SUMMARY
UCS proteins, such as UNC-45, influence muscle contraction and other myosin-dependent motile processes. We report the first x-ray crystal structure of a UCS domain-containing protein, the UNC-45 myosin chaperone from Drosophila melanogaster (DmUNC-45). The structure reveals that the Central and UCS domains form a contiguous arrangement of seventeen consecutive helical layers that arrange themselves into five discrete armadillo repeat subdomains. Small-angle x-ray scattering data suggest that free DmUNC-45 adopts an elongated conformation and exhibits flexibility in solution. Protease sensitivity maps to a conserved loop that contacts the most carboxy-terminal UNC-45 armadillo repeat sub-domain. Amino acid conservation across diverse UCS proteins maps to one face of this carboxy-terminal sub-domain and the majority of mutations that affect myosin-dependent cellular activities lie within or around this region. Our crystallographic, biophysical, and biochemical analyses suggest that DmUNC-45 function is afforded by its flexibility and by structural integrity of its UCS domain.
INTRODUCTION
Myosin is a superfamily of motor proteins whose function is required for muscle contraction, cell division, cell motility, intracellular transport, and signal transduction (Coluccio, 2008). Classical genetic studies in the model organism C. elegans revealed a host of mutations that restrict worm movement (Brenner, 1974). Subsequent investigations identified mutant alleles of a gene called unc-45 that disrupt myofilament organization and hinder movement in developing worms (Epstein and Thomson, 1974; Venolia and Waterston, 1990). Analysis of adult worms of one temperature sensitive mutant strain grown at the restrictive temperature revealed a 50% decrease in thick filaments and scrambling of myosins A and B (Barral et al., 1998). These studies suggested a necessary role for the unc-45 gene product in muscle assembly during embryogenesis. Molecular genetic studies permitted isolation of the C. elegans unc-45 gene (Barral et al., 1998; Venolia et al., 1999).
Homologs of the UNC-45 protein have since been identified in diverse eukaryotic species. Two genes encoding distinct UNC-45 isoforms are encoded within the genomes of fish and mammals (Price et al., 2002). UNC-45A, also known as the general cell isoform, is expressed in all tissues while the striated muscle isoform UNC-45B is produced exclusively in skeletal and cardiac muscle. Injection of morpholino oligonucleotides directed against unc-45b gene transcripts in developing zebrafish embryos results in a lack of myosin-containing thick filaments and consequent muscular paralysis as well as severe cardiac and yolk sac edema and craniofacial abnormalities (Wohlgemuth et al., 2007). In zebrafish, UNC-45 localizes to sarcomeric Z-disks, but re-localizes to myosin-containing A-bands during stress (Etard et al., 2008). These results suggest that UNC-45B could play roles in human myopathies and stress response, though at present it is only predicted to do so (Walker, 2001). Overexpression of UNC-45A has been shown to be associated with ovarian cancer (Bazzaro et al., 2007).
UNC-45 is part of a larger family of myosin-interacting proteins that include the Cro1p protein from the filamentous fungus P. anserina, She4p from the budding yeast S. cerevisiae, and the fission yeast S. pombe protein Rng3p. Each of these shares sequence homology throughout a carboxy-terminal region of roughly 400 amino acids referred to as the UCS (UNC-45, Cro1p, She4p) domain (Hutagalung et al., 2002; Yu and Bernstein, 2003). Deletion of Cro1p leads to multiple cytoskeletal defects including failure to form septa between daughter nuclei following mitotic cell division (Berteaux-Lecellier et al., 1998). She4p is required for endocytosis in yeast and is necessary for transport of ASH1 mRNA to daughter cells through a myosin-dependent process required for mating-type switching (Bobola et al., 1996; Jansen et al., 1996; Wendland et al., 1996). Furthermore, She4p is the only UCS protein that has been shown to interact with unconventional myosins (Toi et al., 2003). Rng3p was originally characterized as a factor that is required for actomyosin contractile ring assembly during fission yeast cell division (Wong et al., 2000). A series of elegant biochemical and genetic experiments has since revealed that Rng3p is required for in vitro actin filament gliding by the conventional myosin-II gene product Myo2p (Lord and Pollard, 2004). Together, these and other studies implicate critical roles for UCS domain-containing proteins in facilitating diverse cellular processes that require myosin motor proteins (Barral et al., 2002).
Structurally, UNC-45 is unique among these UCS domain-containing proteins (referred to throughout this report as UCS proteins) as it contains an amino-terminal tetra-tricopeptide repeat (TPR) domain (Barral et al., 1998; Venolia et al., 1999). TPR repeats are a helical tandem repeat motif common to many proteins that participate in protein-protein interactions (Groves and Barford, 1999; Scheufler et al., 2000). The TPR domain of UNC-45 has been shown to bind the Hsp90 protein-folding chaperone suggesting that UNC-45 might function as a myosin-specific co-chaperone (Barral et al., 2002; Liu et al., 2008; Srikakulam et al., 2008). Bridging the TPR and UCS domains in UNC-45 is a stretch of approximately 400 amino acids in length that is not appear to harbor protein domains of known structure but has been suggested to contain regions of limited homology to β-catenin (Srikakulam et al., 2008). This region is referred to as the Central domain (Hutagalung et al., 2002). UNC-45 on its own is capable of preventing aggregation and heat-dependent fusion of globular heads in dimers of purified skeletal muscle myosin in vitro (Barral et al., 2002; Melkani et al., 2010).
As part of an effort to improve the understanding of molecular mechanisms by which UCS proteins interact with and influence the function of myosin motor proteins, we have begun structural and in vitro biochemical studies on purified recombinant UNC-45 from the fruit fly Drosophila melanogaster (DmUNC-45) (Melkani et al., 2010). Here we report the x-ray crystal structure of DmUNC-45 determined to a resolution of 3.0 Å. The crystallographic model reveals that the Central and UCS domains form a contiguous series of stacked armadillo repeats. Small-angle x-ray scattering data indicate that DmUNC-45 adopts a more elongated conformation and exhibits flexibility in solution. Sequence homology between functionally diverse UCS proteins suggests that they share a conserved surface for interaction with myosin.
RESULTS
DmUNC-45 crystal structure
Single protein crystals suitable for x-ray diffraction were prepared by vapor diffusion of concentrated preparations of recombinant DmUNC-45. Integrity of the crystallized full length histidine-tagged DmUNC-45 was confirmed both by SDS-PAGE and Western blot analyses of washed and dissolved crystals (Figures S1 and S2). The x-ray crystal structure of DmUNC-45 was solved by single anomalous dispersion (SAD) using seleno-methionine-substituted protein crystals and refined against native data collected to a limit of 3.0 Å resolution (Table 1). The resulting DmUNC-45 crystallographic model includes the entire Central and UCS domains (amino acids 138–923). The folded structure is composed almost entirely of stacked alpha-helical motifs known as armadillo (ARM) repeats (Figure 1). Therefore, it is not obvious from the model alone where the Central domain ends and the UCS domain begins. A prior amino acid sequence comparison of UNC-45 isoforms from diverse species and other UCS proteins suggested that the conserved UCS domain begins at amino acid 524 in C. elegans UNC-45 (Hutagalung et al., 2002). The analogous position in DmUNC-45 is amino acid 511, which lies within a loop connecting two ARM repeats (Figure 1A). This suggests that the myosin-interacting element shared by the UCS proteins is a single domain consisting of nine ARM repeats.
Table 1.
