of illness associated with seasonal influenza A and B virus infection.
Results. Trends in symptom scores more closely matched changes in molecular viral loads measured with RT-PCR for influenza A than for influenza B. For influenza A virus infections, the replicating viral loads determined with cultures decreased to undetectable levels earlier after illness onset than did molecular viral loads. Most viral shedding occurred during the first 2–3 days after illness onset, and we estimated that 1%–8% of infectiousness occurs prior to illness onset. Only 14% of infections with detectable shedding at RT-PCR were asymptomatic, and viral shedding was low in these cases.
Conclusions. Our results suggest that “silent spreaders” (ie, individuals who are infectious while asymptomatic or presymptomatic) may be less important in the spread of influenza epidemics than previously thought.
Influenza virus is associated with substantial mortality and morbidity worldwide through seasonal epidemics and occasional emergence of novel strains that lead to pandemics [1]. Volunteer challenge studies have provided detailed data on viral shedding from the respiratory tract through the course of experimental influenza virus infections and the time lines and duration of illness after infection [2]. However, in volunteer challenge studies, the participants are typically young adults who have been screened to ensure that they have low levels of preexisting immunity to the influenza strain of interest; thus, the findings may not generalize to the overall population annually at risk for naturally acquired infections [2]. There are few data on viral shedding after naturally acquired influenza virus infection in a community setting. The proportion of infections that are asymptomatic or subclinical and the degree to which these are contagious, as well as the proportion of shedding that occurs prior to onset of symptoms, affects the potential impact of control measures. We studied trends in clinical illness and viral shedding associated with naturally acquired influenza virus infections in a community setting.
Methods
We conducted a community-based study of the effectiveness of face masks and hand hygiene to prevent influenza transmission in households during 2008 [3]. As part of that study, index cases in patients of all ages who presented with ⩾2 of a range of signs and symptoms consistent with influenza virus infection were recruited from outpatient clinics and private hospital emergency rooms from January through September 2008. A positive result for influenza virus infection on a rapid antigen test was used to determine eligibility of the index cases' households for additional follow-up. Index cases and household contacts were eligible regardless of preexisting conditions or treatment prescribed, but index cases who had household members with concurrent influenza-like illness were excluded [3, 4]. There was no recorded use of antiviral prophylaxis. Symptom diaries were provided to all household members at an initial home visit within 24–48 h of index case recruitment to record respiratory and systemic symptoms daily; 82% of initial home visits were conducted within 12 h of index case recruitment [3]. Compliance with symptom reporting was high; all symptom diaries were complete with the exception of 12 (0.6%) of 2170 entries. We provided each household with a digital tympanic thermometer and asked all household members to record their body temperature daily. Household members were followed up for ∼7 days to observe secondary infections. Pooled nose and throat swab (NTS) specimens were collected from all household members regardless of illness at the initial home visit and at 2 followup visits 3 (±1) and 6 (±2) days later. Because index cases are likely to have higher viral loads, given their positive result on a rapid test [5], and are less likely to have mild illness given that they sought outpatient care, all analyses here focus only on the household contacts to avoid selection bias.
Laboratory methods. NTS specimens from household contacts were tested by quantitative reverse transcription polymerase chain reaction (RT-PCR) to detect influenza A or B virus infection and determine molecular viral loads. Total nucleic acid was extracted from specimens by using the NucliSens easyMAG extraction system (bioMérieux, Boxtel, the Netherlands) according to the manufacturer's instructions. Twelve microliters of extracted nucleic acid was used to prepare complementary DNA by using an Invitrogen Superscript III kit (Invitrogen) with random primer as described elsewhere [6].
For detection of influenza A virus, 2 µL of complementary DNA was amplified in a LightCycler 2.0 system (Roche Diagnostics), with a total reaction-mix volume of 20 µL reaction containing FastStart DNA Master SYBR Green I Mix reagent kit (Roche Diagnostics), 4.0 mmol/L MgCl2 and 0.5 mmol/L of each primer. The forward primer (5′-CTTCTAACCGAGGTCGAAACG- 3′) and the reverse primer (5′-GGCATTTTGGACAAAKCGTCTA- 3′) were used for amplification of the matrix gene of influenza A virus [7]. Cycling conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 3 s, and 72°C for 12 s, with ramp rates of 20°C/s. At the end of the assay, polymerase chain reaction (PCR) products were subjected to a melting-curve analysis to determine the specificity of the assay. The lower limit of detection of the RT-PCR assay was 23 virus gene copies per reaction (ie, ∼900 copies/mL).
