Abstract
Mounting evidence suggests that autophagy is a more selective process than originally anticipated. The discovery and characterization of autophagic adapters, like p62 and NBR1, has provided mechanistic insight into this process. p62 and NBR1 are both selectively degraded by autophagy and able to act as cargo receptors for degradation of ubiquitinated substrates. A direct interaction between these autophagic adapters and the autophagosomal marker protein LC3, mediated by a so-called LIR (LC3-interacting region) motif, their inherent ability to polymerize or aggregate as well as their ability to specifically recognize substrates are required for efficient selective autophagy. These three required features of autophagic cargo receptors are evolutionarily conserved and also employed in the yeast cytoplasm-to-vacuole targeting (Cvt) pathway and in the degradation of P granules in C. elegans. Here, we review the mechanistic basis of selective autophagy in mammalian cells discussing the degradation of misfolded proteins, p62 bodies, aggresomes, mitochondria and invading bacteria. The emerging picture of selective autophagy affecting the regulation of cell signaling with consequences for oxidative stress responses, tumorigenesis and innate immunity is also addressed.
Key words: p62, ubiquitin, LIR, selective autophagy, cell signaling, protein aggregates
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved catabolic process that involves the sequestration and transport of organelles and macromolecules to the lysosomes for degradation.1,2 Following lysosomal degradation, recycling occurs to replenish the cell with nutrients and building blocks for anabolic processes.3 Autophagy is initiated by formation of the phagophore or isolation membrane, a crescent-shaped double membrane that expands and fuses to form a double-membrane vesicle, the autophagosome. Autophagosomes eventually fuse with lysosomes to degrade their content (Fig. 1). Autophagy has most likely evolved both as a pathway for restoring intracellular nutrient supply during starvation and as a quality control mechanism to protect the cell against damage caused by toxic macromolecules and damaged organelles. The process is also mobilized in the innate immune response against invading microbes.1,4,5
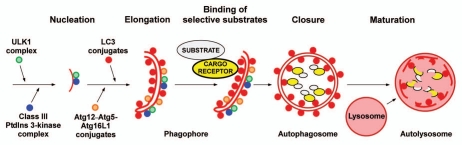
Figure 1.
Model for selective autophagy in mammalian cells. Autophagosome formation is initiated (nucleation step) by the ULK1 complex and the class III PtdIns 3-kinase complex located at the phagophore. Additional ATG proteins are required for growth of the phagophore (elongation step), that depends on two Ub-like conjugation reactions. First, conjugation of ATG12 to ATG5 results in the formation of an oligomeric complex between the ATG12-ATG5 conjugate and ATG16L. This complex then acts as an E3 ligase assisting the E2 ATG3 in the lipidation of ATG8 family proteins at the phagophore. Selective autophagy depends on binding of substrates to the inner surface of the growing phagophore, and this can be achieved by cargo receptors that are associated both with the substrate and with lipidated ATG8 family proteins anchored to the phagophore. Aggregation of the substrate and/or cargo receptor is required for efficient sequestration. Closure results in the formation of a double-membrane autophagosome. Fusion of autophagosomes with late endosomes or lysosomes (maturation step) is then required for the formation of autolysomes where the substrates are degraded.
Yeast genetics has been vital for the elucidation of the molecular machinery involved in autophagy processes.2,6 So far, 34 AuTophaGy-related (ATG) genes have been reported in yeast and 15 of these are “core” ATG genes commonly required for the different autophagy pathways.6,7 Two ubiquitin (Ub)-like protein conjugation systems involving the two Ub-like modifiers Atg8 and Atg12 constitute part of the evolutionarily conserved autophagic machinery. Both systems use the E1 enzyme Atg7. Atg10 acts as an E2 for the conjugation of Atg12 to Atg5 to form a high-molecular-weight complex with Atg16, which then in turn assists the E2 Atg3 in the conjugation of phosphatidylethanolamine (PE) to Atg8. Very recently, mammalian ATG12 was also shown to conjugate to ATG3. This conjugation is not involved in starvation-induced autophagy but is important for regulation of mitochondrial homeostasis and cell death.8 In addition to the two conjugation systems, four more protein complexes make up the remainder of the molecular machinery required for autophagy (Fig. 1). In mammalian cells the serine-threonine protein kinase ULK1 (unc-51-like kinase 1, a homologue of yeast Atg1) forms a complex together with ATG13, FIP200 (focal adhesion kinase family interacting protein of 200 kD) and ATG101. The ULK1 complex regulates the initial events of autophagosome formation together with the class III phosphatidylinositol (PtdIns) 3-kinase complex containing Beclin 1, ATG14L and the PtdIns 3-kinase subunits Vps34 and Vps15. The WIPI (WD-repeat protein interacting with phosphoinositides) family proteins, notably WIPI1 and WIPI2, are Atg18 homologues acting in early steps of autophagosome formation.9,10 Finally, the ATG9 transmembrane protein is involved in membrane trafficking and is required for autophagy, but its function is still unknown.11
In yeast there is a single Atg8 protein; Drosophila and C. elegans have two, while mammals have at least seven ATG8 proteins that can be grouped into two subfamilies containing at least three MAP1 light chain 3 (LC3A, B and C) and four gamma-aminobutyrate receptor-associated protein (GABARAP) and GABARAP-like proteins (ATG8L/GEC-1/GABARAPL1, GATE-16/GABARAPL2 and GABARAPL3).12,13 ATG8 proteins are specifically cleaved at their C-termini by the ATG4 pro- teases to expose a C-terminal glycine producing the form I of the ATG8 molecule (i.e., LC3-I). This glycine residue is then conjugated to PE.14–17 The resulting PE-conjugated form II of ATG8 proteins (i.e., LC3-II) is tightly bound to the autophagosomal membranes and serves as an autophagic marker protein.14,16,18,19 The conjugation of ATG8 to PE is essential for the hemifusion of lipid membranes and is suggested to drive the expansion of the autophagosome.20 ATG8-PE is deconjugated by ATG4 protease to recycle ATG8 from the outer autophagosomal membrane.21 This step can be regulated by reactive oxygen species acting on ATG4.22 In mammals, LC3B is thought to act as the main ATG8 homologue involved in starvation-induced autophagy, but also GABARAP, GABARAPL2 and GABARAPL1 are conjugated to PE and involved in autophagy.16,23 Recent data suggest that both the LC3 and GABARAP subfamilies of ATG8s are indispensable for autophagy in mammalian cells and that there is a division of labor among them during formation of the autophagosome.24
Chaperone-mediated autophagy (CMA) is a selective form of autophagy where autophagosomes do not form, and soluble cytosolic proteins containing a degenerate short sequence motif related to the pentapeptide KFERQ are degraded by the lysosomes. Degenerate variants of this sequence motif are found in 30% of cytosolic proteins. CMA substrate proteins with a KFERQ-like motif bind to a complex of Hsc70, Hsp90 and several co-chaperones associated with the lysosomal receptor LAMP-2A and are transported directly through the membrane into the lumen of the lysosome for degradation.25,26
Another pathway involving transport of selective cargo to the lysosome with participation of part of the autophagic machinery is the biosynthetic cytoplasm-to-vacuole targeting pathway in yeast.7 In the Cvt pathway, precursors of two lysosomal enzymes, aminopeptidase I (Ape1) and α-mannosidase, are selectively delivered to the vacuole in a double-membrane vesicle after recognition of the cargo and of yeast Atg8 by the cargo receptor Atg19. Atg19 binds to an aggregate of dodecameric precursor Ape1 (prApe1) and to α-mannosidase and forms the Cvt complex. In addition, Atg11 binds to Atg19 and is required for transport of the Cvt complex to the phagophore assembly site (PAS) in yeast where the double membrane is recruited and surrounds the Cvt complex.7
In contrast to the selectivity seen in the CMA and Cvt pathways, (macro)autophagy has been considered a rather unselective process for bulk degradation of long-lived proteins and organelles that during nutrition deprivation can recycle building blocks and help restore the energy balance of the cell. However, a number of recent reports present mounting evidence of selective autophagic degradation of protein inclusions caused by aggregate-prone or misfolded proteins (aggrephagy),27–35 of organelles such as peroxisomes (pexophagy),36,37 mitochondria (mitophagy),38–43 bacteria and virus (xenophagy),44–48 surplus ER (reticulophagy),49 and ribosomes (ribophagy).50 In the following sections we will discuss recent developments in the field of selective autophagy with particular focus on the roles played by the autophagic adaptors p62/SQSTM1 (sequestosome 1) and NBR1 (neighbor of Brca1 gene). p62 and NBR1 are both selective autophagy substrates and cargo receptors for degradation of ubiquitinated substrates by autophagy.31,33,51 As discussed further below and in separate reviews in this issue of Autophagy such substrates may include protein aggregates,29,31–33,35,51 soluble proteins,37,52 midbody rings,53 damaged mitochodria,39–41 peroxisomes,37 intracellular bacteria,46–48 phagocytic membrane remnants,44 bacteriocidal precursor- and viral capsid proteins.45,54
p62 and NBR1 are Substratesfor Selective Autophagy
The p62 cDNA was originally cloned and named p62 Lck-ligand because the encoded protein bound to the tyrosine kinase Lck.55 Shin, who originally cloned p62, noted the ability of p62 to form aggregates and coined the name sequestosome 1 (SQSTM1) based on this property.56 Later, the mouse and rat orthologues were named A170 and ZIP, respectively.57,58 The human p62 protein is 440 amino acids long and contains an N-terminal PB1 domain followed by a ZZ-type zinc finger domain, nuclear localization signals (NLS), nuclear export signal (NES), LIR (LC3-interacting region) and KIR (KEAP1 interacting region) motifs and a C-terminal UBA (Ub-associated) domain (Fig. 2A). The PB1 domain is a protein-protein interaction domain enabling p62 to interact with the protein kinases PKCζ, PKCλ/ι, MEKK3 and MEK5 as well as to NBR1.59–62 Importantly, p62 is able to homopolymerize via its PB1 domain.59,61 Interestingly, p62 harbors active nuclear import and export signals and shuttles between the nucleus and cytoplasm.63 The UBA domain of p62 binds both mono- and poly-Ub.64 p62 seems to have a preference for binding to monoUb in vitro.65 Since K63-linked Ub chains have a more open and extended conformation than K48-linked Ub chains, this is consistent with a reported in vivo preferential binding to K63-linked Ub chains.66 However, the isolated UBA domain binds with low affinity,65,67 whereas full-length p62 binds much more strongly to both mono Ub and 4x Ub when assayed in vitro using GST-pulldown assays.31 The UBA domain of p62 forms homodimers in vitro and this inhibits Ub binding.68 The importance of the UBA domain is underscored by the fact that adult onset Paget disease of the bone is frequently associated with dominant-acting mutations in one of the p62 alleles that either lead to deletion of the UBA domain or destroys its ability to bind Ub.69 Likely, p62 mutants without functional UBA increase osteoclastogenesis by potentiating receptor activator of nuclear factor-kappaB ligand (RANKL)-induced activation of nuclear factor-kappaB (NFκB).70–72 This effect may involve TRAF6 and/or atypical protein kinase Cs, and it will be interesting to see whether selective autophagy can be involved in regulating RANKL signaling. As discussed by Sumpter and Levine in another review in this issue of Autophagy, there may also be a link between mutant p62 and accumulation of paramyxovirus nucleocapsids in bone lesions of Paget disease patients.