Data collection and refinement statistics
| Native | Se-Met | L63M | |
|---|---|---|---|
| Data collection | |||
| X-ray source | ALS 8.2.2 | ALS 8.2.1 | ALS 8.2.2 |
| Wavelength (Å) | 1.00000 | 0.97950 | 0.97940 |
| Space group | P23 | P23 | P23 |
| Unit cell (Å)a,b,c | 184.079 | 184.577 | 188.016 |
| Monomers/asymmetric unit | 1 | 1 | 1 |
| Resolution rangea | 50.00–3.00 (3.11–3.00) | 50.00–3.30 (3.42–3.30) | 50.00–3.20 (3.31–3.20) |
| Rsym (%) | 11.5 (81.9) | 13.7 (53.6) | 11.8 (78.5) |
| Observations | 400,191 | 291,598 | 406,585 |
| Unique reflections | 42,099 | 31,539 | 36,692 |
| Completeness (%) | 99.8 (97.8) | 99.7 (99.1) | 100 (100) |
| Redundancy | 9.5 (6.5) | 9.2 (7.3) | 11.1 (10.1) |
| Average intensity (<I/σ>) | 19.2 (2.0) | 15.2 (3.1) | 15.7 (2.1) |
| Structure solution by SAD | |||
| Figure of meritb | |||
| Centric | 0.422 | 0.432 | |
| Acentric | 0.128 | 0.138 | |
| Overall | 0.374 | 0.387 | |
| Refinement | |||
| Resolution range (Å) | 50.0–3.0 | 50.0–3.2 | |
| Reflections used | 39,354 | 35,519 | |
| Protein atoms | 6,077 | 6,077 | |
| Rcryst (%)/Rfree (%)c | 20.1/23.6 | 20.6/24.4 | |
| Geometry | |||
| R.m.s. bond lengths (Å) | 0.013 | 0.012 | |
| R.m.s. bond angles (°) | 1.293 | 1.491 | |
| Mean B (Å2) | 63.0 | 78.3 | |
| Ramachandran plotd | |||
| Most Favorable (%) | 93.8 | 88.1 | |
| Allowed (%) | 5.8 | 8.5 | |
| Disallowed (%) | 0.4 | 3.4 | |
| PDB accession code | 3NOW | ||
Data in parentheses are for the highest resolution shell
PHENIX (Adams et al., 2010)
Calculated against a cross-validation set of 5% of the data selected at random prior to refinement
MOLPROBITY (Davis et al., 2007)
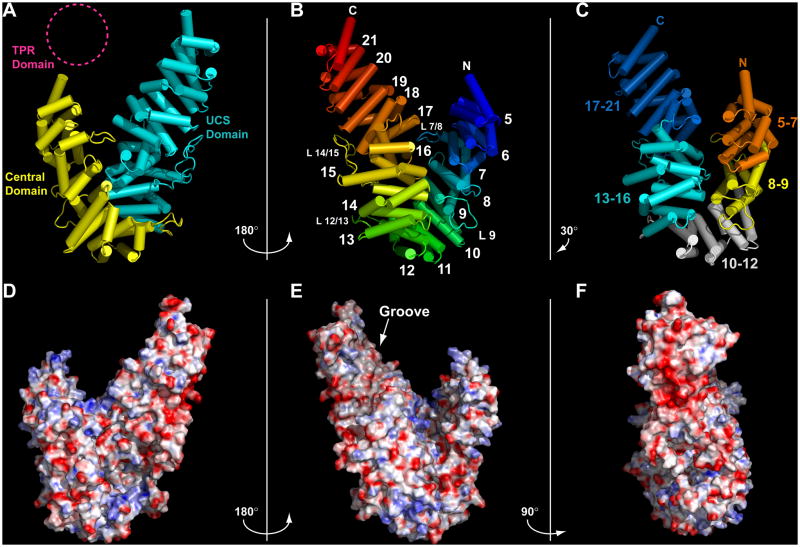
Figure 1.
The DmUNC-45 x-ray crystal structure. A) DmUNC-45 ribbon diagram depicted with cylindrical helices. The Central and UCS domains are colored yellow and cyan, respectively, and the disordered TPR domain is represented by a dashed magenta circle. B) The same DmUNC-45 model rotated 180° about the y-axis. The protein is colored and numbered by helical layer. The amino- and carboxy-termini of the protein are noted along with the locations of four loops (see text). C) DmUNC-45 rotated by 150° about the y-axis relative to panel A. Five individual ARM repeat sub-domains are colored individually and labeled. D) A surface rendering of DmUNC-45 oriented as in A and colored according to negative (red) and positive (blue) electrostatic surface potential. E) Electrostatic surface of DmUNC-45 rotated 180° about the y-axis from view in D (oriented as in B). The concave surface created by ARM repeat 16–20 helices H3 (labeled “Groove”) displays a decrease in electrical potential relative to the rest of the protein surface. F) A 90° rotation of DmUNC-45 relative to E reveals an acidic patch (red) near the UCS domain carboxy-terminus (see also Figures S1–S3).
ARM repeats are alpha helical tandem repeat motifs of approximately 40 amino acids in length that were first described structurally in the proteins β-catenin and importin-α (Conti et al., 1998; Huber et al., 1997). Successive repeats stack in layers to form a continuous scaffold. We number the seventeen ARM repeat-containing layers within the DmUNC-45 crystal structure in order from 5 to 21 (Figure 1B). This numbering scheme allows for the assignment, based upon the solution NMR structure of the human UNC-45A isoform TPR domain (PDB ID: 2DBA), of three additional helical layers and an individual helix 4 within the amino-terminal 137 amino acids of the DmUNC-45 protein (Tochio et al.).
Several of the DmUNC-45 ARM repeats vary from the consensus structure in that they contain non-consensus ordered loops. Most notable among these are: the loop connecting layers 7 and 8 (L7/8), a loop within ARM repeat layer 9 (L9), a loop between layers 12 and 13 (L12/13), and the 30 amino acid long insertion between ARM repeat layers 14 and 15 (L14/15). Interestingly, each of these loops interferes with the continuous stacking of successive ARM repeats and introduces small “breaks” in the overall ARM domain structure. For example, superposition of repeat 6 onto 7 requires that ARM repeat 6 be rotated 23° and translated 11Å, which are typical values (Huber et al., 1997). Due to the interference of ARM repeat stacking by loop 7/8, a similar overlay of repeats 7 and 8 requires a 12 Å translation and rotation of −34°. As a consequence of these four loops disrupting normal ARM repeat stacking, the DmUNC-45 structure is better described as consisting of five ARM repeat sub-domains that articulate through novel interfaces rather than as one continuous domain of seventeen ARM repeats (Figure 1C). The five ARM repeat sub-domains are composed of layers 5–7, 8–9, and 10–12 from the Central domain and layers 13–16 and 17–21 in the UCS domain.
The continuous stack of ARM repeats present in the DmUNC-45 x-ray crystal structure measures roughly 180 Å in length. However, it turns back upon itself so that the longest dimension in the structure, which corresponds roughly to the UCS domain, is approximately 90 Å. Although the Central domain is nearly the same overall length, due to two sharp bends in the structure at the ARM 9/10 and ARM 12/13 interfaces, its longest dimension is roughly 55 Å. Overall, the DmUNC-45 x-ray crystal structure is horseshoe-shaped with one leg longer than the other. The Central domain makes up the shorter leg and curvature while the longer leg contains the UCS domain.
DmUNC-45 ARM repeats
The determination that the DmUNC-45 Central and UCS domains consist entirely of ARM repeats was somewhat surprising at the time, as this motif was not predicted for UNC-45 by structure prediction software. Srikakulam et al. have since predicted b-catenin-like structure for portions of the Central domain (Srikakulam et al., 2008). After-the-fact analysis and comparison to the ARM consensus sequence proposed by Andrade et al. revealed that each of the ARM repeats contains many of the hydrophobic amino acid residues in positions that mediate important intra- and inter-repeat interactions (Andrade et al., 2001) (Figure 2). By contrast, significantly fewer of the remaining consensus residues, including a Gly at position 11, Pro at 14, and Asn at 37, occupy similar positions in the DmUNC-45 ARM repeats. This suggests that these positions are less important for ARM repeat folding and that their presence within the consensus sequence might be the result of bias within the models used to predict ARM repeat domain structures. Rather, the DmUNC-45 structure suggests that the placement of appropriately sized hydrophobic side chains at strategic positions to mediate intra- and inter-repeat interactions is the true ARM repeat signature.