For detection of influenza B virus, forward (5′-GCATCTTTTGTTTTTTATCCATTCC) and reverse (5′-CACAATTGCCTACCTGCTTTCA) primers and 5′ nuclease probe (Fam-TGCTAGTTCTGCTTTGCCTTCTCCATCTTCT- TAMRA) were used for amplification of the matrix gene [8]. Testing was performed by using the TagMan EZ RT-PCR system. Core reagent kit (Applied Biosystems), with 0.8 µmol/L of forward and reverse primers and 0.2 µmol/L of probe in a total reaction volume of 25 µL, comprising 4 µL of nucleic acid extract. Amplification and detection was performed on an ABI StepOneTM real-time PCR system (Applied Biosystems) under the following conditions: initial hold at 50°C for 20 min and 95°C for 15 min, followed by 45 cycles at 95°C for 15 s and 60°C for 1 min.
NTS specimens were additionally tested by quantitative viral dilutions to detect median tissue culture infectious dose (TCID50) and determine replicating viral load. Madin-Darby canine kidney (MDCK) cells were grown on microtiter plates. The cells were rinsed in serum-free minimum essentialmedium (Gibco). The NTS specimen was diluted initially by 1 in 5 and then in 10-fold steps in serum-free minimum essential medium containing tosylsulfonyl phenylalanyl chloromethyl ketonetreated trypsin (2 µg/mL) (Sigma Aldrich). One hundred microliters of the undiluted NTS specimen as well as each of the specimen dilutions was added in quadruplicate to the MDCK cell monolayers. Medium alone was added to control cells. An additional 100 µL of serum-free minimum essential medium with 2 µg/mL trypsin was added to each well, and the plates were incubated at 33°C for 7 days. The plates were evaluated for cytopathic effect daily. The TCID50 was determined according to the Reed and Muench Method. The lower limit of detection was ∼100.3 TCID50.
Statistical analysis. We calculated daily scores grouped as systemic signs and symptoms (fever of ⩾37.8°C, headache, and myalgia), upper respiratory symptoms (sore throat and runny nose), and lower respiratory symptoms (cough and phlegm) by summing the presence versus absence of each symptom or sign and dividing by 3, 2, and 2, respectively [2]. We define acute respiratory illness (ARI) as ⩾2 of the 7 symptoms listed above [9]. We plotted average symptom scores by time since ARI onset, which was defined as the first day when the subject reported ⩾2 of the 7 symptoms. We plotted quantitative viral loads by time since ARI onset and calculated daily geometric means, imputing half the lower limit of detection (ie, 450 copies/ mL and 0.15 log10 TCID50) for the undetectable values. As a sensitivity analysis, we plotted viral shedding by time since onset of fever of ⩾37.8°C. We investigated the associations between tympanic temperature and viral shedding, and between symptom scores and viral shedding, by day since ARI onset.