Figure 2.
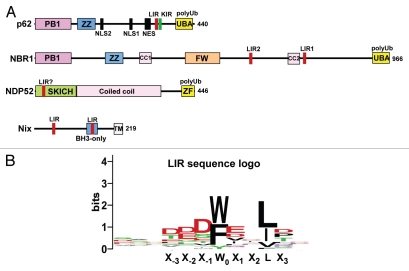
(A) Domain architecture of the hitherto characterized mammalian autophagic cargo receptors p62, NBR1, NDP52 and Nix. The FW domain has been named NBR1 box in a review by Kraft et al.79 (B) A consensus sequence logo for the LIR motif. The logo is based on 25 different LIR motifs from 21 different proteins that all bind directly to ATG8 family proteins.
p62 accumulates in Ub-positive inclusions in neurodegenerative diseases and proteinopathies of the liver.73–76 Immunostaining of p62 has been recommended as histochemical diagnostic marker, together with Ub and cytokeratins, for various human protein aggregation diseases.77 p62- and Ub-positive protein bodies form in response to stress including amino acid starvation, oxidative stress, accumulation of defective ribosomal products or inhibition of autophagy.28,32,33 While studying the PB1 domain-mediated interaction between p62 and atypical PKC we noticed that p62 displayed a perinuclear, punctate distribution in cells and that overexpression increases the number and size of these structures.59 We could subsequently show that p62 is a selective autophagy substrate, continuously degraded by autophagy.29,33 Our group and Komatsu's group independently showed that p62 binds directly to LC3 via the LIR motif (named LC3 recognition sequence; LRS, by Komatsu) and this motif is required for autophagic degradation of p62.33,51 The LIR motif mediates binding to all the members of the mammalian ATG8 family of proteins (both LC3- and GABARAP subfamilies).31,33,51
In a collaborative effort involving the groups of Dikic, Komatsu and Elazar we identified NBR1 as another autophagic cargo receptor that is selectively degraded by autophagy.31 The 966-amino acids long NBR1 is one of the PB1-domain containing interaction partners of p62 and has a similar domain organization as p62. They both share the arrangement of PB1, ZZ and UBA domains,78 but the twice-as-long NBR1 also has other domains not found in p62. Among these is a coiled coil domain required for dimerization of NBR1 and a novel, evolutionarily conserved domain of unknown function we denote the FW (Four W (tryptophan)) domain (Fig. 2A). This domain was recently called the NBR1 box by Kraft et al.79 We prefer the name FW domain based on the four conserved tryptophan residues since this domain is also found in another evolutionarily conserved eukaryotic protein. We found that the protein level of NBR1 is regulated by autophagy, and NBR1 is, together with p62 and Ub-proteins, located to inclusion bodies associated with human pathologies. NBR1 cannot polymerize via its PB1 domain, like p62 can. Instead it binds to p62 via this domain and can be part of, or the chain terminator of, a polymeric chain of p62 molecules. We mapped two LIR motifs in NBR1 of which LIR1, centered at Y732, is most important for autophagic degradation of NBR1. In contrast to p62, the isolated UBA domain of NBR1 binds very well to both K48- and K63-linked diUb.31 The role of NBR1 as an autophagy receptor interacting with LC3 and Ub was confirmed by a study from the group of Mathias Gautel.80
In contrast to p62, NBR1 has not been extensively studied. However, this is now changing and the protein has already been implicated in quite diverse biological contexts. NBR1 binds directly to the giant sarcomeric protein kinase titin and to p62 in the M line of the sarcomere of skeletal muscles. Mutations in titin that disrupt the binding to NBR1, cause hereditary muscle disease in humans.81 Upon loss of binding to titin, both NBR1 and p62 are dispersed from the M lines into many punctate structures probably being so-called p62 bodies (see below). It is not known if autophagy plays any role in this disease. NBR1 interacts and colocalizes with Spred2 on intracellular vesicles.82 This interaction with NBR1 is essential for the role of Spred2 as an attenuator of fibroblast growth factor signaling resulting in lysosomal degradation of activated receptors. A recent study of transgenic mice expressing a C-terminally truncated mutant of NBR1 suggests a role for NBR1 in bone remodeling. Loss of NBR1 function leads to perturbation of p62 levels and hyperactivation of p38 MAPK favoring osteoblastogenesis.83 Hence, both p62 and NBR1 are involved in bone remodeling processes and may cooperate or have antagonistic roles. More studies are needed to elucidate their relative contributions. Jorge Moscat's group has knocked out NBR1 in the T cells of mice and found that NBR1 is a critical mediator of T-cell activation and acts in the control of Th2 differentiation and allergic airway inflammation.84 Interestingly, NBR1 is involved in T cell polarity and recruited to the immunological synapse.
NBR1 displays a wider evolutionary distribution than p62. NBR1 is found in plants and fungi and in metazoans, but the gene is lost in several animal lineages including model organisms like Drosophila and C. elegans. p62 is not found in plants and fungi and seems confined to the metazoans.
Mechanistic Basis for Selective Autophagy: The LIR-ATG8 (LC3/GABARAP) Interaction
Autophagic degradation of p62 is dependent on an 11-amino acid long, linear motif (LC3-interacting region; LIR) mediating an interaction with LC3/GABARAP family proteins.33,51,85 Our group initially mapped the LIR motif to a 22 amino acid-long sequence of human p62 harboring an evolutionarily conserved motif of three acidic residues followed by a tryptophan (DDDW in p62). A triple mutation of the three aspartic acid residues to alanine or a single mutation of the W to A abolished the interaction with LC3.33 Komatsu's group subsequently showed that the 11-amino acid-long conserved motif is sufficient to confer binding to LC3.51 The groups of Komatsu and Inagaki reported x-ray and NMR structures, respectively, of the LIR peptide of p62 docked onto LC3.51,85 The p62 LIR with its core sequence DDDWTHL appears as an extended β-strand and forms an intermolecular parallel β-sheet with the β2 strand of LC3. LIR is positioned in the interface between the N-terminal arm and the C-terminal Ub-like domain of the ATG8 protein. The side chains of the W and L residues are bound in two hydrophobic pockets (the “W-site” and “L-site”) in the Ub-like domain of LC3.51,85,86 In addition to these hydrophobic interactions, electrostatic interactions involving the three D residues of the LIR motif and basic residues in the N-terminal arm and Ub-like domain of LC3 (R10, R11, K49 and K50) are also important for the p62-LC3 interaction. The importance of both the WXXL core motif and the acidic residues of the p62 LIR motif was verified by substitution of these residues with alanine.33,85,87 The crucial role of the interaction with the N-terminal arm of LC3 for binding and autophagic degradation of p62 was demonstrated by Elazar's group.88
We have compiled a sequence logo from 25 different LIR motifs from 21 different proteins that all have been tested for binding to ATG8 family proteins (Fig. 2B). The LIR motifs come from published LC3B-, GABARAP- and/or GABARAPL1-interacting proteins,52,89–92 as well as 14 unpublished LIR motifs from 12 different proteins mapped and verified by our group. As is evident from the sequence conservation revealed by the logo, the LIR motif seems to be eight amino acids long. Adopting the nomenclature of Inagaki's group (X−3X−2X−1W0X1X2 LX3),86 acidic residues (D and E) are prevalent in the X−3X−2X−1 positions and in the X1 position. The residue binding to the W-site is usually W or F although the main LIR in NBR1 harbors a Y at that position,31 as does a LIR identified in human ATG4B.92 The L-site can be occupied by either L, I or V (Fig. 2B). Based on the current knowledge, a consensus LIR motif could be written as D/E-D/E-D/E-W/F/Y-X-X-L/I/V where there is not an absolute requirement for acidic residues at all three X−3X−2X−1 positions but usually there is at least one at these positions. If there are no acidic residues at these positions there are usually acidic residues at X1 or X2, or at both, like in yeast Atg19. The requirement for aromatic residues in the W-site seems absolute as is the requirement for large, hydrophobic residues in the L-site (L/I/V). As more LIR motifs are being characterized it is likely that we will see a pattern where proteins binding preferentially to the LC3 subfamily have somewhat different residue preferences at certain positions of the eight amino acid core relative to those that prefer binding to the GABARAP subfamily of mammalian ATG8 proteins.
Following the characterization of the LIR motif in p62, similar motifs were identified in other ATG8-interactors, including yeast Atg19,85 the cargo receptor in the yeast Cvt pathway. NBR1, Nix, calreticulin and clathrin heavy chain use LIR motifs to bind to LC3 and/or GABARAP subfamily members.31,38,93–95 We recently reported that a novel protein of hitherto unknown function(s) named FYCO1 (FYVE and coiled-coil domain-containing protein 1) binds to LC3, Rab7 and PtdIns-3-phosphate (PtdIns(3)P) on autophagosomes and autolysosomes.90 FYCO1 binds to LC3 via a LIR motif located adjacent to the FYVE domain. Our results suggest that FYCO1 forms an adaptor complex on the autophagosomes that is involved in kinesin-mediated microtubule plus end-directed transport of autophagosomes and other Rab7-positive vesicles. FYCO1 is recruited to the outer membrane of autophagosomes and is as such not an autophagy substrate or cargo receptor for autophagic degradation. Lately, an important adaptor protein in the Wnt signaling pathway, dishevelled (Dvl), was shown to interact with LC3 via a LIR motif and to be degraded by autophagy.52
The groups of Inagaki and Ohsumi have collaborated on showing that both the core autophagy components yeast Atg3 and human ATG4B interact with yeast Atg8 and human LC3B, respectively, via LIR motifs.91,92 In the x-ray crystal structure of ATG4B-LC3B the sequence motif YDTL in the N-terminal arm of ATG4B is bound to the LIR docking site (LDS) of the adjacent LC3B molecule where Y occupies the W-site and L the L-site.91 The biological relevance of this interaction is not yet understood. The yeast Atg3 LIR motif (DWEDL) is required for transfer of Atg8 from the Atg8-Atg3 thioester intermediate to PE, but not for the formation of the intermediate. Interestingly, the Atg3 LIR is required for the Cvt pathway, but not for starvation-induced autophagy, and is important for lipidation of Atg8 that is bound to the Cvt pathway cargo receptor Atg19.92
The list of LIR-containing ATG8 interactors is rapidly expanding. The group of Wade Harper conducted a proteomics screen of the autophagy interaction pathway in human embryonic kidney cells (HEK293T) under conditions of basal autophagy.96 Using immunoprecipitation experiments of tagged proteins and mass spectrometry (LC-MS/MS) to identify co-precipitated proteins, Berhends and coworkers also tested the six ATG8 orthologues LC3A, B and C, GABARAP, GABARAPL1 and GABARAPL2, and found 67 so-called high-confidence candidate interaction proteins.96 Among these only p62, NBR1, FYCO1, ATG3 and ATG4B were known from before. Interestingly, more components of the conjugation machinery such as ATG7, ATG10, ATG5 and ATG16L were identified as ATG8 interactors and many proteins in the ATG8 interaction network contain domains implicated in vesicle function including a number of Rab GTPase activating proteins. Also several protein kinases feature in this network.