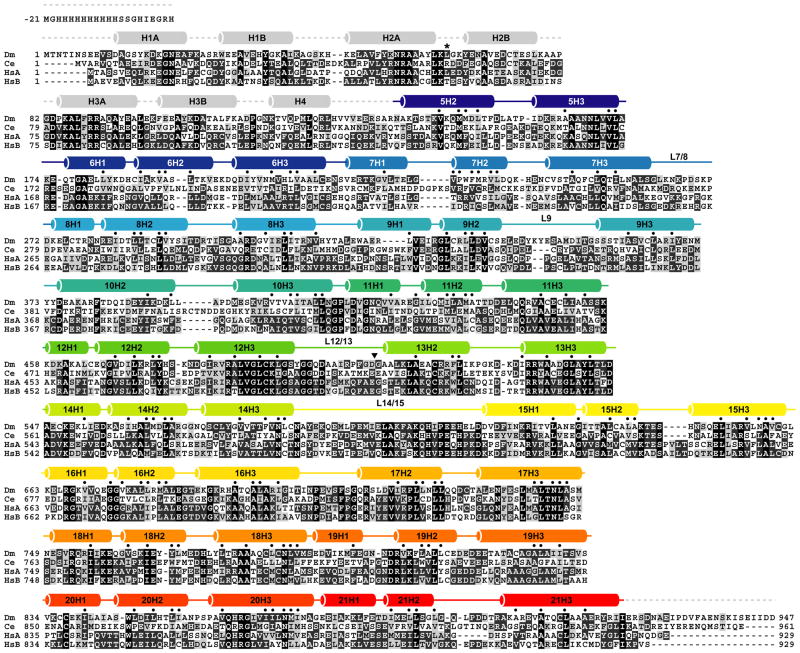
Figure 2.
Primary amino acid sequence and secondary structure of DmUNC-45. DmUNC-45 (accession no. AAK93568) was aligned with the UNC-45 proteins from C. elegans (Ce; accession no. AAD01976) and both H. sapiens general cell (HsA; accession no. AAH45635) and striated muscle (HsB; accession no. AAI01064) isoforms. Black boxes highlight identical positions while dark grey boxes indicate conserved positions and light grey boxes signify substitution of a homologous amino acid. DmUNC-45 secondary structure is indicated with helices represented as cylinders above their corresponding amino acids. Each stacked helical layer is assigned a principal number and individual ARM repeat helices are numbered H1, H2, and H3. Secondary structure of the amino-terminal TPR domain, which is present but disordered in the crystal, is depicted in light grey and assigned by homology to other TPR domain-containing proteins. Other colors correspond to helices depicted in Figure 1B. Leu63, marked with an asterisk underneath, was mutated to Met for SAD data collection and structure solution of the L63M mutant. Small dots are placed over amino acid positions that fit the ARM repeat consensus sequence and structural requirements proposed by Andrade et al. (Andrade et al., 2001).
Kippert and Gerloff recently described an iterative algorithm for ARM as well as HEAT repeat detection based on analysis of multiple aligned sequences of homologous proteins by HHPRED and COACH (Kippert and Gerloff, 2009). This approach was used to successfully predict 14 ARM repeats from the primary sequence of DmUNC-45 alone. Interestingly, the three repeats that were not identified correspond to ARM repeat layers 5, 8, and 9. Layer 5 is the first in the Central domain stack and, consequently, does not require all the consensus hydrophobic residues to contact its neighbors. Layers 8 and 9 constitute the second ARM repeat sub-domain within the Central domain. These interact with layers 7 and 10, respectively, through unique inter-repeat interfaces involving non-consensus amino acid positions. Finally, it is interesting to note that the Kippert and Gerloff analysis suggests that ARM repeat 13 begins at amino acid residue Asp501. In the DmUNC-45 crystal structure this residue lies within the loop L12/13 that links the Central and UCS domains. It seems reasonable, therefore, that in solution this loop might fold into an H1 helix and that the observed L12/13 configuration is more a consequence of the overall bent conformation of DmUNC-45 in the crystal and the direct involvement of loop L12/13 residues in crystal packing.
DmUNC-45 surface analysis
To gain insight into the surface characteristics of the DmUNC-45 protein, we calculated the electrostatic surface potential and qualitatively analyzed surface polarity. Calculation of the surface electrostatic potential for a solvated model of the DmUNC-45 x-ray crystal structure was carried out with the program APBS (Baker et al., 2001). The results revealed that, with two notable exceptions, electrostatic potential is fairly randomly distributed over the surface of the protein (Figure 1D–F). First among the exceptions is the surface created by stacking of the H2–H3 inter-helix portions of ARM repeats 17–21 in the UCS domain, which is highly acidic (Figure 1F). Second, the groove that is created by the stack of helices H3 from ARM repeats 17–21 displays noticeably less electrostatic potential than is observed in the rest of the model (Figure 1E).
The unique surface properties of the UCS domain groove are even more pronounced when colored according to polarity of surface amino acids in PyMol (DeLano, 2002). Using a qualitative routine to render the DmUNC-45 surface according to the amino acid polarity classifications it is clear that the UCS domain groove presents a strikingly hydrophobic surface devoid of charged amino acid residues (Figure S3).
The “missing” TPR domain
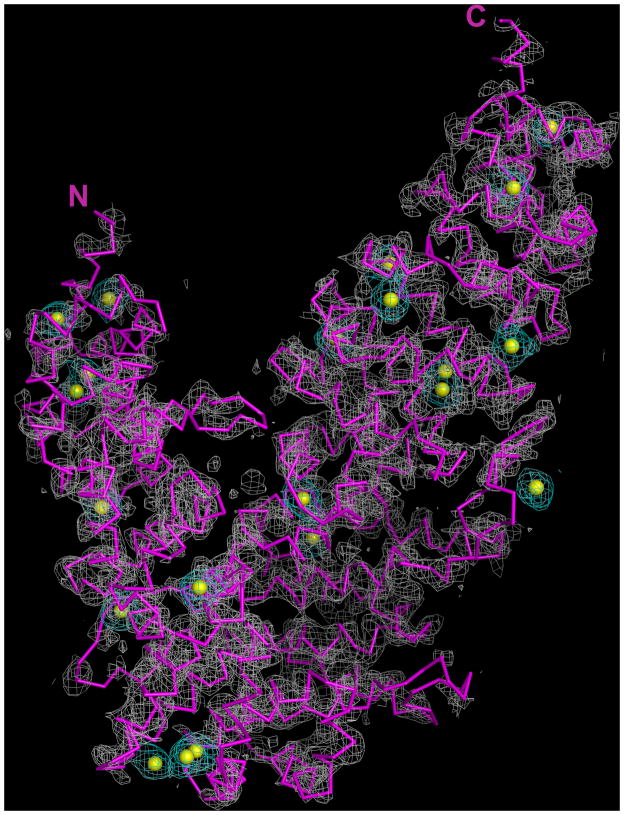
Upon determination by SAD techniques of the sub-structure containing 19 unique Se sites, it was discovered that the relatively large unit cell of the DmUNC-45 crystal (~184 Å on each edge) is nearly 80% solvent by volume (Figure 3). Large solvent-filled channels run orthogonally in three-dimensions throughout the crystal (Figure S4A). Furthermore, within each unit cell, DmUNC-45 monomers pack with cubic symmetry to form a hollow solvent-filled structure (Figure S4B). Early in the process of DmUNC-45 refinement, model building was focused upon the clearly helical portions in the experimental electron density map and the correct placement of Met side chains at the 19 Se sites (Met594 was the only non-amino-terminal Se site not identified by SAD phasing). Non-helical connecting loops were then carefully built within 2|FO| − |FC| and |FO| − |FC| electron density maps. Refinement and stereochemistry statistics were closely monitored throughout this process.