We used a Bayesian approach to fit alternative parametric forms to the viral shedding trajectories and selected between models using the Bayesian information criterion [10]. We used the models to quantify the proportion of infectiousness remaining at ARI onset and 1, 2, or 3 days after onset, assuming lognormal, Weibull, or gamma-form associations between molecular viral shedding and infectiousness. Additional technical details are provided in the Appendix, which is not available in the print edition of the Journal. All analyses were conducted with R software (version 2.7.1; R Development Core Team) [11] and WinBUGS software (version 1.4) [12]. Additional information about the study design, raw data from the study, and R syntax to permit reproducible statistical analyses are available on the authors' Web site at http://www.hku.hk/bcowling/influenza/HK_NPI_study.htm.
Results
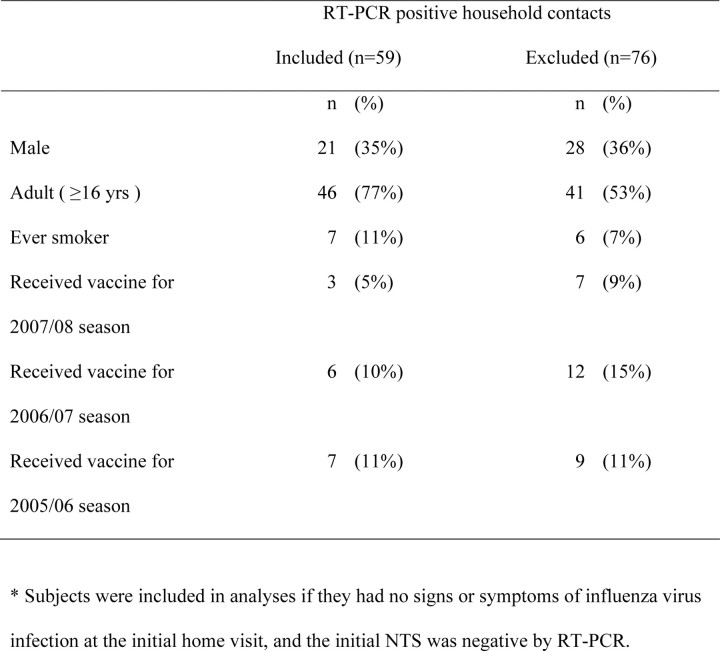
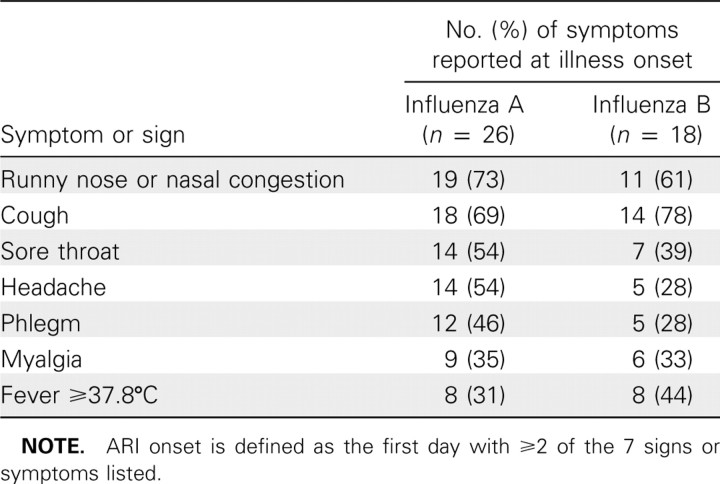
We followed up 1015 household contacts in 322 households. A total of 135 (13%) contacts were confirmed with RT-PCR to have influenza virus infection. Because we recorded symptoms prospectively and were particularly interested in patterns of viral shedding and symptoms around illness onset, for this analysis we excluded 59 contacts for whom the first NTS specimens collected were RT-PCR positive regardless of whether ARI was reported at the first home visit, and an additional 17 contacts who met the criteria for ARI onset at the first home visit with influenza virus infection subsequently confirmed with RT-PCR. Characteristics of the excluded 76 contacts were similar to those of the 59 retained contacts (Table 1). Of the 59 secondary infections analyzed here, 16 were caused by influenza A/H1N1 viruses, 17 by influenza A/H3N2 viruses, and 1 by both influenza A/H1N1 and A/H3N2 confection, with the remainder by influenza B virus. Forty-six (78%) of 59 subjects were persons aged 16 years or older. At ARI onset, the most common symptoms were cough, nasal congestion, runny nose, and sore throat (Table 2). Of the 34 subjects infected with influenza A viruses, 15 (44%) reported a temperature of ⩾37.8°C on at least 1 day over the course of illness, whereas 21 (62%) reported having a temperature of ⩾37.8°C on at least 1 day, taking antipyretic medication, or both. The corresponding proportions for influenza B virus infections were 8 (32%) of 25 subjects and 13 (52%) of 25 subjects. Thirty (51%) of 59 subjects reported seeking medical care.
Table 1.
Comparison of Characteristics of 59 Secondary Influenza Virus Infections with 76 Excluded Infections
Table 2.
Initial Symptoms and Signs of Naturally Acquired Influenza A and B Virus Infections Reported at Acute Respiratory Illness (ARI) Onset
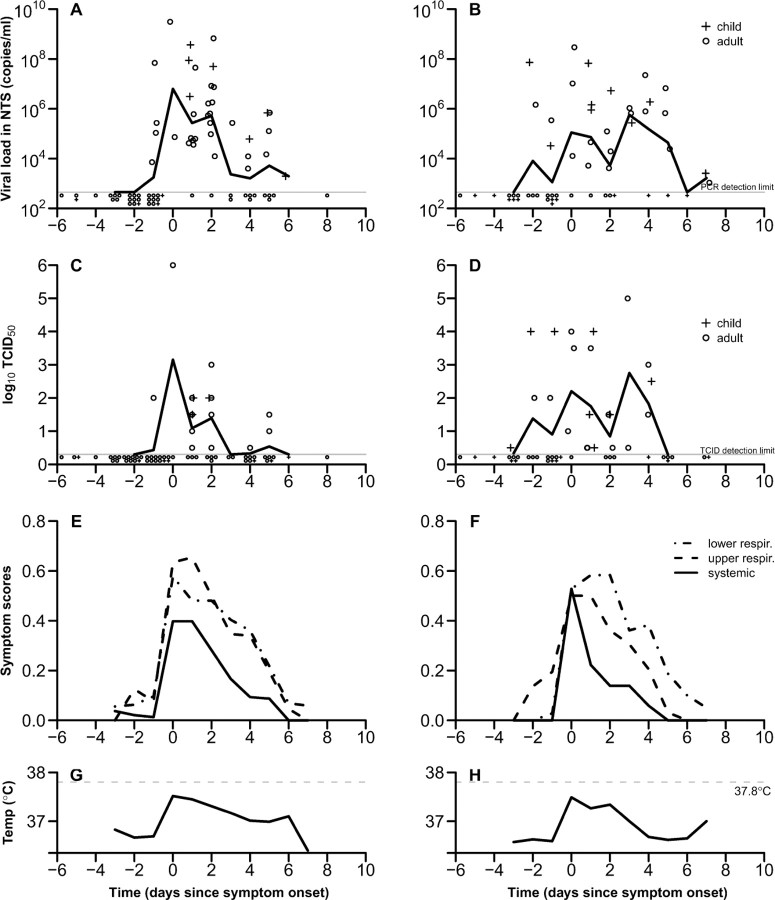
Peak molecular viral shedding for influenza A virus infection as assessed with RT-PCR occurred on the day of ARI onset, followed by a steady decrease in viral shedding through the following 7 days (Figure 1). The pattern of viral shedding for A/H1N1 and for A/H3N2 subtypes were similar (data not shown). Viral shedding was detected with RT-PCR in 4 (27%) of 15 subjects (95% confidence interval [CI], 8%–55%), with a specimen collected 1 day before ARI onset. Influenza B viral shedding was more variable over time and a clear peak was not observed; initial shedding was detected at 1–2 days before ARI onset in 4 (29%) of 14 subjects (95% CI, 8%–58%) and persisted for ∼6 days before subsiding. We did not have sufficient sample size to explore differences in viral shedding between children and adults.