Using mutants in the LIR docking site (LDS) Behrends et al. found a significant fraction of ATG8 interactors to show LIR-dependent binding.96 However, the presence of many proteins that could interact in vitro with ATG8 proteins mutated in their LDS strongly suggests that ATG8 family proteins harbor another interaction surface. Roughly one third of the interactors did not show subfamily specificity, whereas a third each associated with either the LC3 or GABARAP subfamilies. When this was tested using in vitro binding assays, the specificity was more relaxed. All proteins only associated with the LC3 subfamily by the proteomics approach could bind to GABARAP proteins in vitro. Hence, in vivo there may be more specific complexes formed than revealed by the specificities observed in in vitro binding assays.
The presence of a LIR motif is not sufficient for a protein to be efficiently degraded by selective autophagy. Analogous to the oligomerization and aggregation of preApe1 in the Cvt pathway, PB1 domain-mediated polymerization of p62 is absolutely required for its degradation by autophagy.51 Clearly, in addition to LIR-dependent binding, aggregation/polymerization is required. In order to form p62 bodies that are subsequently degraded by autophagy, p62 needs, in addition to the PB1 domain and the LIR motif, also the UBA domain that binds to Ub-labeled proteins.29,33
The Autophagic Adapter Hypothesis: p62 and NBR1 as Cargo Receptors or Adapters for Autophagic Degradation of Ubiquitinated Substrates
What are the signals or labels used to target substrates to a selective autophagy pathway? As already mentioned, there is increasing evidence that Ub is used as a signal for autophagic degradation of protein aggregates and other cellular substrates.78 A simplified general model for p62-mediated selective autophagy can be proposed (Fig. 3A): First, the substrate likely needs to be recognized by molecular chaperones or other cellular systems sorting it into an autophagy degradation pathway. Second, the substrate is ubiquitinated. Third, p62 is recruited and sequesters the autophagy substrates into larger units or aggregates. Via its interaction with LC3 the phagophore is recruited and the autophagosome forms around the aggregated substrate. The ability of p62 to recognize Ub substrates was nicely illustrated in a paper from the group of Lippincott-Schwartz. They demonstrated that ubiquitination of a long-lived, cytosolic soluble protein (red fluorescent protein; mRFP) and of peroxisomes was sufficient to target these very different substrates for autophagic degradation.37 Strikingly, p62 was required for targeting both types of substrates. The authors used a strategy where they labeled peroxisomes with monoUb by means of a GFP- and monoUb-tagged peroxisomal membrane protein, pMP34. Although this strategy represents an artificial situation, the results showed convincingly that peroxisomes are targeted for autophagy when they display monoUb on their membrane facing the cytosol (Fig. 4A). Clearly, monoUb can here act as a signal for pexophagy or selective autophagy of peroxisomes. Importantly, the authors also demonstrated that siRNA-mediated knockdown of p62 strongly inhibits the autophagic degradation of peroxisomes in the absence of artificial Ub labeling.37
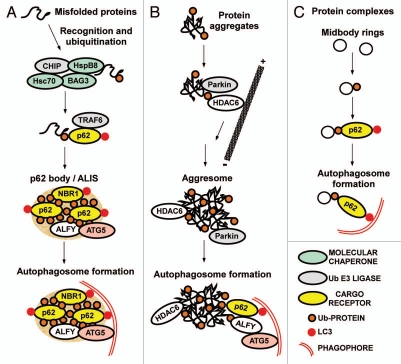
Figure 3.
Cargo receptors involved in degradation of proteins by selective autophagy. (A) Degradation of misfolded proteins via the formation of p62 bodies. Misfolded proteins are first recognized by molecular chaperones. Recognition by a complex of Bag3, HspB8 and Hsc70 and subsequent ubiquitination by an associated E3 ligase such as CHIP, results in the recruitment of p62. Together with its interaction partners NBR1 and ALFY, p62 then mediates the assembly of Ub-labeled misfolded proteins into p62 bodies of variable sizes. The contents of p62 bodies can be degraded by the proteasome or by autophagy and autophagic degradation depends on interactions of p62 and NBR1 with LC3, and ALFY with ATG5. (B) Degradation via formation of aggresomes. Aggresomes are formed by HDAC6-mediated transport of smaller Ub-containing aggregates to the microtubule-organizing center region. Recruitment of p62 and ALFY to the aggresomes may induce their degradation by autophagy. However, it is often unclear if it is the large aggregates or smaller precursors that are most efficiently degraded. (C) Degradation of protein complexes. Midbody rings are selectively degraded by autophagy after mitosis, and the process depends on their ubiquitination and on recruitment of p62.
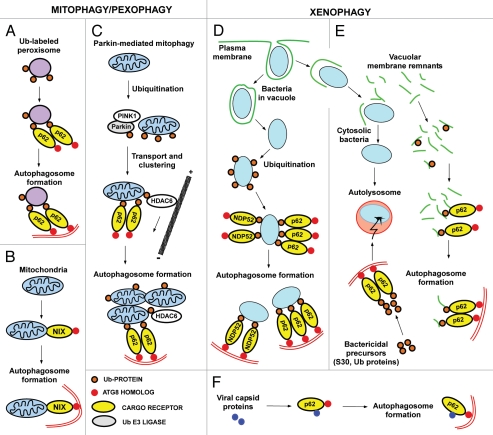
Figure 4.
Cargo receptors involved in mitophagy, pexophagy and xenophagy in mammalian cells. (A) Peroxisomes artificially labeled with monoUb on their surface are specifically recognized by p62 and degraded by autophagy, illustrating that Ub is a signal for p62-mediated autophagic degradation. (B) Nix-dependent mitophagy during development. Nix interacts with ATG8 family proteins, and acts as a cargo receptor for the delivery of mitochondria to the phagophore. The process is Ub-independent. (C) Removal of depolarized mitochondria by Parkin-mediated autophagy. Recruitment of Parkin to the mitochondria is regulated by PINK1. Parkin-mediated ubiquitination is essential and it recruits p62 and HDAC6. These proteins are responsible for a transport of depolarized mitochondria to the perinuclear region and for their assembly into clusters. It is not clear what cargo receptors are directly involved in the delivery of mitochondria to the phagophore. (D) Removal of intracellular bacteria by selective autophagy. Many intracellular bacteria such as S. enterica can reside and replicate within vacuolar structures, but they also leak out in the cytosol and are then recognized and ubiquitinated by an unknown machinery. Both p62 and NDP52 are recruited to ubiquitinated bacteria resulting in their delivery to the phagophore. (E) Some specialized pathogens have a cytosolic lifestyle. These bacteria may have developed specific systems to avoid ubiquitination, and fewer bacteria are then delivered to the autophagic system. p62 acts as a cargo receptor for the delivery of bactericidal precursors to the autolysosomes via selective autophagy, and for some pathogens such as M. tuberculosis, this may be essential for the killing of the bacteria. p62 also acts as a cargo receptor for autophagy of vacuolar membrane remnants after a bacterium escapes from the phagosome after cell entry.
A functional role of the Ub chains added in various forms of selective autophagy is obviously to recruit essential adapter proteins such as p62 and HDAC6. For most Ub-labeled autophagy substrates, it is neither known which Ub ligases are involved nor which specific proteins become ubiquitinated, or the type of Ub modifications involved. Both p62 and HDAC6 seem to have a preference for K63-linked polyUb chains,66,97 and the addition of K63-linked Ub chains has been reported to correlate with the clearance of protein inclusions by autophagy.98 However, different types of Ub modifications may be added to autophagy substrates and more studies are needed to clarify the in vivo binding preferences of p62, either alone or as part of a complex with NBR1 and ALFY (autophagy linked FYVE protein).31,99 It also seems plausible to ask if recruitment of p62 creates a positive feedback loop where p62 through interaction with E3 ligases induces further ubiquitination leading to recruitment of more p62. Such a positive feedback loop would help to explain the strong correlation between p62 and Ub-proteins observed for most autophagy substrates. p62 interacts with the E3 ligase TRAF6,100 and may also via KEAP1 be responsible for recruitment of the E3 ligase cullin 3.101 Ub ligases are present in most protein inclusions and the Ub chains present in mature p62 bodies or aggresomes may differ from the Ub chains initially used to recruit p62 or HDAC6. It is experimentally very difficult to distinguish a role for Ub in recognition versus a role for Ub in the construction of autophagy competent structures.
An important role for both p62 and HDAC6 is to sequester the autophagy substrates into larger units or aggregates before they become degraded by autophagy. Aggregation is very likely itself a signal for degradation by autophagy with no need for other labels as long as the aggregate is recognized directly by a cargo receptor or adapter that also can bind to ATG8 family proteins.