Figure 3.
Experimental electron density map (grey mesh) contoured at 1.5 σ. The positions of the Se sites determined by SAD are represented by yellow spheres and those portions of the map that surround the Se sites are depicted in cyan. As a reference, the final refined DmUNC-45 x-ray structure is shown as a C-alpha trace in magenta oriented as in panel A of Figure 1 (see also Figures S4 and S5).
During the course of structural refinement it became clear that, though present in the crystal, the amino-terminal and presumably alpha-helical TPR domain remained unaccounted for while the Central and UCS domains present in the model exhibited a structure composed almost entirely of alpha-helical stacks. Consequently, in an effort both to confirm the disorder of the predicted TPR domain and correctness of the refined DmUNC-45 crystallographic model we prepared crystals of a seleno-methionine-substituted DmUNC-45 protein in which amino acid Leu63 was replaced by a Met residue.
X-ray diffraction data were collected for the DmUNC-45 L63M mutant crystal at the anomalous edge of Se and the structure was solved by SAD and refined to 3.2 Å resolution (Table 1). No additional Se site was detected nor was there novel TPR domain electron density observed in the L63M model. This experiment supports our conclusion that the amino-terminal TPR domain, which is projected toward the vast solvent channels by each of the DmUNC-45 monomers in our crystal, exhibits disorder and/or flexibility relative to the Central and UCS domains. This flexibility might result from disorder of the entire TPR domain or it could be afforded by the stretch of ten amino acids that link the end of the predicted TPR domain structure and the first amino acid that could be refined within electron density (Asn138).
The 3.2 Å DmUNC-45 L63M x-ray structure shows little change from the native sequence structure (Figure S5). However, the cubic unit cell did exhibit a 6.6% increase in volume from 184 to 188 Å on each edge. This is likely a consequence of differences in stabilization and cryo-preservation of the crystals prior to data collection. The UCS domains (amino acids 511–923) of the two structures overlay with a root-mean-squared deviation (rmsd) for C-alpha positions of 1.257 Å. However, after UCS domain superposition the Central domains (amino acids 138–510) overlay with an rmsd of 2.489 Å. When the two domains are overlaid separate from one another, the same Central domains superpose with an rmsd of 1.631 Å. These differences suggest that the Central and UCS domains are capable of some degree of relative motion and that one point about which they hinge might be focused upon loop 12/13 at the domain interface.
DmUNC-45 conformation in solution
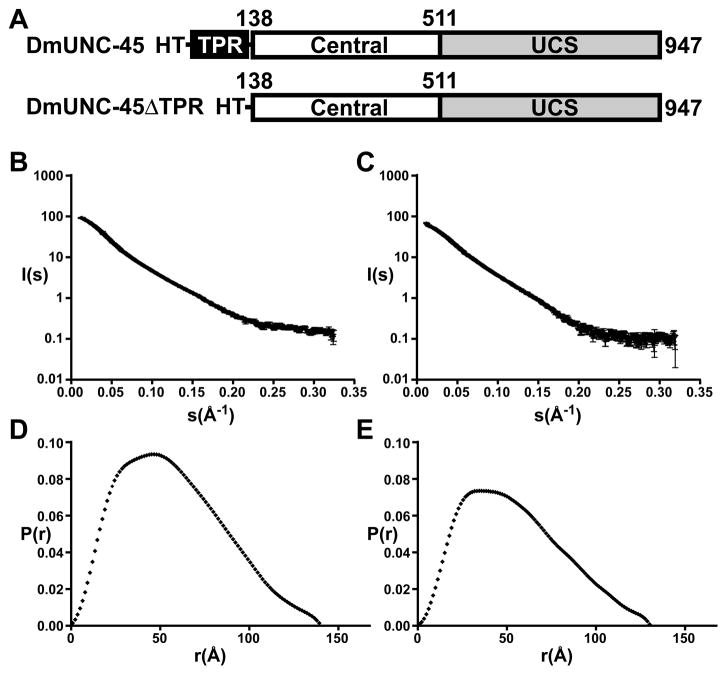
The dimensions of monomeric DmUNC-45 in the crystal are roughly 100 × 60 × 40 Å. Electron microscopy on purified samples of FLAG-tagged murine UNC-45B contrasted by rotary shadowing revealed a long dimension of 190 Å (Srikakulam et al., 2008). In an effort to address this disparity we carried out an analysis of the oligomerization state and dimensions of free DmUNC-45 protein in solution by small angle x-ray scattering (SAXS) (Putnam et al., 2007).
SAXS data were collected on purified full-length DmUNC-45 and mutant protein lacking the amino-terminal TPR domain (DmUNC-45ΔTPR) (Figure 4A). Porod analysis of the scattering curves (Porod volume × 1.2/2 ≈ MW) revealed an approximate molecular weight of 110 kDa for DmUNC-45, which is in excellent agreement with the histidine-tagged DmUNC-45 monomer (theoretical molecular weight 107.6 kDa). Some modest radiation damage was revealed by comparing the low-angle scattering plots at increasing exposure times. To ameliorate this effect and obtain quality high angle data, we merged the low angle (0.011–0.072 Å−1) short exposure and high angle (0.072–0.32 Å−1) long exposure data to generate the final scattering curves (Figure 4B,C). Guinier plot analysis carried out with the program AutoRg indicates little or no aggregation with a quality value of 91%.
Figure 4.
Small-angle x-ray scattering (SAXS) analysis. A) Schematic domain map of DmUNC-45 protein constructs used in SAXS experiment. HT refers to the amino-terminal histidine-tag. B) SAXS intensity profile I(s) as a function of the magnitude of the scattering vector s for DmUNC-45 and C) DmUNC-45ΔTPR. D) Experimental P(r) function calculated from SAXS data for DmUNC-45 and E) DmUNC-45ΔTPR (see also Figure S6).
The program GNOM was used to generate pair-wise distance distribution functions by indirect Fourier transform analysis of the raw scattering data (Figure 4D,E) (Svergun, 1992). The radii of gyration (Rg) were determined at 44.8 ± 0.1 Å for DmUNC-45 and 42.3 ± 0.1 Å for DmUNC-45ΔTPR, which are in good agreement with the Guinier analysis. The maximum intramolecular distance was determined to be approximately 140 Å for DmUNC-45 and 130 Å for DmUNC-45ΔTPR (Table 2). Interestingly, these dimensions are intermediate to our crystal structure and the values derived from electron microscopy studies on the homologous murine UNC-45B. Furthermore, Kratky plot analysis is consistent with an elongated and well-folded protein (Figure S6). Taken together, these data suggest that DmUNC-45 exhibits significant conformational flexibility, rather than disorder, in solution
Table 2.
DmUNC-45 structural parameters from SAXS data and models
| Rg (Å) | Dmax (Å) | χ2 | |
|---|---|---|---|
| Experimental data | |||
| DmUNC-45 | 44.8 ± 0.1 | 140 | N/A |
| DmUNC-45ΔTPR | 42.3 ± 0.1 | 130 | N/A |
| Model analysis | |||
| DmUNC-45 crystal structure | 32.4 | 115.1 | 116.96 |
| DmUNC-45 crystal structure + TPRa | |||
| Best | 38.43 | 130.0 | 10.87 |
| Ensemble | 39.4 ± 0.3 | 126.7 ± 12.9 | 13.49 |
| DmUNC-45 elongated model + TPRb | |||
| Best | 38.62 | 136.3 | 23.02 |
| Ensemble | 38.9 ± 0.4 | 136.5 ± 0.9 | 83.06 |
| DmUNC-45 crystal structure + Flexc | |||
| Best | 36.35 | 121.2 | 15.09 |
| Ensemble | 36.4 ± 0.4 | 116.2 ± 3.3 | 27.71 |
| DmUNC-45 crystal structure + TPR + Flexd | |||
| Best | 41.60 | 139.7 | 5.38 |
| Ensemble | 41.1 ± 1.1 | 137.3 ± 6.8 | 5.48 |
The DmUNC-45 x-ray structure with TPR2A (PDB ID 1ELR) modeled at its amino-terminus. The TPR domain is allowed to move via introduction of flexibility at DmUNC-45 residues 138–142 (see text) (Scheufler et al., 2000).