Figure 1.
Patterns of viral shedding and symptoms and signs in naturally acquired influenza A and B virus infections by day relative to acute respiratory illness (ARI) onset (day 0). Top row: Viral shedding for children (plus signs) and adults (circles), and the geometric mean viral shedding (solid lines) of (A) 26 individuals with influenza A and (B) 18 individuals with influenza B virus infections. The lower limit of detection of the reversetranscription polymerase chain reaction assay was ∼900 copies/mL (gray line). Second row: Median tissue culture infectious dose (TCID50) of specimens collected from children (plus signs) and adults (circles), and the geometric mean TCID50 of (C) influenza A and (D) influenza B. Third row: Symptoms and signs for subjects with (E) influenza A and (F) influenza B virus infection are split into lower respiratory (dotted line), upper respiratory (dashed line), and systemic (solid line). Bottom row: Mean tympanic temperature associated with (G) influenza A and (H) influenza B virus infections. ARI onset is defined as at the first day with ⩾2 of the 7 signs or symptoms listed in Table 2. Individuals with asymptomatic or subclinical infections were excluded. NTS, nose and throat swab specimen.
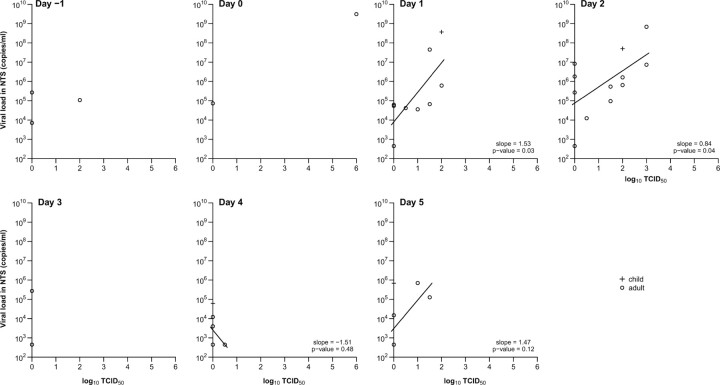
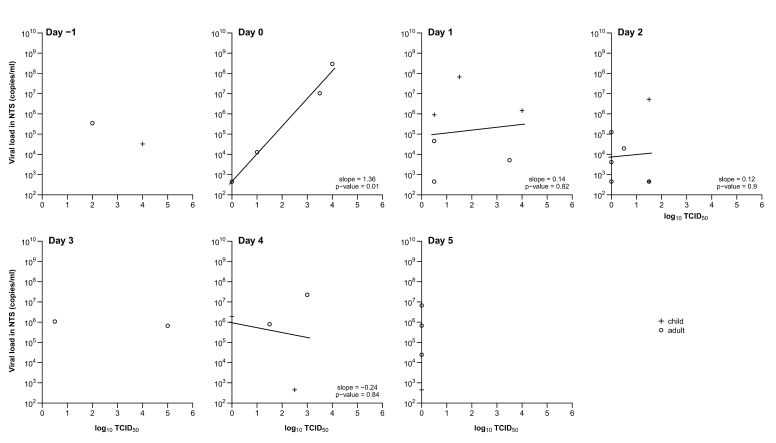
In influenza A virus infections the replicating viral load determined by viral culture peaked on the day of ARI onset and steadily decreased over ∼5 days (Figure 1). For influenza virus B infections, the TCID50 levels initially peaked on the day of ARI onset and were more variable over time. Molecular viral shedding was most closely correlated with replicating viral load on the day after ARI onset. As replicating viral load decreased more rapidly with time, the association between molecular viral shedding and replicating viral load became weaker later in the course of illness (Figures 2 and 3). Replicating viral load determined with viral culture was detected in 1 adult with influenza A infection and 2 adults and 3 children with influenza B infection before ARI onset.