Selective Autophagy of Misfolded Proteins and p62 Bodies
Structures similar to p62 bodies were first described and characterized in dendritic cells by the group of Philippe Pierre and named DALIS (dendritic cell aggresome-like inducible structures).102 Using puromycin to induce the formation of defective ribosomal products (DRiPs), they were able to show that DALIS are used for storage and ubiquitination of newly synthesized defective proteins.103 Their data also indicated that DALIS have an ordered structure. Puromycin-labeled DRiPs were recruited to the center of DALIS as early as 5 min after their formation. Their data also indicated that DRiPs are ubiquitinated after their entry into DALIS, and Ub conjugation enzymes such as E1, E2-UBC4/UBC5 and CHIP (E3 ligase) are found in the center of DALIS.103 Later studies revealed that structures similar to the DALIS can be formed in many cell types and the term ALIS was therefore introduced as a more general name for these structures.28 We subsequently showed that p62 is a major protein in ALIS and concluded that p62 bodies and ALIS are indistinguishable structures.33 The formation of p62 bodies is normally a transient event and their contents are degraded by autophagy or by the proteasome.28,33,63 These structures are highly ubiquitinated and are distinct from aggresomes and have a characteristic rounded appearance when visualized by light microscopy. They can adopt variable sizes (mostly 0.5–1 µm, but some can be several µm in diameter) and they are formed in response to various stressors.28,33 It should be noted that LC3 is efficiently recruited to p62 bodies.28,29,104,105 GFP-LC3-positive dots, commonly used as a marker for autophagosomes, may in addition to autophagosomes also represent some nonmembrane-bound p62 bodies. However, most of the p62 bodies visualized by fluorescence microscopy in the size range of 0.5–1 µm are sequestered into autophagosomes.29
The transient accumulation of p62 bodies is associated with stress conditions such as the accumulation of misfolded proteins or induction of autophagy.28 However, using inhibitors of autophagosomal maturation, we have observed that Ub-proteins are continuously delivered to autophagic vesicles also under normal growth conditions. What is interesting is that no accumulation of Ub-proteins in vesicular structures was observed in cells depleted for p62, indicating that p62 is essential for constitutive autophagy of misfolded proteins.33 We recently reported that two other p62 interaction partners, NBR1 and ALFY, are essential components of puromycin-induced p62 bodies, and both proteins are also needed for constitutive autophagy of Ub-proteins.31,99 In conclusion, we believe that p62, NBR1 and ALFY constitute a complex responsible for degradation of Ub-proteins via constitutive autophagy and for the stress-induced formation of p62 bodies (Fig. 3A).99
ALFY is a 400 kDa scaffold protein that was initially shown to be redistributed from the nucleus or nuclear envelope to autophagic structures in response to amino acid starvation or proteasomal inhibition.106 The protein was later found to interact with p62, and redistribution of ALFY in HeLa cells depends on the ability of p62 to shuttle between the cytoplasm and nucleus.99 Unlike p62 and NBR1, ALFY is not a selective autophagy substrate, and ALFY is probably degraded by autophagy only when associated with p62-bodies or other types of aggregates.35,99 Support for a role of ALFY in constitutive autophagy of Ub-labeled proteins also comes for studies of Drosophila lacking the ALFY homologue Blue Cheese (bchs). These flies have a reduced life span and accumulate Ub-positive inclusions and display neurodegeneration.107 The C-terminal part of ALFY contains a p62-interacting BEACH domain,99 an ATG5-interacting WD40 repeat region,35 and a PtdIns(3)P-binding FYVE domain.106 The large size of ALFY also makes it a candidate scaffold involved in the assembly of p62 bodies. However, more studies are needed to clarify the individual roles of p62, NBR1 and ALFY in the formation and autophagic degradation of p62 bodies.
The formation of p62 bodies under stress conditions is in part due to the increased expression of the p62 gene. In addition, the decision to make p62 bodies depends on the expression of specific molecular chaperones (Fig. 3A). Increased expression of the Hsp/Hsc70 co-chaperone Bag3 was recently shown to induce the formation of p62 bodies.30 The amount of misfolded proteins gradually increases during cell aging, and a shift from Bag1 to Bag3 expression during aging correlates with a shift from proteasomal degradation of misfolded proteins into autophagic degradation. Co-precipitation of Bag3 with p62 indicates that these proteins are in the same complex.30 An important interaction partner of Bag3 in selective autophagy of misfolded proteins is the small heat shock protein HspB8.108 HspB8 expression is induced by proteasomal inhibition.109 A complex consisting of Bag3 and HspB8 was initially implicated in autophagic clearance of mutant Huntingtin.108 Later a similar complex including CHIP was shown to be important for selective autophagy of SOD in amyotrophic lateral sclerosis (ALS).109 Furthermore, a complex of Bag3, HspB8, Hsc70 and CHIP, or a similar Drosophila complex containing the Bag3 homologue Starvin, were shown to be needed for constitutive autophagy in muscles.110 The complex is required for maintenance of Z disks and facilitates selective autophagy of damaged components such as filamin.110
Another group of proteins involved in the sorting and degradation of misfolded proteins are the ubiquilins (ubiquilin 1–4). Their domain architecture with an N-terminal ubiquilin (UbL) domain and a C-terminal UBA domain have clear similarities with that of p62 (Fig. 2A). The PB1 domain of p62 and the UbL domain of ubiquilin both interact with the S5A/Rpn10 subunit of the 26S proteasome, indicating a role in the shuttling of ubiquitinated substrates to the proteasome.66,111 Ubiquilin is involved in ER-associated degradation (ERAD) as part of a complex also containing Erasin and the AAA+ ATPase chaperone p97/VCP (valosin-containing protein).112 p97/VCP has an important role in ERAD in the delivery of retranslocated proteins to the UPS. ERAD substrates may then be ubiquitinated and shuttled to the 26S proteasome by ubiquilin. p97/VCP is a multifunctional protein that also binds to HDAC6 to regulate delivery of Ub aggregates to aggresomes.113 Ubiquilin was recently implicated in both CMA and autophagy and it is degraded by both pathways.114 For the latter pathway, ubiquilin is recruited to an LC3-containing complex and stimulates formation of autophagosomes. Although the interaction between LC3 and ubiquilin appears to be indirect, this suggests that ubiquilin may act as an autophagy receptor. It is not known whether p62 and ubiquilin have redundant roles or are associated with different subfractions of misfolded proteins.
While we have shown that p62 is required for the delivery of Ub-proteins in autophagic vesicles,33 Ub-labeled autophagosomes accumulate in cells either lacking or expressing disease-associated mutants of p97/VCP.115 Hence, loss of p62 or p97/VCP seems to block different steps in the same autophagy pathway. This is particularly interesting since mutations in p62 or p97/VCP can both lead to Paget disease of the bone.
Inefficient Autophagic Degradation of p62 Leads to the Accumulation of Ubiquitinated Aggregates
The importance of constitutive autophagy of misfolded proteins has been strongly supported by autophagy knockout studies. Loss of ATG5 or ATG7 in mice results in the accumulation of Ub aggregates in neurons and hepatocytes although there is no evidence that proteasomal degradation is affected.116,117 Knockdown of autophagy in flies similarly results in the accumulation of Ub aggregates.118,119 The most likely explanation is that a significant amount of Ub-labeled proteins are degraded by constitutive autophagy under normal conditions. Constitutive autophagy may in particular be important in differentiated, non-proliferating cells. Tissue-specific knockout of ATG genes causes accumulation of p62-positive protein aggregates in neurons,116,117,120 hepatocytes,32 skeletal muscle,121,122 cardiac muscles,123 pancreatic β-cells,124,125 and podocytes in the kidneys.126
It has been demonstrated, both in cell culture and in vivo, that mammalian p62 and the Drosophila homologue Ref(2)P are required for formation of ubiquitinated protein aggregates.29,32,33,127 Strikingly, Komatsu and coworkers found that the observed accumulation of Ub-positive protein aggregates in hepatocytes upon liver-specific knockout of the Atg7 gene in mice is dependent on p62. The hepatomegaly and liver dysfunction is strongly ameliorated by simultaneous knockout of p62.32 The high level of Ub aggregates observed in hepatocytes of autophagy-deficient mice indicates that this organ may be particularly sensitive to p62 accumulation.
p62 and Ub are also major constituents of two types of aggregates found in chronic liver diseases, i.e., intracellular hyaline bodies (IHBs) found in hepatocellular carcinoma, and Mallory-Denk bodies (MBs) characteristic for alcoholic and nonalcoholic steatohepatitis.128 p62 is needed for the formation of both these structures.74 In particular, IHBs seem to resemble p62 bodies, whereas MBs are distinguished from IHBs in that they contain abnormal keratins in addition to p62 and Ub.128
One discrepancy that has been noted is the much lower level of Ub proteins in Atg7/p62 double-knockout tissues as compared to Atg7-deficient tissues.32 One likely scenario is that Ub-proteins are less aggregated in p62-deficient cells and therefore can be more easily degraded by CMA or the proteasome. There is crosstalk between different degradation pathways for misfolded proteins, and the loss of one degradation system may result in the activation of other systems.129 However, a high constitutive level of p62 caused by autophagy inhibition may itself contribute to an increased formation or decreased degradation of Ub-proteins. It is suggested that p62 accumulating due to autophagy inhibition delays the delivery of ubiquitinated proteins to the proteasome.130 Also, at least part of the pathophysiological effects seen in the autophagy-deficient liver can be attributed to chronic activation of Nrf2 caused by high levels of p62.131 In the brain of p62 knockout mice, K63-linked Ub-proteins increase and the mice display an Alzheimer-like phenotype.132,133 Distinct from the liver, the presence of p62 does not exaggerate neurodegeneration in autophagy-deficient mice.32
Selective Autophagy of Aggregated Protein Substrates—Aggrephagy
The term aggresome denotes a general type of protein aggregate that is induced by the transport of smaller Ub-aggregates along microtubule tracks into the MTOC region where the structure is formed usually encaged by intermediate filaments.134 This is believed to be a protective mechanism since small and diffuse aggregates are more toxic than aggresomes.135 HDAC6 is required for the formation of aggresomes (Fig. 3B). It binds to Ub and dynein molecular motors, and this enables HDAC6 to shuttle soluble Ub-aggregates to aggresomes.136,137 HDAC6 binds Ub via a C-terminal BUZ domain and has preference for K63-linked chains.97,113 Lewy bodies in Parkinson disease are characterized by the presence of aggregated DJ-1 or α-synuclein and are among the best studied aggresomal structures. Mutations in the E3 ligase Parkin are common in recessive forms of Parkinson. Parkin was, in a recent study, reported to cooperate with the heterodimeric E2 enzyme UbcH13/Uev1a to mediate K63-linked polyubiquitination of misfolded DJ-1, and this is essential both for HDAC6-mediated transport of DJ-1 aggregates and formation of Lewy bodies.97 Another essential role of HDAC6 is to promote the creation of an actin network enhancing fusions between aggresome-containing autophagosomes and lysosomes.138 HDAC6 may also be important for the transport of the autophagy machinery to the aggresomes.136 Both in mammals and flies, HDAC6 promotes autophagic degradation of polyglutamine inclusions.136,139 Autophagy is important for degradation of aggresomes since proteins must be unfolded before they can be degraded by the proteasome or CMA.27 Several studies present evidence for an autophagic degradation of polyglutamine inclusions.136,140–142 However, dependent on the specific aggregating protein involved, not all types of aggresomes can be degraded by autophagy,143 and it is often unclear whether it is the large aggregates or their precursors that are most efficiently degraded.