An elongated model of DmUNC-45 created in FATCAT with flexibly-linked amino-terminal TPR domain (see text) (Ye and Godzik, 2003).
The DmUNC-45 x-ray structure with flexibility in UCS domain residues 705–715.
The DmUNC-45 x-ray structure with flexibly linked TPR domain and flexibility in UCS domain residues 705–715.
To test this hypothesis, we designed a series of flexible DmUNC-45 models using the Ensemble Optimization Method (EOM) and calculated agreement between the theoretical scattering curves of the computer-generated conformers and our experimental data with the program CRYSOL (Svergun et al., 1995). We first prepared a model that included coordinates from the TPR2A x-ray crystal structure flexibly linked to the amino-terminus of the DmUNC-45 crystal structure (Table 2). Using the FATCAT bioinformatics server to flexibly align DmUNC-45 onto β-catenin, a more linear ARM repeat domain-containing protein, we next prepared a DmUNC-45 model that was elongated to a final maximal dimension of 130 Å and flexibly linked the TPR2A domain (Ye and Godzik, 2003). Although theoretical data for both these two new models agreed better with experiment than the x-ray structure, neither was highly convincing. We hypothesized that flexibility within the ARM repeat domains themselves could explain the remaining differences and so tested four models in which flexibility was introduced at each of the interfaces between ARM repeat sub-domains. The model that agreed best with experimental data was a DmUNC-45 x-ray crystal structure with a TPR2A domain linked flexibly to its amino-terminus and flexibility at the interface between ARM repeat sub-domains 13–16 and 17–21 within the UCS domain (Table 2).
Limited proteolysis with chymotrypsin
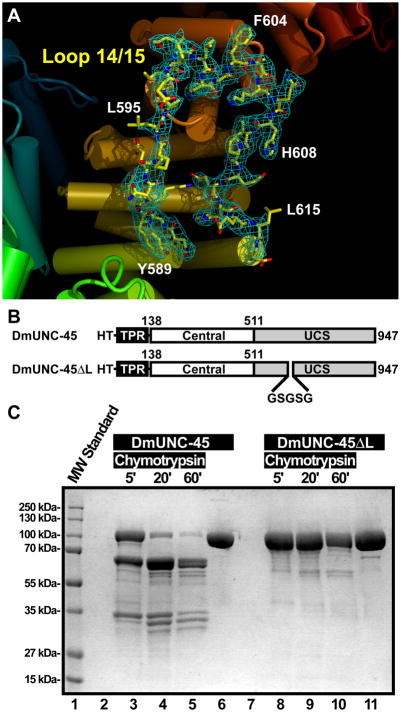
One of the most striking features of the DmUNC-45 x-ray crystal structure is an ordered loop of thirty amino acids positioned between ARM repeats 14 and 15 in the UCS domain (Figure 5A). The loop punctuates the ARM repeat stack of the UCS domain and likely contributes to the break in continuity between ARM repeats 13–16 and 17–21 in the UCS domain. Previous studies have shown that mammalian UNC-45 proteins are sensitive to proteolysis by both trypsin at chymotrypsin at sites that map within this loop (Price et al., 2002; Srikakulam et al., 2008).
Figure 5.
Protease sensitivity of loop L14/15. A) Electron density about the loop L14/15 region of DmUNC-45. Coloring is as in Figure 2B. Loop 14/15 is shown in a stick representation with electron density from a refined 2|FO| − |FC| map contoured at 1.1 σ depicted as a cyan colored mesh. Several individual amino acids are labeled. This electron density is representative of the quality throughout the final refined map. B) Schematic diagram of the full-length histidine-tagged DmUNC-45 protein used in crystallography studies and the DmUNC-45ΔL deletion construct in which amino acids corresponding to loop L14/15 (585–614) are replaced by a linker. C) Coomassie-blue stained 10% SDS-PAGE gel reveals that the full-length DmUNC-45 protein is readily cleaved into 66 and 34 kDa fragments by chymotrypsin (lanes 3–5) while the loop L14/15-deleted protein is resistant to proteolytic cleavage by chymotrypsin (lanes 8–10).
In order to confirm the solvent accessibility of L14/15 and as an independent test of the correctness of our crystallographic model to predict behavior in solution, we performed proteolysis experiments with limiting amounts of chymotrypsin on the recombinant DmUNC-45 proteins both of native sequence and a modified version in which the L14/15 loop amino acids 585–614 are replaced by the pentapeptide linker Gly-Ser-Gly-Ser-Gly (Figure 5B). The modified protein, which we refer to as DmUNC-45ΔL, was expressed and purified similarly to the native protein. However, it proved significantly resistant to proteolysis by chymotrypsin (Figure 5C). The native recombinant DmUNC-45 protein was rapidly cleaved into roughly 66 and 34 kDa fragments as illustrated by SDS-PAGE. Similar analysis of a construct that lacks its TPR domain yielded a similar pattern of proteolysis as the full-length protein, except that the 66 kDa band was replaced by a smaller fragment (data not shown). Finally, it is interesting to note that the TPR domain from the larger amino-terminal proteolytic fragment remains largely intact over the course of the limited proteolysis experiment. This might indicate that the TPR domain is well ordered in solution and that the lack of structural data derives from flexibility of the entire domain afforded by the short stretch amino acids that link it to the rest of the of the DmUNC-45 molecule. Notably, there are no aromatic amino acids present within this linker, which explains why chymotrypsin fails to target this apparently flexible region.
Identification of a conserved surface in the UCS domain
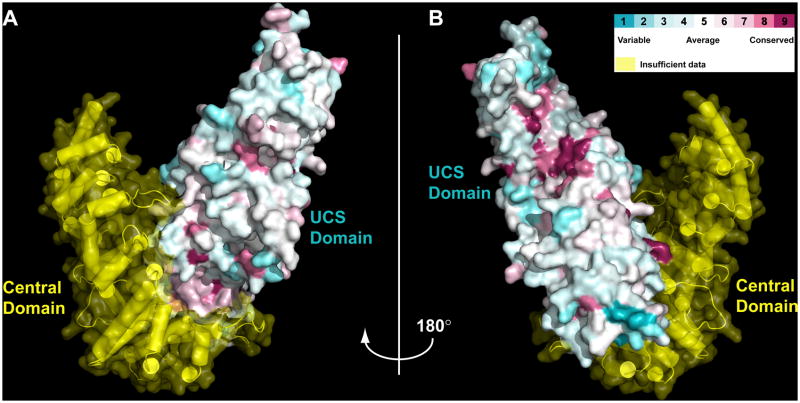
Other than their sequence homology, a physical and/or functional interaction with myosin is the defining feature shared by UCS proteins. In an effort to identify conserved surface amino acids that might be involved in mediating interaction with myosin, we carried out an alignment of the UCS domains from five divergent proteins: DmUNC-45, the human striated muscle UNC-45 protein (HsUNC-45B), P. anserina Cro1p, S. cerevisiae She4p, and S. pombe Rng3p. The results of pair-wise homology alignment are summarized in Table 3. Using the freely available ConSurf server, we assigned values of homology to each position within the UCS domain and then mapped those values onto the surface of the DmUNC-45 model (Landau et al., 2005). The result is a strikingly high degree of identity at amino acid positions that line the hydrophobic groove near the UCS domain carboxy-terminus (Figure 6). As a consequence of this observation, we hypothesize that conserved small hydrophobic amino acids that line the concave surface of DmUNC-45 created by helices H3 from the UCS domain ARM repeats 17–21 are involved in the interaction of these proteins with myosin.