Figure 2.
Association between replicating (by median tissue culture infectious dose [TCID50]) and molecular (by reverse-transcription polymerase chain reaction) influenza A viral shedding by day since acute respiratory illness (ARI) onset for children (plus signs) and adults (circles). Linear regression lines are plotted where there are ⩾3 points. ARI onset is defined as at the first day with ⩾2 of the 7 symptoms or signs listed in Table 2. NTS, nose and throat swab specimen.
Figure 3.
Association between replicating (by median tissue culture infective dose [TCID50]) and molecular (by reverse-transcription polymerase chain reaction) influenza B viral shedding by day since acute respiratory illness (ARI) onset for children (plus signs) and adults (circles). Linear regression lines are plotted where there are ⩾3 points. ARI onset is defined as at the first day with ⩾2 of the 7 symptoms or signs listed in Table 1. NTS, nose and throat swab specimen.
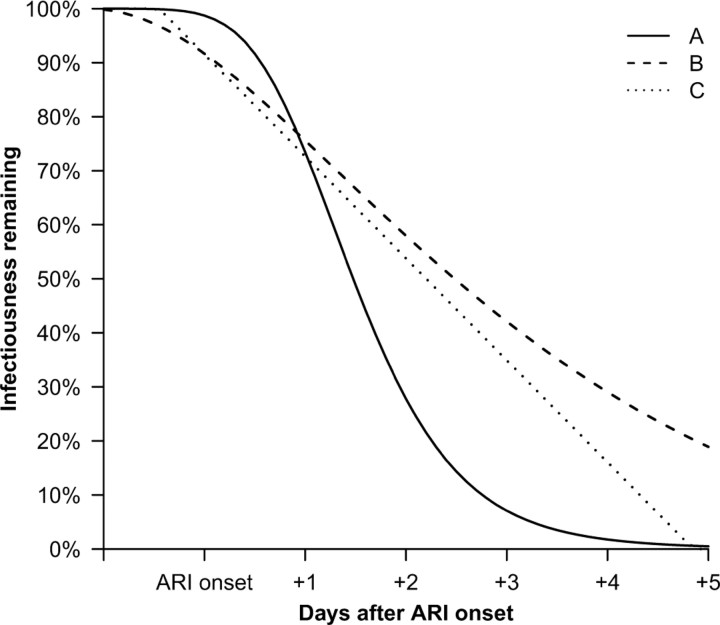
We found that a modified lognormal form provided a good fit to the molecular viral shedding patterns for influenza A virus infections. If infectiousness were proportional to molecular viral shedding, most infectiousness would occur within 2–3 days of ARI onset, whereas if infectiousness were proportional to log10 molecular viral shedding or presence of detectable viral RNA, infectiousness would persist for longer periods (Figure 4). We did not find any parametric forms that provided a good fit to the influenza B molecular viral shedding patterns, or replicating viral load for influenza A or B measured by TCID50.
Figure 4.
Proportion of infectiousness remaining by time since acute respiratory illness (ARI) onset in the course of naturally acquired influenza A virus infection. Infectiousness assumed proportional to (A) molecular viral shedding, (B) log10 molecular viral shedding, and (C) presence of molecular viral shedding in excess of 900 copies/mL under the fitted curve.
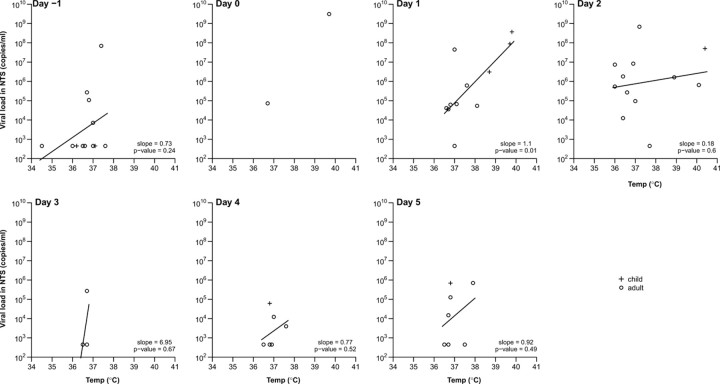
In both influenza A and B virus infections, mean tympanic temperature and trends in average scores for systemic signs and symptoms peaked at the day of ARI onset and then steadily subsided to baseline after 3–5 days (Figure 1). On average, upper respiratory and lower respiratory symptom scores peaked at ARI onset; however, respiratory symptoms resolved less quickly than systemic symptoms and signs (Figure 1). For influenza A virus infections. tympanic temperature was significantly positively correlated with viral RNA shedding on the day following ARI onset (P = .01), although correlations were less clear later in the course of illness (Figure 5). There was also a positive correlation between number of symptoms and molecular viral shedding (data not shown). Data were insufficient to repeat these analyses for influenza B virus infections.