The presence of p62 is a common feature of most cytoplasmic and nuclear protein aggregates found in human diseases.74 Most pathological aggregates are induced by a specific protein other than p62 and the recruitment of p62 is a later event induced by the ubiquitination of the aggregates (Fig. 3B). Hence, p62 is for example not needed for the formation of polyglutamine inclusions of Huntington disease,144 and the presence of p62 in aggregates is believed to reflect a role of p62 in autophagic degradation. In support of this, LC3B mutants defective in p62 binding are defective in autophagy-mediated clearance of polyglutamine inclusions.145 What should be noted is that NBR1 and ALFY are likely to be present together with p62 in protein inclusions.31,35,99 Do these three proteins collaborate to enhance the autophagic degradation of aggresomes, analogous to their role in the formation and degradation of the transient p62 bodies? ALFY is recruited to polyglutamine inclusions as part of a complex containing p62, NBR1, LC3, ATG5, ATG12 and ATG16L.35 ALFY is required for efficient autophagic clearance of polyglutamine or α-synuclein inclusions in mammalian cells dependent on a direct interaction between ALFY and ATG5. Furthermore, overexpression of ALFY promotes degradation of polyglutamine inclusions in a neuronal lentiviral model, and overexpression of the Drosophila homologue Bchs diminishes neurotoxicity in a Drosophila eye model of polyglutamine toxicity.35 For further discussions on ALFY, see the article by Yamamoto and Simonsen in this issue of Autophagy.
One intriguing observation is that nuclear p62 and ALFY also collaborate in the formation of nuclear aggregates that in several ways seem to be similar to the p62 bodies.99 These structures are associated with nuclear PML bodies.35,63,99 PML bodies have been proposed to function as sites for proteasomal degradation of misfolded proteins.146 The recruitment of p62 and ALFY to PML bodies is needed for accumulation of Ub-proteins in PML bodies.63,99 p62 also contributes to the recruitment of PML bodies and proteasomes to, or close to, nuclear inclusions formed by polyglutamine-expanded ataxin1Q84, and our data suggest a role for p62 in degradation of these inclusions.63
Selective Autophagy of Mitochondria—Mitophagy
The idea of selective removal of damaged or surplus mitochodria by autophagy (mitophagy) has recently received support from studies lending some mechanistic insight into this complex process. Using yeast genome-wide screening, the groups of Ohsumi and Klionsky independently discovered the mitophagy receptor, Atg32.40,41 The Atg32 protein is a single-spanning mitochondrial outer membrane protein with a free N-terminal region facing the cytosol. Atg32 mutant yeast cells are defective in mitophagy. Atg32 binds both to the selective autophagy factor Atg11 and to Atg8, and functions specifically in mitophagy. Atg32 has a LIR motif important for the interaction with Atg8.40 In yeast there seems to be no involvement of Ub in mitophagy.
In mammalian cells, loss of membrane potential of damaged mitochondria was proposed to lead to autophagic degradation, as suggested from studies of hepatocytes from GFP-LC3 transgenic mice.147 Mitophagy is important during the developmental elimination of mitochondria in reticulocytes. Nix/BNIP3L is essential for this clearance of mitochondria occurring during erythrocyte maturation.148,149 The Dikic group recently showed that Nix/BNIP3L recruits the ATG8 family Ub-like modifier GABARAPL1 to damaged mitochondria via a LIR motif in its N-terminal region.38 Ablation of the Nix:LC3/GABARAP interaction slows mitochondrial degradation in murine reticulocytes. Also, Nix recruits GABARAPL1 to depolarized mitochondria. Hence, Nix acts as a mitophagy receptor for selective clearance of mitochondria during erythrocyte differentiation (Fig. 4B). Using a phage display screening, the Wilbold group also identified Nix as binding to GABARAP via a LIR motif.95 The role of Nix/BNIP3L in mitophagy is covered in detail in the article by Novak and Dikic in this issue of Autophagy.
Several recent reports have described a specific mitophagy pathway that is Ub-dependent and has been referred to as Parkin-mediated mitophagy (Fig. 4C). Richard Youle's group provided the first evidence for a role of Parkin in mitophagy of damaged mitochondria showing that Parkin is recruited to and important for the degradation of damaged mitochondria in response to the treatment of cells with ROS or the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP).42 They also subsequently showed that PINK1 rapidly accumulates on damaged mitochondria and that PINK1 stabilization on the mitochondria is both necessary and sufficient for recruitment of Parkin.150 The Springer group reported that PINK1, Ub, p62 and the voltage dependent anion channel 1 (VDAC1) are required for Parkin-mediated mitophagy in both neuronal and non-neuronal cells.39 PINK1 interacts with Parkin and is needed for the translocation of Parkin to the membrane of damaged mitochondria. Parkin-mediated ubiquitination then results in accumulation of p62, clustering of mitochondria and autophagic degradation.39 See also the article by Springer and Kahle in this issue of Autophagy. The group of Tso-Pang Yao demonstrated that Parkin-mediated ubiquitination of damaged mitochondria also serves to recruit HDAC6, and that HDAC6 is needed for efficient degradation. They compared the role of HDAC6 in mitophagy with the role of HDAC6 in aggresome formation, and showed that HDAC6-mediated transport of damaged mitochondria results in the formation of perinuclear mito-aggregates resembling aggresomes.151 Other Parkin substrates relevant for mitophagy are the mitofusins which become ubiquitinated in a PINK1- and parkin-dependent manner following mitochondrial damage.152,153 The mitofusins are transmembrane GTPases located in the mitrochondrial outer membrane involved in regulating fusion of mitochondria. Ubiquitination of mitofusins may be involved in segregating damaged mitochondria for mitophagy, via an inhibition of mitochondrial fusion events, and/or the ubiquitinated mitofusins may act as tags for autophagic cargo receptors.152,153
Nix is also important for Parkin-mediated mitophagy as reported in a study on CCCP-induced mitophagy, concluding that Nix is required for translocation of Parkin to the damaged mitochondria.154 Recent papers from the groups of Komatsu and Youle disagree with the Springer group on the exact role of p62 in mitophagy.155,156 Both the Komatsu and Youle groups demonstrate that p62 is required for Parkin-induced mitochondrial clustering, but using p62 knockout murine embryo fibroblasts they claim p62 is not required for mitophagy. The Youle group shows this also using siRNA knockdown in HeLa cells and finds that VDAC1 is dispensable for both the clustering and autophagic degradation of mitochondria following CCCP-treatment of cells.156 However, it seems clear that Parkin (but not a pathogenic mutant) promotes K63-linked polyubiquitination of mitochondrial substrate(s) leading to recruitment of p62. Thereafter, p62 aggregates dysfunctional mitochondria dependent on its PB1-domain-mediated homopolymerization similar to its aggregation of polyubiquitinated proteins. In the study by Komatsu and coworkers, a delay in mitochondrial degradation was observed in p62 KO MEFs complemented with a LIR mutant of p62 (W340A) suggestive of involvement of p62 in Parkin-mediated mitophagy. A possible explanation for the different results on the role of p62 in Parkin-mediated mitophagy can be functional redundancy.155 NBR1 is a likely candidate that may compensate for the absence of p62. More studies are needed to resolve this conundrum. The essential substrates ubiquitinated by Parkin in Parkin-mediated autophagy remain to be identified. It is also important to clarify the differences between mitophagy pathways utilized in degradation of damaged versus surplus mitochondria.
Selective Autophagy of Bacteria and Viruses—Xenophagy
Autophagy used as a defense system against infection has been termed “xenophagy”.4,157 Several articles in this thematic issue of Autophagy discuss the selective autophagy of bacteria (See Shahnazari et al., Cemma et al., Ogawa et al., Randow, and Ponpuak and Deretic). Autophagic degradation of invading bacteria is important in order to protect cells from bacterial colonization. Invading Salmonella enterica serovar Typhimurium (S. typhimurium) reside and replicate within intracellular compartments termed Salmonella-containing vacuoles (SCVs), but the bacteria can also be released into the cytosol and are then targeted by the host Ub conjugation system. Two different autophagy receptors were recently shown to be involved in autophagic degradation of Ub-labeled S. typhimurium.46,48 The group of John Brumell implicated p62 in this process (Fig. 4D). They showed that p62 and LC3 are recruited to the surface of Ub-labeled bacteria, and the recruitment of p62 is needed for efficient autophagy of the bacteria. The UBA domain is critical for the recruitment of p62 to Ub-positive S. typhimurium, and both the UBA and LIR domains are required for autophagy of the bacteria.48 In this study siRNA knockdown of endogenous NBR1 suggested that NBR1 does not play any major role in the autophagy targeting of the bacteria. In a separate study, the group of Felix Randow characterized another autophagy receptor, NDP52 (nuclear dot protein 52 kDa) (Fig. 2A), which interacts directly with LC3 and is needed for efficient autophagy of S. typhimurium (Fig. 4D). NDP52 uses a C-terminal zinc finger to bind to Ub chains located on cytosolic bacteria.46 Both p62 and NDP52 restrict intracellular replication of S. typhimurium. Interestingly, p62 and NDP52 are recruited independently to distinct microdomains surrounding the bacteria and cooperate for efficient autophagy of the intracellular bacteria (See article by Cemma et al. in this issue of Autophagy). It remains to be clarified whether a putative LIR motif in the N-terminal region of NDP52 mediates the binding to LC3.79
Other intracellular pathogens like Shigella flexneri and Listeria monocytogenes are adapted to a cytosolic lifestyle and they break the vacuolar membrane after entry into host cells (Fig. 4E). Whereas S. enterica become ubiquitinated and degraded by autophagy, pathogens that replicate in the cytosol seem to avoid host ubiquitination.158 An interesting study on L. monocytogenes illustrates how this bacterium depends on the bacterial protein ActA to escape from autophagic degradation.47 ActA-mediated recruitment of the Arp2/3 complex, Ena/VASP and actin prevents ubiquitination of the bacteria and thereby protects the bacteria against autophagic recognition. In contrast, ActA deletion mutants are ubiquitinated, recognized by p62, and degraded by autophagy.47 Another study on S. flexneri showed that vacuolar membrane remnants are polyubiquitinated, recognized by p62 and targeted for autophagic degradation (Fig. 4E).44 These membrane remnants contain the p62-interacting Ub ligase TRAF6. TRAF6 activates NEMO and both NEMO and p62 act in the NFκB signaling pathway promoting inflammatory and prosurvival pathways.44 It is not known if NBR1 is acting together with p62 in xenophagy of ActA-deleted Listeria and Shigella membrane remnants.
A very different role of p62 in xenophagy has been reported by the group of Vojo Deretic. In a recent study, they found that in addition to the delivery of Mycobacterium tuberculosis into autolysosomes, efficient killing of the bacterium depends on a p62-mediated uptake of specific ribosomal and bulk ubiquitinated cytosolic proteins into the same autolysosomes.54 Interestingly, p62 is in this case not needed for the autophagic delivery of the bacterium, but only for the delivery of the bactericidal precursors that are processed to active compounds within the autolysosomes (Fig. 4E).