Table 3.
Sequence homology within the UCS domains of functionally diverse UCS proteins
| HsUNC-45B | Cro1p | She4p | Rng3p | |
|---|---|---|---|---|
| DmUNC-45 | 41 (66) | 29 (51) | 20 (36) | 24 (43) |
| HsUNC-45B | 29 (48) | 20 (38) | 24 (42) | |
| Cro1p | 20 (42) | 26 (48) | ||
| She4p | 20 (39) |
Analysis was carried out using EMBOSS pairwise alignment (Rice et al., 2000). Sequence identity and similarity (in parentheses) are expressed as percentages.
Amino acid numbers: DmUNC-45 (511–923); HsUNC-45B (506–925); Cro1p (270–702); She4p (342–791); Rng3p (324–746).
Figure 6.
Conservation of amino acid positions from diverse UCS proteins mapped onto the surface of DmUNC-45. A) In this molecular surface representation of the DmUNC-45 x-ray crystal structure the Central domain is depicted in semitransparent yellow. The amino acid sequences of the five UCS proteins listed in Table 3 have been aligned and a numeric value of homology (1–9) has been assigned to each position. This value has then been mapped onto the surface. This view is the same as in Figure 1A. B) UCS domain surface amino acid conservation displayed on the DmUNC-45 surface viewed after 180° rotation about the y-axis from part A. Note the higher than normal degree of conservation within the groove of the UCS domain.
Mapping UCS domain mutation sites on DmUNC-45
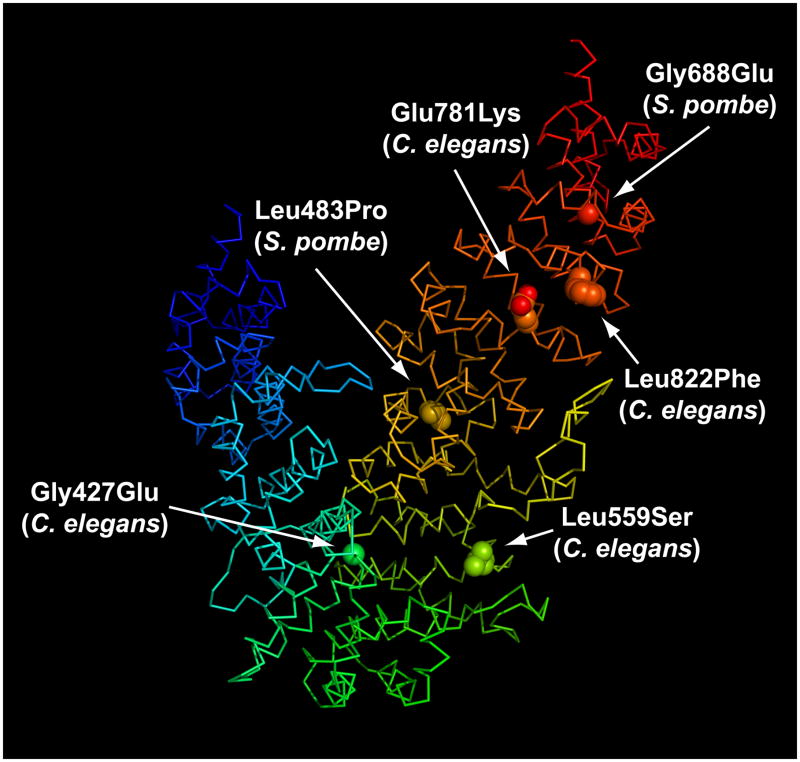
Genetic and physiological studies have uncovered mutations that can affect the ability of UCS proteins to function as essential modulators of myosin activity. Barral et al. identified two lethal alleles resulting from the introduction of in-frame STOP codons early in the Central domain and five temperature sensitive alleles caused by four unique single amino acid point mutations (Barral et al., 1998). Mapping the sites of these temperature sensitive mutations onto the DmUNC-45 structure reveals that they are distributed throughout the Central and UCS domains (Figure 7). Mutation of Gly427 in C. elegans UNC-45 (equivalent to Gly414 in DmUNC-45) to Glu likely disrupts the tight turn between ARM repeat layers 9 and 10 in the Central domain. The consequence on protein structure of mutation at C. elegans Leu559 (Leu545 in DmUNC-45) to Ser is less obvious. Mutation of C. elegans Glu781 (DmUNC-45 Glu766) to Lys would likely disrupt stability of the UCS domain since this residue in ARM repeat 18 participates in a conserved inter-repeat ion pair interaction with Arg803 from ARM repeat 19. Inter-repeat ion pairs between amino acid residues at consensus positions frequently contribute to stabilization of the closely related HEAT repeat domain fold (Andrade et al., 2001). The fourth C. elegans UNC-45 mutation identified by this screen was Leu822 to Phe. Interestingly, the identical position in DmUNC-45 is Phe806. The significance of this position is unclear from the structure alone, although its proximity to the Glu781-Arg803 ion pair suggest that stability in this region of UNC-45 is paramount for its function.
Figure 7.
Mutation sites mapped onto DmUNC-45. DmUNC-45 is depicted as a C-alpha trace with rainbow coloring as in Figure 1B. DmUNC-45 amino acid side chains that occupy mutation sites in other UCS proteins are rendered as space-filling spheres and labeled and numbered according to the original source species. See text for corresponding residues in DmUNC-45.
In studying the UCS protein Rng3p from S. pombe, Wong et al. identified two point mutations that impeded growth of fission yeast above the restrictive temperature (Wong et al., 2000). Mutation of Leu483 (analogous to DmUNC-45 Val669) to Pro is likely to introduce changes into helix H1 of ARM repeat layer 16 and, consequently, disrupt the unique inter-repeat interface at the border of the UCS ARM repeat 13–16 and 17–21 sub-domains. Mutation of Rng3 Gly688 (DmUNC-45 Gly865) to Glu introduces a charged amino acid into the hydrophobic groove created by helices H3 of the ARM repeat 17–21 sub-domain. On the whole, the mutational analyses suggest that the entire Central-UCS domain structure must be intact for UNC-45 to function. The higher prevalence of deleterious mutations within and around the carboxy-terminal ARM repeat 17–21 sub-domain lends added support to the hypothesis that this region is vital for proper function of UCS proteins.
DISCUSSION
Following up on earlier observations of UNC-45 protease sensitivity, Srikakulam et al. recently determined that trypsinolysis of a human UNC-45B protein yielded a carboxy-terminal 37 kDa fragment that fully retained the ability to interact with its amino-terminal 60 kDa fragment and to pull down myosin in vitro (Barral et al., 1998; Srikakulam et al., 2008). Lord et al. showed that the inability of Myo2p to function in actin gliding assays could be rescued through the addition of the UCS domain of Rng3p (Lord and Pollard, 2004). These and many other studies suggest that the UCS domain mediates the physical interaction between UCS proteins and myosins.
Though all UCS proteins studied thus far share the ability to influence myosin-mediated processes in cells, our understanding of the mechanisms by which they function remains incomplete. In support of efforts to uncover these mechanisms we have used x-ray crystallography to determine the structure of the UNC-45 protein from the fruit fly Drosophila melanogaster. The DmUNC-45 crystal structure reveals that the Central and UCS domains form a contiguous stack of armadillo repeats in a highly bent conformation. Though present in the crystals, electron density for the amino-terminal TPR domain was not observed throughout the course of structure refinement (Figure S1C, D). We conclude that the short polypeptide segment of ten amino acids in length that connects TPR and Central domains is flexible and that the lack of crystal contacts between the TPR domain and neighboring molecules in the high solvent DmUNC-45 crystal prevents its detection in x-ray diffraction experiments.