Figure 5.
Association between tympanic temperature and molecular influenza A viral shedding by reverse-transcription polymerase chain reaction by day since acute respiratory illness (ARI) onset for children (plus signs) and adults (circles). Linear regression lines are plotted where there are ⩾3 points. ARI onset is defined as at the first day with ⩾2 of the 7 signs or symptoms listed in Table 1. NTS, nose and throat swab specimen.
We detected viral shedding with RT-PCR in the absence of any reported signs or symptoms in 8 (14%) of 59 subjects (95% CI, 6.0%–25%), 5 of whom were infected with influenza A virus. In 2 of the 8 subjects, only NTS samples collected at the final home visit were positive with RT-PCR; we may have identified presymptomatic rather than asymptomatic shedding because these subjects may have subsequently developed symptoms. The geometric mean of peak viral titers in the NTS specimens of these 8 asymptomatic subjects was 3.2×103 copies/ mL compared with 3.6×107 copies/mL in symptomatic subjects. Among the 8 individuals with asymptomatic infections, 3 had specimens that were positive on quantitative viral culture with a geometric mean TCID50 of 100.7. Subclinical infections were identified in an additional 7 of 59 subjects who reported at most 1 of 7 signs or symptoms on each day of the study period, and viral shedding in the NTS specimens of these 7 subjects had geometric mean 3.4×103 copies/mL. Three of 7 specimens from subjects with subclinical infection were positive on viral culture with a mean TCID50 of 100.5.
Viral shedding and replication trends assessed with RT-PCR and TCID50, respectively, were also assessed relative to time since fever resolution, along with symptom scores and tympanic body temperature time lines for influenza A (data not shown). Viral shedding measured with RT-PCR peaked 1 day before fever resolution. Shedding was detected in 4 (44%) of 9 subjects (95% CI, 14%–79%) after fever resolution. Symptom trends, mean tympanic temperature, and replicating viral load measured by TCID50 all followed a similar trend.
In a sensitivity analysis, we investigated viral shedding relative to onset of fever of ⩾37.8°C in the subset of 22 subjects who reported a fever and found that trends in viral shedding were generally consistent with the main analysis based on ARI onset (data not shown).
Discussion
There are few data in the literature on the patterns in infectiousness in influenza virus infections over time, and how infectiousness may be related to viral shedding or symptoms. We investigated 3 alternative simple models for infectiousness in influenza A virus infections and found that if infectiousness is proportional to viral RNA shedding (or log10 viral shedding or presence/absence of shedding), then the majority of infectiousness occurs within 1–2 days (or 3–4 days) after ARI onset (Figure 4). Given the more rapid decrease in replicating viral load compared with molecular viral shedding (Figure 1), we may have overestimated the duration of infectiousness. In previous work, we estimated the mean serial interval of influenza to be 3.6 days [9], given an incubation period of 1.5–2 days [13]. This implies that the average time between ARI onset and subsequent transmission to household contacts was ∼2 days, which is consistent with infectiousness profiles in between a proportional relationship with log10 viral shedding and viral shedding. From a detailed model of influenza transmission in households in a French household study [14], Ferguson et al estimated that infectiousness was highest around the time of illness onset, and that the infectiousness profile correlated closely with viral shedding [15], whereas in our data viral shedding peaked within 1–2 days of symptom onset (Figure 1). Similarly, Pitzer et al [16] found that moderate increases in transmissibility of severe acute respiratory syndrome 5–10 days after illness onset appeared more consistent with changes in log viral shedding rather than viral shedding over the same period [17]. Because the majority of viral shedding measured with RT-PCR or TCID50 occurred within 2–3 days of ARI onset, isolation of patients or other interventions must be applied very soon after ARI onset; otherwise, they could not have any substantial effectiveness in reducing onwards transmission.
Our data are consistent with data from previous studies that documented the time lines of influenza viral shedding from volunteer challenge studies [2]. We found that influenza A viral shedding measured with RT-PCR peaked around the same day as ARI onset before decreasing. Whereas some viral shedding was detectable with RT-PCR before ARI onset, this occurred in a minority of cases. Daily replicating viral shedding measured with TCID50 for influenza A virus infections decreased faster than viral shedding measured with RT-PCR. Patterns in viral shedding associated with influenza B virus infections were more variable and followed a pattern similar to that of data observed in volunteer challenge studies, with shedding at substantial levels from ARI onset through to 5 days after ARI onset [2]. We found that levels of replicating virus measured by TCID50 had high correlation with molecular viral loads measured with RTPCR soon after ARI onset but diverged as the infection progressed when TCID50 decreased more rapidly (Figures 2 and 3). A previous study also found that viral culture is less sensitive than RT-PCR later in the course of illness [18], perhaps because inactivated virus or viral DNA persist in the respiratory tract towards the end of illness.