Evidence for the use of p62 as an adaptor for selective autophagy also in antiviral defense comes from the group of Beth Levine.45 They studied the role of autophagy in mice where the central nervous system was infected with a lethal Sindbis (SIN) virus and showed that SIN capsid proteins are cleared by selective autophagy involving p62. The clearance of surplus capsid proteins somehow protects against cell death although viral replication is not affected. p62 and SIN proteins accumulate in mouse neurons lacking ATG5 function, and they could co-immunoprecipitate p62 with SIN capsid protein in virus-infected HeLa cells. Knockdown experiments show that p62-mediated selective autophagic degradation of the capsid protein reduces virally-induced cell death. It is presently not known if there is a direct interaction between p62 and SIN capsid protein (Fig. 4F). See the review on the role of p62 in the degradation of the Sindbis virus capsid in the article by Sumpter and Levine in this issue of Autophagy.
Ubiquitin-Independent Selective Autophagy
In yeast, Atg11 acts as an adaptor protein for Ub-independent selective autophagy. Atg11 is responsible for the recruitment of autophagy substrates to the PAS where phagophores are made (Fig. 5). In the Cvt pathway, the delivery of the Ape1 complex to Atg11 is mediated by the cargo receptor Atg19 that interacts with the Ape1 complex, Atg11 and Atg8.7 The cytosolic cysteine protease Lap3 is selectively degraded by autophagy utilizing the Cvt pathway components because it binds to the prApe1 oligomer and is transported to the vacuole under nitrogen starvation in a manner dependent on both Atg19 and Atg11.159 Similar to Lap3, yeast Ald6 is degraded by selective autophagy upon induction of bulk autophagy by nutrient limitation.160 However, in contrast to Lap3, the cytosolic acetaldehyde dehydrogenase Ald6 is transported by autophagosomes to the yeast vacuole by an unknown mechanism. Because overexpression of Ald6 is toxic during nutrient starvation, its activity may need to be regulated by specific autophagic degradation to ensure cell survival upon nutrient limitation.160
Figure 5.
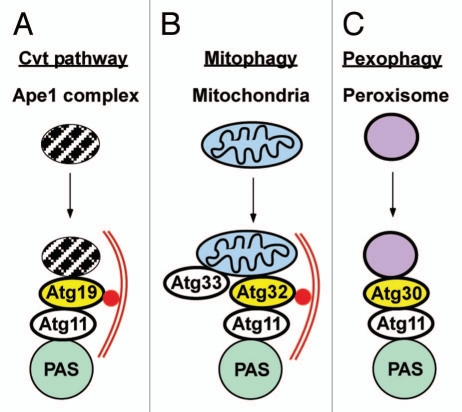
Selective autophagy in yeast. (A) Atg19 is acting as a cargo receptor in the Cvt pathway. It binds to the Ape1 complex, and delivery to the phagophore is mediated by interactions with Atg11 located at the phagophore assembly site (PAS), and Atg8 attached to the phagophore membrane. (B) Atg32 is the cargo receptor in mitophagy. Atg32 is a mitochondrial outer membrane protein, and it interacts with Atg11 and Atg8 to mediate the delivery of mitochondria to the phagophore. (C) Atg30 is the cargo receptor in pexophagy. It is a peroxisomal membrane protein and interacts with PpAtg11, but does not interact with PpAtg8.
Mitophagy in yeast can be induced by a reduction in membrane potential, nitrogen starvation or entry into stationary phase. There seems to be a single mitophagy pathway in yeast that depends on the two outer membrane proteins Atg32 and Atg33. Similar to Atg19, Atg32 contains a LIR motif that binds to Atg8 and it also interacts with Atg11.40,41 In contrast, Pichia pastoris Atg30, the cargo receptor for pexophagy in this methylotrophic yeast, has no LIR motif, but is located on peroxisomal membranes and interacts with PpAtg11.161 A detailed discussion of mitophagy in yeast is found in the article by Wang and Klionsky in this issue of Autophagy.
In mammals, there seems to be both Ub-dependent and Ub-independent mitophagy pathways, and the current belief is that Nix/Bnip3L, but not p62, is essential for Ub-independent mitophagy (see above). There is also a recent report indicating that p62 can be involved in Ub-independent selective autophagy of misfolded proteins. A mutant of superoxide dismutase 1 (SOD1) causing amyotrophic lateral sclerosis (ALS) is degraded by autophagy in a Ub-independent way due to a direct interaction with p62.162 It is also likely that ubiquitination is not involved in p62-mediated selective autophagy of Sindbis virus capsid proteins (see above). Hence, this may be another example of Ub-independent selective autophagy like the one reported for the mutant SOD1 protein. It remains to be determined how this protein is recognized by p62.
Similar to p62 and NBR1, the SEPA-1 protein in C. elegans oligomerizes and forms cytoplasmic aggregates that are selectively removed by autophagy during embryogenesis. Furthermore, SEPA-1 acts as a cargo receptor or adaptor for autophagic degradation of a specialized type of protein aggregates called germ P granules by binding directly to the P granule component PGL-3 and to the ATG8 homologue LGG-1.163 This way the P granules become sequestered in autophagosomes and degraded by autophagy. Although Ub is not involved, this is very similar to the selective autophagy mediated by p62.
Regulation of Cell Signaling by Selective Autophagy
Both p62 and NBR1 are involved in cellular signaling processes that seem unrelated to autophagy. In these cases selective autophagy can be an important mechanism to regulate the level and lifetime of the signaling complexes they participate in. The groups of Jorge Moscat and Sylvia Stabel found p62 to bind to atypical protein kinase C,58,60 and it was subsequently demonstrated that p62 acts as a scaffold or adapter protein in NFκB signaling pathways.164–166 In addition to the atypical protein kinase Cs, PKCζ and PKCλ/ι, p62 also binds to the kinases MEKK3, MEK5 and ERK1.59–62,167 Clearly, many of the phenotypes observed in p62 knockout mice can be attributed to defects or perturbations in cellular signaling pathways.168 Studies involving knockout, knockin and transgenic mice show that p62 plays important roles in bone remodelling, obesity, oxidative stress response and cancer development.70,131,169–172
An antagonistic relationship between p62 and ERK1 activation has been established,167,170 and increased autophagy of p62 under hypoxia is responsible for activation of the ERK1/2 pathway.173 This shows that removal of p62 by selective autophagy can have an impact on signaling (Fig. 6A). A recent study performed with both fly and mammalian macrophages show that constitutive, p62-mediated selective autophagy is required for cell spreading and Rho1-induced cell protrusions. It is suggested that p62 may mediate selective autophagic degradation of a regulator of the Rho pathway.174 In the Wnt signaling pathway the adaptor protein Dvl is degraded by selective autophagy to downregulate the pathway.52 The E3 ligase VHL (Von Hippel-Lindau) ubiquitinates Dvl2 which is then bound by p62, which in turn facilitates aggregation and LC3-mediated autophagosome recruitment of Dvl2 under starvation conditions. The details and biological roles are further discussed in the article by Gao and Chen in this issue of Autophagy.
Figure 6.
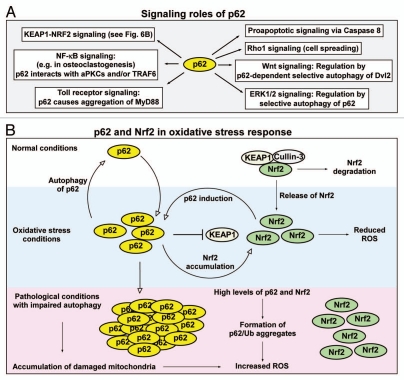
Signaling roles of p62 involving selective autophagy. (A) Signaling roles of p62 that may, at least in part, be regulated by autophagy. (B) p62 and Nrf2 as regulators of the oxidative stress response. Under normal conditions, there is a low level of p62 and Nrf2 due to selective autophagy of p62 and KEAP1-mediated proteasomal degradation of Nrf2. Under oxidative stress conditions, there is an elevated level of p62 and Nrf2. This results in the establishment of a feedback loop where Nrf2 induces expression of p62, and p62 inhibits KEAP1-mediated degradation of Nrf2. The net effect is induction of the intracellular antioxidant response. Under pathological conditions associated with inhibition of autophagy, there is a constitutive high level of p62 and Nrf2. This potentially induces ROS production, inhibits proteasomes and acts as a tumor-promoting factor.
The p62 bodies have been suggested to act as signaling hubs where p62 interacts with TRAF6 and caspase-8, and may affect both prosurvival and pro-apoptotic signaling pathways.175,176 Along the same lines, the polyubiquitinated vacuolar membrane remnants resulting from Shigella entry that are recognized by p62 and targeted for autophagic degradation, also act as signaling hubs promoting inflammatory and prosurvival pathways.44 Analogous to p62, another autophagic adaptor or cargo receptor recognizing Ub-labeled invading bacteria, NDP52, is part of a TBK1 signaling complex containing the kinases TBK1 and IKKε as well as the scaffold proteins Nap1 and Sintbad.46 Hence, both p62 and NDP52, which both may help clear Ub-labeled intracellular bacteria by autophagy,46,48 act as true innate immune receptors involved in cytosolic surveillance. Interestingly, p62 is induced by lipopolysaccharide (LPS) and is also involved in regulation of aggregation of the MyD88 adaptor in Toll receptor signaling, suggesting that p62 may be regulated by, and act as a regulator of, innate immunity signaling pathways.177,178
p62 is a proteotoxic stress response protein and is induced at the mRNA and protein level by oxidants, sodium arsenite, cadmium, ionophores, proteasomal inhibitors or overexpression of polyglutamine-expanded proteins.144,179 The transcription factor Nrf2 induces a battery of genes upon oxidative stress as part of a protective response against oxidative damage to organelles and macromolecules. Under conditions without oxidant or electrophile stress, Nrf2 is bound to KEAP1, an adaptor for the Cul3 E3 ligase complex, and rapidly degraded by the proteasome. But upon oxidative stress KEAP1 is modified on specific cysteine residues, and Nrf2 is stabilized and turns on a set of antioxidant response genes.180–182 p62 was recently shown to be able to induce Nrf2 when overexpressed, and the induction of p62 is itself strongly inhibited in Nrf2 knockout mice.183,184 Earlier this year, five different groups reported that p62 binds to KEAP1 and thereby mediates the induction of Nrf2.101,131,185–187 Three groups mapped a so-called KIR motif in p62 that binds to the Kelch repeat domain of KEAP1.101,131,187 The LIR and KIR motifs are located directly adjacent to each other, and the binding to LC3/GABARAP family proteins can be competed by KEAP1 in vitro and slows the degradation of GFP-p62, in a reporter cell system to monitor autophagic degradation of p62,188 suggesting that there is a mutually exclusive binding to either the LIR or the KIR in a single p62 molecule.187 However, since p62 is a polymer, both KEAP1 and LC3 may be bound by a polymeric p62.