Small-angle x-ray scattering experiments carried out on DmUNC-45 suggest that in solution the protein rapidly samples a broad ensemble of folded conformations that are, on average, significantly elongated relative to the x-ray structure. We propose ARM repeat 13 as a point where a break in the canonical ARM repeat motif observed in the x-ray crystal structure is likely a consequence of the participation of the L13 loop in crystal packing contacts. Sequence analysis predicts that this loop contains the ARM repeat consensus residues and, therefore, supports our hypothesis that UNC-45 adopts a more linear conformation of stacked ARM repeats throughout this region. Besides the TPR domain, which clearly exhibits flexibility relative to the rest of the protein, our modeling data suggest the interface between ARM repeats 16 and 17 at the center of the UCS domain as another likely source of flexibility. Structurally, this region harbors a clear break in the continuous stack of UNC-45 ARM repeats that is mediated through unique inter-repeat contacts between ARM repeat layers 16 and 17 and the L14/15 loop. Furthermore, these elements represent regions of amino acid sequence that are among the most highly conserved throughout the family of diverse UCS proteins. Finally, limited proteolysis data presented in this and previous studies support our SAXS data in suggesting that, despite its apparent flexibility, DmUNC-45 is well folded in solution. Flexibility in the folded structure of the helical repeat protein importin-β has been observed in molecular dynamics simulations (Kappel et al., 2010).
The fundamental ARM repeat motif is a short alpha-helix (H1) followed by two longer helices (H2 and H3). The two longer helices run nearly anti-parallel to one another. ARM repeats connect one to another to generate a right-handed superhelix. Successive repeats stack linearly with their neighbors and the stacks exhibit a slightly right-handed superhelical twist such that successive helices H3 generate a right-handed spiral concave groove reminiscent of a corkscrew. This surface has been implicated as a binding site in the interaction of importin-α with diverse nuclear localization signal polypeptides (Conti and Kuriyan, 2000). Analysis of the DmUNC-45 calculated electrostatic surface potential and qualitative assessment of its surface polarity reveal that within the ARM repeat 17–21 sub-domain this groove is strikingly nonpolar in character. Even more intriguing, when the amino acid sequences from the five most evolutionarily divergent UCS proteins are compared, the highest degree of sequence identity maps to this surface. These data strongly suggest that this portion of the UNC-45 UCS domain is involved in mediating interactions with myosin.
One somewhat surprising conclusion from the DmUNC-45 crystal structure is the ARM repeat structure of the entire Central domain. This portion of UNC-45 is less well conserved at the amino acid level. It is possible that the role played by the Central domain in UNC-45 is one of providing the appropriate arrangement in space for the amino-terminal TPR and carboxy terminal UCS domains. In light of the curvature supported by the Central domain within the conformation of DmUNC-45 observed in its crystal structure, another function of the Central domain might include a contribution to the overall conformational flexibility of the molecule. The absence of TPR domains in the fungal UCS proteins suggest that Central domain-like regions within these proteins might carry out additional functions.
While our DmUNC-45 crystallographic model provides a structural template for future genetic and biochemical studies aimed at deducing the molecular mechanisms of UCS proteins, such mechanistic details are speculative at present. However, one can imagine a scenario in which binding to Hsp90 through a flexibly linked TPR domain on the amino-terminal end of UNC-45 and contact with myosin through the ARM repeat 17–21 sub-domain on the carboxy-terminal end might facilitate an interaction between the motor protein and the Hsp90 protein-folding chaperone. The flexibility we observe in DmUNC-45 in solution might support interaction between the two bound factors in a manner akin to cullin family E3 ubiquitin-protein ligase complex scaffolds that modulate the interaction of E2 ubiqutin-conjugating enzymes and their substrates (Petroski and Deshaies, 2005). The modular arrangement of five ARM repeat sub-domains throughout the larger Central and UCS domains of DmUNC-45 suggests the possibility that each of these ARM repeat stacks might be capable of arranging itself in multiple conformations relative to one another. Furthermore, it is possible that each sub-domain mediates different weak binding interactions with unique portions on the specific myosin target or other binding partner. Finally, the presence of conserved loops within and between individual ARM repeats leads to the intriguing possibility that they might be involved in binding partner recognition or in mediating allosteric regulation through interaction with other factors or small molecule modifiers.
During the review process Shi and Blobel have reported the x-ray crystal structure of the S. cerevisiae She4p protein (Shi and Blobel, 2010).
EXPERIMENTAL PROCEDURES
Protein expression, purification, and crystallization
Details regarding the design and construction of recominant DmUNC-45 expression plasmids are available as Supplemental Information. Expression and purification of DmUNC-45 in E. coli has been described elsewhere (Melkani et al., 2010). Se-Met substituted proteins were expressed in 1X M9 salts augmented with amino acids and Se-Met. Peak protein fractions from size exclusion chromatography were pooled and concentrated by centrifugation to 20 mg/mL, flash cooled in liquid nitrogen, and stored at −80°C. DmUNC-45 crystals were grown by the hanging drop vapor diffusion method. 2 μL of protein were mixed with 1 μL of mother liquor solution (0.1 M sodium citrate pH 5.6, 0.2 M ammonium acetate, 15% (w/v) polyethylene glycol 4000, and 1% (v/v) ethylene glycol) and sealed over 1 mL mother liquor. Single crystals of up to 0.4 × 0.4 × 0.4 mm grew at 18°C after 2–4 days.
Data collection and processing
Native and Se-Met crystals of approximately 0.15 mm on each edge were transferred by nylon loop directly into a mother liquor solution augmented with 18% glycerol for 1 minute before flash cooling and storage in liquid nitrogen. The 0.125 mm L63M Se-Met crystal was transferred stepwise into stabilizer solutions containing 15% and 30% dextrose for ten minutes each prior to flash cooling. Synchrotron data were collected at 100 K on an ADSC q315 CCD detector and processed in HKL2000 (Otwinowski and Minor, 1997).
Structure solution, model building, and refinement
The processed Se-Met DmUNC-45 data was run under the single anomalous dispersion (SAD) routine within the AutoSol module of Phenix (Adams et al., 2010). The Phenix AutoBuild module was run using all of the reflection intensities collected in the native 3.0 Å data set. The resulting initial model contained 501 amino acids (469 of which were Ala) with an R-factor of 0.432 and R-free of 0.459. All polypeptides in the computer-generated model exhibited extended conformations despite the preponderance of electron density that was clearly alpha-helical. Consequently, manual model building commenced by maximizing alpha-helical polypeptide structure and was later influenced by recognition of ARM repeat motifs in the structure. The 19 unique Se sites were referred to extensively in establishing and maintaining the proper register during ARM repeat model building. Finally, manual building and fitting of connecting loops into 2|FO| − |FC| electron density maps was carried out in Coot (Emsley et al., 2010). The L63M structure was determined independently by SAD in Phenix, built using the native DmUNC-45 structure as a guide, and refined.
SAXS data collection
SAXS data were collected with synchrotron x-rays (λ=1Å) at room temperature on the SIBYLS beamline (12.3.1) at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory on purified full-length DmUNC-45 and the TPR domain-lacking mutant (DmUNC-45ΔTPR) at three concentrations (5, 2.5, and 1.25 mg/mL) in 20 mM Tris-HCl (pH 7.5), 500 mM NaCl, 1 mM DTT and at four exposure times (0.5, 1, 8, and 0.5 s) (Putnam et al., 2007). The beamline was equipped with a MarCCD 165 detector positioned at a distance of 1.5 m. Data were collected over an s-range of 0.0113-0.323 Å−1. All scattering data were averaged radially and buffer signal was subtracted prior to analysis using orgeNEW.