In both influenza A and B virus infections, systemic symptoms and signs subsided more rapidly than respiratory symptoms [2]. The decrease of systemic signs and symptoms, primarily fever, correlated closely with the decrease in viral shedding, most likely due to the subsiding of immune response with the gradual clearing of virus from the body [19]. Resolution of fever could also be affected by use of antipyretic medication. The correlation between higher tympanic temperature and higher viral shedding (Figure 5) suggests that individuals with a higher fever could be more infectious in general.
Eight (14%) of the 59 individuals (95% CI, 6.0%–25%) with RT-PCR—confirmed secondary infections did not report any clinical signs or symptoms, and in total 15 (25%) of 59 subjects (95% CI, 15%–38%) were asymptomatic (reporting 0 of 7 signs or symptoms) or subclinical (reporting 1 of 7 signs or symptom). Our upper bound for the frequency of asymptomatic infection is somewhat lower than that identified in volunteer challenge studies [2] or from longitudinal studies including preand post-season serology with illness recall [20]. It is possible that we failed to detect infections associated with lower levels of viral shedding or shedding for a shorter period, because swab samples were collected on average only at 3-day intervals, and proportionally more such infections may be asymptomatic [2, 21]. Some individuals may have been infected without shedding virus. It is still unknown whether or to what degree asymptomatic individuals could transmit infection to others [22], although mathematical models typically assume that 33%–50% of infections are asymptomatic or subclinical, and these individuals are approximately half as infectious as symptomatic persons [23, 24]. Our results suggest the possibility that “silent spreaders” (ie, individuals who are infectious while asymptomatic or presymptomatic) may be less important in the spread of epidemics than was previously thought.
US pandemic guidelines suggest sick individuals should remain home for a minimum of 24 h after fever resolution in the absence of fever-reducing medication [25]. In our study of seasonal influenza predominantly among adults, viral shedding resolved within the recommended exclusion period for 10 (83%) of 12 individuals with febrile influenza A virus infection, suggesting that the exclusion period covers the majority of infectiousness for febrile infections if the patterns of fever and viral shedding are similar to that in pandemic influenza.
A strength of our study is that we analyzed laboratory-confirmed, naturally acquired influenza virus infections in a community setting, which should allow broad generalizability. It is important also to note the limitations of our study. First, because of sample-size limitations, our study was underpowered to explore in detail the differences by age and other characteristics. Second, because our study design tended to exclude households with >1 case at the recruitment stage, subjects in our study could be biased toward those having a lower risk of infection or illness [3]. Third, although we have investigated alternative models for the relationship between infectiousness and viral shedding, it is possible that specific symptoms also play an important role in infectiousness. Fourth, we have a relatively small sample size, and more data would be valuable to more precisely characterize patterns in viral shedding and illness, and particularly viral shedding associated with asymptomatic infections and before illness onset. Finally, we did not record symptoms in household contacts retrospectively, so we were unable to include in this analysis household contacts with evidence of influenza virus infection at the initial home visit; however, the general characteristics of excluded contacts were similar to those of contacts who were included (Table 1).
One research gap highlighted by our work is the need for studies that collect paired serologic data as well as detailed viral shedding data. This would allow more accurate estimates of the proportion of infections that are asymptomatic or subclinical, and the characteristics of viral shedding in asymptomatic infections. Future studies of naturally acquired infections with increased frequency of clinical specimens and a longer followup period would also contribute valuable knowledge about the shape of the peak and duration of viral shedding. If combined with data on transmission to contacts, these studies may be able to provide additional information on the relations between viral shedding, illness, and infectiousness, which would facilitate optimal application of interventions and control measures. Volunteer challenge studies can provide useful data, but large detailed studies of naturally acquired infections are essential to inform differences in time lines and trends by age and other characteristics.
Acknowledgments
We thank all the physicians, nurses, and staff of participating centers for facilitating recruitment and Rita Fung, Lai-Ming Ho, Winnie Wai, and Eileen Yeung for research support.
Footnotes
Potential conflicts of interest: none reported.
Financial support: US Centers for Disease Control and Prevention (grant 1 U01 CI000439-02), the Research Fund for the Control of Infectious Disease, Food and Health Bureau, Government of the Hong Kong SAR (grant 08070632), the Harvard Center for Communicable Disease Dynamics from the US National Institutes of Health Models of Infectious Disease Agent Study program (grant 1 U54 GM088558), and the Area of Excellence Scheme of the Hong Kong University Grants Committee (grant AoE/M-12/06). The funding agencies had no role in data collection and analysis, or the decision to publish, but the CDC was involved in study design and preparation of the manuscript. This work represents the views of the authors and not their institutions, including the Centers for Disease Control and Prevention.