A pertinent question to ask is if KEAP1 is degraded by autophagy. We found that overexpressed KEAP1 is degraded by autophagy in a p62-dependent manner. However, due to lack of good antibodies for immunofluorescence and EM studies of endogenous KEAP1 we have been unable to determine whether endogenous KEAP1 is a selective autophagy substrate or not. Copple et al. found that knockdown of p62 leads to increased protein levels of endogenous KEAP1 caused by a decrease in its degradation rate.185 This could be due to reduced degradation of KEAP1 via autophagy. On the other hand, Fan et al. suggest that KEAP1 is not itself an autophagy substrate but forms a complex with p62 and LC3 under oxidative stress conditions to facilitate efficient removal of protein aggregates via autophagy.186 Clearly more studies are needed to determine if endogenous KEAP1 is degraded by selective autophagy or by the proteasome, or both. We also demonstrated that Nrf2 binds together with a small Maf protein to an antioxidant response element in the p62 upstream region and activates transcription of the gene. This way p62 sets up a positive feedback loop in the KEAP1-Nrf2 pathway (Fig. 6B).187
These findings connect the earlier reports about p62 as a stress response protein directly to an oxidative stress response signaling pathway. Related to this, Eileen White's group showed that under conditions of metabolic stress and autophagy deficiency the failure to properly regulate p62 levels leads to oxidative stress and increased tumorigenicity.189 High levels of p62 due to autophagy defects is sufficient to alter NFκB regulation and gene expression and to promote tumorigenesis. Previously, it had been shown that p62 is required for efficient transformation of murine embryo fibroblasts by the activated Ha-Ras oncogene, and for tumor formation in a murine model mimicking human lung adenocarcinoma.169 The groups of Moscat and White report opposite effects on the regulation of NFκB signaling. The experiments performed are not directly comparable, so more studies are needed to elucidate the reasons for this discrepancy. Depending on the level of p62 there may clearly be different outcomes. On one hand, p62-mediated selective autophagy is part of the antioxidative response that protects cells against ROS-induced damage of organelles and genome integrity. On the other hand, blockade of autophagy leads to p62-mediated accumulation of Ub aggregates that increases ROS and leads to DNA damage, thereby augmenting tumorigenesis.190 It is also highly likely that selective autophagy may help tumor cells survive under conditions of metabolic stress and that efficient transformation by many oncogenes may depend on selective autophagy.
Future Directions
An important focus for future studies will be to identify and characterize more cargo receptors for selective autophagy and identify the nature of their substrates. Such proteins must be able to interact with ATG8 family proteins and with their substrate as well as being able to multimerize to form an aggregated structure. However, a self-association property is not needed for cargo receptors sitting in the membranes of organelles, like Nix and Atg32. It will be interesting to see whether all cargo receptors depend on a LIR motif for their interaction with ATG8 family proteins or if there are other motifs mediating binding to another surface(s) of the ATG8 family proteins. An obvious first place to look for novel cargo receptors will be among the recently characterized ATG8-interacting proteins. In addition, other screens, like yeast two-hybrid screens, are likely to yield novel cargo receptors. Surprisingly few human cargo receptors in addition to p62 have so far been characterized. These are NBR1, Nix and NDP52, and in yeast there is Atg19 and Atg32. This is in stark contrast to the large number of LC3/GABARAP-interacting proteins that have now been identified. Many of these interacting proteins can be assumed to have roles in vesicle transport, fusion processes or other processes that may be distinct from autophagy. However, analyses of these proteins will very likely result in the identification of more proteins important for autophagosome formation, autophagosome maturation and/or selective autophagy. To improve our understanding of selective autophagy, it is clearly also very important to elucidate the different roles played by the different members of the mammalian LC3 and GABARAP subfamilies.
It is important to understand how autophagy substrates are selected and labeled for degradation. For Ub-labeled substrates, are there distinct Ub linkages responsible for sorting substrates for degradation by the proteasome or by selective autophagy? For misfolded proteins that can be degraded by several systems, it will be important to further analyze the roles of molecular chaperones, and how they respond to signaling events regulating the different degradation pathways. There is likely to be a fine-tuned regulation between different degradation pathways. Aggregation versus degradation is another interesting layer of regulation that is particularly relevant for autophagy substrates. The detection limit of the light microscope has restricted our ability to investigate the fate of small aggregates. However, new developments in live cell imaging and high resolution fluorescence microscopy will shed new light on this problem highly relevant for protein aggregation diseases.
It will be particularly interesting and likely challenging to elucidate roles of selective autophagy in different signaling pathways. The roles played by p62 in tumorigenesis are clearly linked to the autophagy status of the tumor cells as well as to the environmental stress that they may experience. The role played by p62 in mitophagy may be linked to tumorigenesis because of the tumorigenic effect of ROS produced by uncleared damaged mitochondria. However, as mentioned above there are unresolved issues that need to be clarified about the role of p62 in Parkin-mediated mitophagy. Is it a cargo receptor here or only an “aggregator”?
The contribution of autophagic cargo receptors, like p62 and NDP52, in innate immunity and signaling during xenophagy is another field where future research is expected to give important insight. In general, for several of the complicated selective autophagy processes the level of redundancy of autophagic cargo receptors is important to decipher. Related to this, we know that p62 interacts with NBR1 and ALFY, but we do not know much about under what conditions these three proteins collaborate and when/where they have distinct roles.
In conclusion, research on selective autophagy has the potential to contribute strongly to increased understanding of, and identification of, drug targets for important diseases like cancer, neurodegenerative diseases, diabetes and infections. Although initiated in the early 1960s, the research on different aspects of autophagic processes, including all variants of autophagy, has grown almost exponentially since the start of this century and it will be very interesting to see the future development of this fascinating field of research.
Acknowledgements
This work was supported by grants from the FUGE program of the Norwegian Research Council, the Norwegian Cancer Society, the Aakre Foundation and the Blix Foundation to T.J.
Abbreviations
- (MAP)LC3
microtubule-associated protein 1 light chain 3
- ATG
AuTophaGy-related
- CMA
chaperone-mediated autophagy
- Cvt
cytoplasm-to-vacuole-targeting
- GABARAP
gamma-aminobutyrate receptor associated protein
- LAMP-2A
lysosome-associated membrane protein 2A
- LIR
LC3 interacting region
- NFκB
nuclear factor kappaB
- PE
phosphatidylethanolamine
- PtdIns(3)P
phosphatidylinositol(3)phosphate
- ROS
reactive oxygen species
- SOD
superoxide dismutase
- Ub
ubiquitin
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 4.Deretic V. Autophagy in infection. Curr Opin Cell Biol. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 7.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 10.Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 11.Webber JL, Tooze SA. New insights into the function of Atg9. FEBS Lett. 2010;584:1319–1326. doi: 10.1016/j.febslet.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 12.He H, Dang Y, Dai F, Guo Z, Wu J, She X, et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003;278:29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 13.Xin Y, Yu L, Chen Z, Zheng L, Fu Q, Jiang J, et al. Cloning, expression patterns and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics. 2001;74:408–413. doi: 10.1006/geno.2001.6555. [DOI] [PubMed] [Google Scholar]
- 14.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 16.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 17.Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem. 2004;279:47704–47710. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 18.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP and GATE-16. J Biol Chem. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 20.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakrama FZ, Seguin-Py S, Le Grand JN, Fraichard A, Delage-Mourroux R, Despouy G, et al. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy. 2010;6:495–505. doi: 10.4161/auto.6.4.11819. [DOI] [PubMed] [Google Scholar]
- 24.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP sub-families are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 26.Kon M, Cuervo AM. Chaperone-mediated autophagy in health and disease. FEBS Lett. 2010;584:1399–1404. doi: 10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 28.Szeto J, Kaniuk NA, Canadien V, Nisman R, Mizushima N, Yoshimori T, et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006;2:189–199. doi: 10.4161/auto.2731. [DOI] [PubMed] [Google Scholar]
- 29.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009 doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 32.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 33.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 34.van der Vaart A, Mari M, Reggiori F. A picky eater: exploring the mechanisms of selective autophagy in human pathologies. Traffic. 2008;9:281–289. doi: 10.1111/j.1600-0854.2007.00674.x. [DOI] [PubMed] [Google Scholar]
- 35.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 37.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss JC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 47.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 48.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 49.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 51.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 52.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 53.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat Cell Biol. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- 54.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joung I, Strominger JL, Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc Natl Acad Sci USA. 1996;93:5991–5995. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin J. p62 and the sequestosome, a novel mechanism for protein metabolism. Arch Pharm Res. 1998;21:629–633. doi: 10.1007/BF02976748. [DOI] [PubMed] [Google Scholar]
- 57.Ishii T, Yanagawa T, Kawane T, Yuki K, Seita J, Yoshida H, et al. Murine peritoneal macrophages induce a novel 60 kDa protein with structural similarity to a tyrosine kinase p56lck-associated protein in response to oxidative stress. Biochem Biophys Res Commun. 1996;226:456–460. doi: 10.1006/bbrc.1996.1377. [DOI] [PubMed] [Google Scholar]
- 58.Puls A, Schmidt S, Grawe F, Stabel S. Interaction of protein kinase C zeta with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci USA. 1997;94:6191–6196. doi: 10.1073/pnas.94.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamark T, Perander M, Outzen H, Kristiansen K, Øvervatn A, Michaelsen E, et al. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 60.Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol. 1998;18:3069–3080. doi: 10.1128/mcb.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12:39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 62.Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NFκB activation. J Biol Chem. 2010;285:2077–2089. doi: 10.1074/jbc.M109.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pankiv S, Lamark T, Bruun JA, Øvervatn A, Bjørkøy G, Johansen T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285:5941–5953. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vadlamudi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 65.Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, Layfield R, et al. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. 2008;283:5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- 66.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 68.Long J, Garner TP, Pandya MJ, Craven CJ, Chen P, Shaw B, et al. Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NFκB signalling. J Mol Biol. 2010;396:178–194. doi: 10.1016/j.jmb.2009.11.032. [DOI] [PubMed] [Google Scholar]
- 69.Cavey JR, Ralston SH, Hocking LJ, Sheppard PW, Ciani B, Searle MS, et al. Loss of ubiquitin-binding associated with Paget's disease of bone p62 (SQSTM1) mutations. J Bone Miner Res. 2005;20:619–624. doi: 10.1359/JBMR.041205. [DOI] [PubMed] [Google Scholar]