SAXS data analysis
Extrapolation of the scattering profiles to zero angle for both DmUNC-45 and DmUNC-45ΔTPR over the concentration range described revealed no concentration dependent oligomerization. Data processing and Guinier analysis were performed using PRIMUS. Indirect Fourier transformations of the scattering data were performed with GNOM using data sets consisting of 507 data points over the s-range 0.0149–0.3232 for DmUNC-45 and 512 data points over the s-range 0.0112–0.3219 for DmUNC-45ΔTPR (Svergun, 1992). The maximum intramolecular scattering distance, Dmax, was determined iteratively using the PERL script RunGNOMRun. Pair distribution functions were plotted as a function of intramolecular distances in EXCEL. Theoretical scattering curves from the atomic coordinates of the DmUNC-45 crystal structure and associated DmUNC-45 flexible conformer models were generated and compared to the experimental scattering data curves using CRYSOL (Svergun et al., 1995). To generate atomic models with engineered flexibility between adjacent ARM sub-domains in the DmUNC-45 crystal structure (with and without the TPR domain derived from PDB 1ELR), the Ensemble Optimization Method (EOM) was used. From a large number of random conformers (15,000) generated by EOM, a subset of 50 best-fitting conformers were selected using a genetic algorithm to give an ensemble of conformers that best fit the experimental scattering data.
Sequence and structural analyses
ARM repeat detection was carried out according to previously published methods (Kippert and Gerloff, 2009). Multiple sequence alignment of the five most divergent UCS proteins was performed using EMBOSS (Rice et al., 2000). Positional amino acid conservation was then scored by the ConSurf server (Landau et al., 2005). Figures were made in PyMol (DeLano, 2002).
Proteolysis assay
2 μg chymotrypsin (EMD) was added to 100 μg native full-length DmUNC-45 and DmUNC-45ΔL in 100 μL size exclusion buffer. Reactions were incubated at room temperature and 10 μL samples were taken at 5, 20, 60, 180 minutes, quenched with 1 μL 250 mM PMSF and 4 μL of 4X Laemmli buffer, heated to 96°C for 2 minutes, centrifuged, and stored at −20°C. The collected samples were separated by SDS PAGE and analyzed by Coomassie blue stain.
Supplementary Material
Acknowledgments
The authors thank Y. Suzuki for assistance in the wet lab, C. Ralston and P.H. Zwart for support during synchrotron data collection, G.L. Hura for assistance with SAXS data collection and analysis, F. Kippert and D. Gerloff for ARM repeat analysis, and D.-B. Huang for helpful discussion. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This research is funded by NIH/NIAMS grant R01-AR055958 to S.I.B. T.H. is the recipient of an American Cancer Society grant RSG-08-287-01-GMC. Research in the Department of Chemistry & Biochemistry at SDSU is supported in part by the California Metabolic Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, Petosa C, O'Donoghue SI, Müller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Bauer CC, Ortiz I, Epstein HF. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J Cell Biol. 1998;143:1215–1225. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 2002;295:669–671. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- Bazzaro M, Santillan A, Lin Z, Tang T, Lee MK, Bristow RE, Shih Ie M, Roden RB. Myosin II co-chaperone general cell UNC-45 overexpression is associated with ovarian cancer, rapid proliferation, and motility. Am J Pathol. 2007;171:1640–1649. doi: 10.2353/ajpath.2007.070325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berteaux-Lecellier V, Zickler D, Debuchy R, Panvier-Adoutte A, Thompson-Coffe C, Picard M. A homologue of the yeast SHE4 gene is essential for the transition between the syncytial and cellular stages during sexual reproduction of the fungus Podospora anserina. EMBO J. 1998;17:1248–1258. doi: 10.1093/emboj/17.5.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio LM. A Superfamily of Molecular Motors. Vol. 7. Dordrecht: Springer; 2008. Myosins. [Google Scholar]
- Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Thomson JN. Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature. 1974;250:579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- Etard C, Roostalu U, Strahle U. Shuttling of the chaperones Unc45b and Hsp90a between the A band and the Z line of the myofibril. J Cell Biol. 2008;180:1163–1175. doi: 10.1083/jcb.200709128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves MR, Barford D. Topological characteristics of helical repeat proteins. Curr Opin Struct Biol. 1999;9:383–389. doi: 10.1016/s0959-440x(99)80052-9. [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Landsverk ML, Price MG, Epstein HF. The UCS family of myosin chaperones. J Cell Sci. 2002;115:3983–3990. doi: 10.1242/jcs.00107. [DOI] [PubMed] [Google Scholar]
- Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Kappel C, Zachariae U, Dolker N, Grubmuller H. An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins. Biophys J. 2010;99:1596–1603. doi: 10.1016/j.bpj.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippert F, Gerloff DL. Highly sensitive detection of individual HEAT and ARM repeats with HHpred and COACH. PLoS One. 2009;4:e7148. doi: 10.1371/journal.pone.0007148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Srikakulam R, Winkelmann DA. Unc45 activates Hsp90-dependent folding of the myosin motor domain. J Biol Chem. 2008;283:13185–13193. doi: 10.1074/jbc.M800757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin–II. J Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkani GC, Lee CF, Cammarato A, Bernstein SI. Drosophila UNC-45 prevents heat-induced aggregation of skeletal muscle myosin and facilitates refolding of citrate synthase. Biochem Biophys Res Commun. 2010;396:317–322. doi: 10.1016/j.bbrc.2010.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction data Collected in Oscillation Mode. In: Carter CW Jr, Sweet RM, editors. Macromolecular Crystallography. part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Price MG, Landsverk ML, Barral JM, Epstein HF. Two mammalian UNC-45 isoforms are related to distinct cytoskeletal and muscle-specific functions. J Cell Sci. 2002;115:4013–4023. doi: 10.1242/jcs.00108. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Shi H, Blobel G. UNC-45/CRO1/She4p (UCS) protein forms elongated dimer and joins two myosin heads near their actin binding region. Proc Natl Acad Sci USA. 2010;107:21382–21387. doi: 10.1073/pnas.1013038107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikakulam R, Liu L, Winkelmann DA. Unc45b forms a cytosolic complex with Hsp90 and targets the unfolded myosin motor domain. PLoS One. 2008;3:e2137. doi: 10.1371/journal.pone.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- Tochio N, Sasagawa A, Koshiba S, Inoue M, Kigawa T, Yokoyama S. PDB ID: 2DBA The solution structure of the tetratrico peptide repeat of human Smooth muscle cell associated protein-1, isoform 2. RIKEN Structural Genomics/Proteomics Initiative (RSGI); [Google Scholar]
- Toi H, Fujimura-Kamada K, Irie K, Takai Y, Todo S, Tanaka K. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:2237–2249. doi: 10.1091/mbc.E02-09-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L, Ao W, Kim S, Kim C, Pilgrim D. unc-45 gene of Caenorhabditis elegans encodes a muscle-specific tetratricopeptide repeat-containing protein. Cell Motil Cytoskeleton. 1999;42:163–177. doi: 10.1002/(SICI)1097-0169(1999)42:3<163::AID-CM1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Venolia L, Waterston RH. The unc-45 gene of Caenorhabditis elegans is an essential muscle-affecting gene with maternal expression. Genetics. 1990;126:345–353. doi: 10.1093/genetics/126.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MG. Pharmaceutical target identification by gene expression analysis. Mini Rev Med Chem. 2001;1:197–205. doi: 10.2174/1389557013407034. [DOI] [PubMed] [Google Scholar]
- Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth SL, Crawford BD, Pilgrim DB. The myosin co-chaperone UNC-45 is required for skeletal and cardiac muscle function in zebrafish. Dev Biol. 2007;303:483–492. doi: 10.1016/j.ydbio.2006.11.027. [DOI] [PubMed] [Google Scholar]
- Wong KC, Naqvi NI, Iino Y, Yamamoto M, Balasubramanian MK. Fission yeast Rng3p: an UCS-domain protein that mediates myosin II assembly during cytokinesis. J Cell Sci. 2000;113(Pt 13):2421–2432. doi: 10.1242/jcs.113.13.2421. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19:ii246–255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- Yu Q, Bernstein SI. UCS proteins: managing the myosin motor. Curr Biol. 2003;13:R525–527. doi: 10.1016/s0960-9822(03)00447-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.