References
- 1.Nicholson KG, Wood JM, Zambon M. Influenza. Lancet. 2003;362:1733–1745. doi: 10.1016/S0140-6736(03)14854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrat F, Vergu E, Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–785. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 3.Cowling BJ, Chan KH, Fang VJ, et al. Facemasks and hand hygiene to prevent influenza transmission in households: a cluster randomized trial. Ann Intern Med. 2009;151:437–446. doi: 10.7326/0003-4819-151-7-200910060-00142. [DOI] [PubMed] [Google Scholar]
- 4.Cowling BJ, Fung RO, Cheng CK, et al. Preliminary findings of a randomized trial of non-pharmaceutical interventions to prevent influenza transmission in households. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002101. 3:e2101.doi: 10.1371/journal.pone.0002101. Published 7 May 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng CK, Cowling BJ, Chan KH, et al. Factors affecting QuickVue Influenza A + B rapid test performance in the community setting. Diagn Microbiol Infect Dis. 2009;65:35–41. doi: 10.1016/j.diagmicrobio.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Peiris JS, Tang WH, Chan KH, et al. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis. 2003;9:628–633. doi: 10.3201/eid0906.030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan KH, Peiris JSM, Lim W, Nicholls JM, Chiu SS. Comparison of nasopharyngeal flocked swabs and aspirates from pediatric patients for rapid diagnosis of respiratory viruses. J Clin Virol. 2008;42:65–69. doi: 10.1016/j.jcv.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Lambert SB, Whiley DM, O'Neill NT, et al. Comparing nose-throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real-time polymerase chain reaction. Pediatrics. 2008;122:e615–e620. doi: 10.1542/peds.2008-0691. doi:10.1542peds.2008-0691. Published 25 August 2008. [DOI] [PubMed] [Google Scholar]
- 9.Cowling BJ, Fang VJ, Riley S, Peiris JSM, Leung GM. Estimation of the serial interval of influenza. Epidemiology. 2009;20:344–347. doi: 10.1097/EDE.0b013e31819d1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz GE. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- 11.R Development Core Team . Vienna, Austria: R Foundation for Statistical Computing; 2005. R: a language and environment for statistical computing. [Google Scholar]
- 12.Spiegelhalter DJ, Thomas A, Best NG, Lunn D. WinBUGS user manual, version 1.4. Cambridge, England: MRC Biostatistics Unit. 2003 [Google Scholar]
- 13.Lessler J, Reich NG, Brookmeyer R, Perl TM, Nelson KE, Cummings DA. Incubation periods of acute respiratory viral infections: a systematic review. Lancet Infect Dis. 2009;9:291–300. doi: 10.1016/S1473-3099(09)70069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viboud C, Boelle PY, Cauchemez S, et al. Risk factors of influenza transmission in households. Br J Gen Pract. 2004;54:684–689. [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson NM, Cummings DA, Cauchemez S, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 16.Pitzer VE, Leung GM, Lipsitch M. Estimating variability in the transmission of severe acute respiratory syndrome to household contacts in Hong Kong,China. Am J Epidemiol. 2007;166:355–363. doi: 10.1093/aje/kwm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee N, Chan PK, Hui DS, et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis. 2009;200:492–500. doi: 10.1086/600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. Local and systemic cytokine responses during experimental human influenza A virus infection: relation to symptom formation and host defense. J Clin Invest. 1998;101:643–649. doi: 10.1172/JCI1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrat F, Sahler C, Rogez S, et al. Influenza burden of illness: estimates from a national prospective survey of household contacts in France. Arch Intern Med. 2002;162:1842–1848. doi: 10.1001/archinte.162.16.1842. [DOI] [PubMed] [Google Scholar]
- 21.Foy HM, Cooney MK, Allan ID, Albrecht JK. Influenza B in households: virus shedding without symptoms or antibody response. Am J Epidemiol. 1987;126:506–515. doi: 10.1093/oxfordjournals.aje.a114683. [DOI] [PubMed] [Google Scholar]
- 22.Patrozou E, Mermel LA. Does influenza transmission occur from asymptomatic infection or prior to symptom onset? Public Health Rep. 2009;124:193–196. doi: 10.1177/003335490912400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson NM, Cummings DA, Fraser C, Cajka JC, Cooley PC, Burke DS. Strategies for mitigating an influenza pandemic. Nature. 2006;442:448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longini IM, Halloran ME, Jr, Nizam A, Yang Y. Containing pandemic influenza with antiviral agents. Am J Epidemiol. 2004;159:623–633. doi: 10.1093/aje/kwh092. [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and Prevention CDC recommendations for the amount of time persons with influenza-like illness should be away from others. http://www.cdc.gov/h1n1flu/guidance/exclusion.htm. Published 5 August 2009. Accessed 19 August 2009.