- 70.Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, et al. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell. 2004;6:303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 71.Kurihara N, Hiruma Y, Zhou H, Subler MA, Dempster DW, Singer FR, et al. Mutation of the sequestosome 1 (p62) gene increases osteoclastogenesis but does not induce Paget disease. J Clin Invest. 2007;117:133–142. doi: 10.1172/JCI28267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yip KH, Feng H, Pavlos NJ, Zheng MH, Xu J. p62 ubiquitin binding-associated domain mediated the receptor activator of nuclear factorkappaB ligand-induced osteoclast formation: a new insight into the pathogenesis of Paget's disease of bone. Am J Pathol. 2006;169:503–514. doi: 10.2353/ajpath.2006.050960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12:2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- 74.Strnad P, Zatloukal K, Stumptner C, Kulaksiz H, Denk H. Mallory-Denk-bodies: lessons from keratin-containing hepatic inclusion bodies. Biochim Biophys Acta. 2008;1782:764–774. doi: 10.1016/j.bbadis.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 75.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, et al. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 77.Kuusisto E, Kauppinen T, Alafuzoff I. Use of p62/SQSTM1 antibodies for neuropathological diagnosis. Neuropathol Appl Neurobiol. 2008;34:169–180. doi: 10.1111/j.1365-2990.2007.00884.x. [DOI] [PubMed] [Google Scholar]
- 78.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 79.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 80.Waters S, Marchbank K, Solomon E, Whitehouse C, Gautel M. Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett. 2009;583:1846–1852. doi: 10.1016/j.febslet.2009.04.049. [DOI] [PubMed] [Google Scholar]
- 81.Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 82.Mardakheh FK, Yekezare M, Machesky LM, Heath JK. Spred2 interaction with the late endosomal protein NBR1 downregulates fibroblast growth factor receptor signaling. J Cell Biol. 2009;187:265–277. doi: 10.1083/jcb.200905118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Whitehouse CA, Waters S, Marchbank K, Horner A, McGowan NW, Jovanovic JV, et al. Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proc Natl Acad Sci USA. 2010;107:12913–12918. doi: 10.1073/pnas.0913058107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang JQ, Liu H, Diaz-Meco MT, Moscat J. NBR1 is a new PB1 signalling adapter in Th2 differentiation and allergic airway inflammation in vivo. EMBO J. 2010 doi: 10.1038/emboj.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 86.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 87.Ichimura Y, Kominami E, Tanaka K, Komatsu M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy. 2008;4:1063–1066. doi: 10.4161/auto.6826. [DOI] [PubMed] [Google Scholar]
- 88.Shvets E, Fass E, Scherz-Shouval R, Elazar Z. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- 89.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 90.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–1350. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamaguchi M, Noda NN, Nakatogawa H, Kumeta H, Ohsumi Y, Inagaki F. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J Biol Chem. 2010;285:29599–29607. doi: 10.1074/jbc.M110.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mohrluder J, Hoffmann Y, Stangler T, Hanel K, Willbold D. Identification of clathrin heavy chain as a direct interaction partner for the gamma-aminobutyric acid type A receptor associated protein. Biochemistry. 2007;46:14537–14543. doi: 10.1021/bi7018145. [DOI] [PubMed] [Google Scholar]
- 94.Mohrluder J, Stangler T, Hoffmann Y, Wiesehan K, Mataruga A, Willbold D. Identification of calreticulin as a ligand of GABARAP by phage display screening of a peptide library. FEBS J. 2007;274:5543–5555. doi: 10.1111/j.1742-4658.2007.06073.x. [DOI] [PubMed] [Google Scholar]
- 95.Schwarten M, Mohrluder J, Ma P, Stoldt M, Thielmann Y, Stangler T, et al. Nix directly binds to GABARAP: A possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–598. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 96.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, et al. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 99.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 100.Geetha T, Wooten MW. Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett. 2002;512:19–24. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- 101.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lelouard H, Gatti E, Cappello F, Gresser O, Camosseto V, Pierre P. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature. 2002;417:177–182. doi: 10.1038/417177a. [DOI] [PubMed] [Google Scholar]
- 103.Lelouard H, Ferrand V, Marguet D, Bania J, Camosseto V, David A, et al. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J Cell Biol. 2004;164:667–675. doi: 10.1083/jcb.200312073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shvets E, Elazar Z. Autophagy-independent incorporation of GFP-LC3 into protein aggregates is dependent on its interaction with p62/SQSTM1. Autophagy. 2008;4:1054–1056. doi: 10.4161/auto.6823. [DOI] [PubMed] [Google Scholar]
- 105.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 106.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 107.Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, et al. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J Neurosci. 2003;23:1254–1264. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Carra S, Seguin SJ, Lambert H, Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008;283:1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- 109.Crippa V, Sau D, Rusmini P, Boncoraglio A, Onesto E, Bolzoni E, et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of mis-folded proteins involved in amyotrophic lateral sclerosis (ALS) Hum Mol Genet. 2010;19:3440–3456. doi: 10.1093/hmg/ddq257. [DOI] [PubMed] [Google Scholar]
- 110.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 111.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 112.Lim PJ, Danner R, Liang J, Doong H, Harman C, Srinivasan D, et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol. 2009;187:201–217. doi: 10.1083/jcb.200903024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boyault C, Gilquin B, Zhang Y, Rybin V, Garman E, Meyer-Klaucke W, et al. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 2006;25:3357–3366. doi: 10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–3232. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, et al. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 117.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 118.Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 120.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285:3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, et al. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 123.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 124.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 125.Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 126.Hartleben B, Godel M, Meyer-Schwesinger C, Liu S, Ulrich T, Kobler S, et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest. 2010;120:1084–1096. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Denk H, Stumptner C, Fuchsbichler A, Muller T, Farr G, Muller W, et al. Are the Mallory bodies and intracellular hyaline bodies in neoplastic and non-neoplastic hepatocytes related? J Pathol. 2006;208:653–661. doi: 10.1002/path.1946. [DOI] [PubMed] [Google Scholar]
- 129.Lamark T, Johansen T. Autophagy: links with the proteasome. Curr Opin Cell Biol. 2010;22:192–198. doi: 10.1016/j.ceb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 130.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 132.Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, et al. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 133.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, et al. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 134.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 136.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 137.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 138.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 140.Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 142.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 143.Wong ES, Tan JM, Soong WE, Hussein K, Nukina N, Dawson VL, et al. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17:2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, et al. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- 145.Tung YT, Hsu WM, Lee H, Huang WP, Liao YF. The evolutionarily conserved interaction between LC3 and p62 selectively mediates autophagy-dependent degradation of mutant huntingtin. Cell Mol Neurobiol. 2010;30:795–806. doi: 10.1007/s10571-010-9507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rockel TD, Stuhlmann D, von Mikecz A. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J Cell Sci. 2005;118:5231–5242. doi: 10.1242/jcs.02642. [DOI] [PubMed] [Google Scholar]
- 147.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Narendra DP, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 158.Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 159.Kageyama T, Suzuki K, Ohsumi Y. Lap3 is a selective target of autophagy in yeast, Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;378:551–557. doi: 10.1016/j.bbrc.2008.11.084. [DOI] [PubMed] [Google Scholar]
- 160.Onodera J, Ohsumi Y. Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J Biol Chem. 2004;279:16071–16076. doi: 10.1074/jbc.M312706200. [DOI] [PubMed] [Google Scholar]
- 161.Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gal J, Strom AL, Kwinter DM, Kilty R, Zhang J, Shi P, et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111:1062–1073. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 164.Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NFκB activation. EMBO J. 1999;18:3044–3053. doi: 10.1093/emboj/18.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NFκB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wooten MW, Seibenhener ML, Neidigh KB, Vandenplas ML. Mapping of atypical protein kinase C within the nerve growth factor signaling cascade: relationship to differentiation and survival of PC12 cells. Mol Cell Biol. 2000;20:4494–4504. doi: 10.1128/mcb.20.13.4494-4504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lee SJ, Pfluger PT, Kim JY, Nogueiras R, Duran A, Pages G, et al. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 2010;11:226–232. doi: 10.1038/embor.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 169.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NFκB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 170.Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, et al. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 171.Hiruma Y, Honjo T, Jelinek DF, Windle JJ, Shin J, Roodman GD, et al. Increased signaling through p62 in the marrow microenvironment increases myeloma cell growth and osteoclast formation. Blood. 2009;113:4894–4902. doi: 10.1182/blood-2008-08-173948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hiruma Y, Kurihara N, Subler MA, Zhou H, Boykin CS, Zhang H, et al. A SQSTM1/p62 mutation linked to Paget's disease increases the osteoclastogenic potential of the bone microenvironment. Hum Mol Genet. 2008;17:3708–3719. doi: 10.1093/hmg/ddn266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pursiheimo JP, Rantanen K, Heikkinen PT, Johansen T, Jaakkola PM. Hypoxia-activated autophagy accelerates degradation of SQSTM1/p62. Oncogene. 2009;28:334–344. doi: 10.1038/onc.2008.392. [DOI] [PubMed] [Google Scholar]
- 174.Kadandale P, Stender JD, Glass CK, Kiger AA. Conserved role for autophagy in Rho1-mediated cortical remodeling and blood cell recruitment. Proc Natl Acad Sci USA. 2010;107:10502–10507. doi: 10.1073/pnas.0914168107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 176.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Into T, Inomata M, Niida S, Murakami Y, Shibata KI. Regulation of MyD88 aggregation and the MyD88-dependent signaling pathway by sequestosome 1 and histone deacetylase 6. J Biol Chem. 2010 doi: 10.1074/jbc.M110.126904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Swearingen KE, Loomis WP, Zheng M, Cookson BT, Dovichi NJ. Proteomic profiling of lipopolysaccharide-activated macrophages by isotope coded affinity tagging. J Proteome Res. 2010;9:2412–2421. doi: 10.1021/pr901124u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ishii T, Yanagawa T, Yuki K, Kawane T, Yoshida H, Bannai S. Low micromolar levels of hydrogen peroxide and proteasome inhibitors induce the 60 kDa A170 stress protein in murine peritoneal macrophages. Biochem Biophys Res Commun. 1997;232:33–37. doi: 10.1006/bbrc.1997.6221. [DOI] [PubMed] [Google Scholar]
- 180.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 181.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 182.Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal. 2010;3:3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, et al. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 184.Liu Y, Kern JT, Walker JR, Johnson JA, Schultz PG, Luesch H. A genomic screen for activators of the antioxidant response element. Proc Natl Acad Sci USA. 2007;104:5205–5210. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, et al. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem. 2010;285:16782–16788. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, et al. Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy. 2010;6:614–621. doi: 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jain A, Lamark T, Sjøttem E, Bowitz Larsen K, Awuh JA, Øvervatn A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Larsen KB, Lamark T, Øvervatn A, Harneshaug I, Johansen T, Bjørkøy G. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy. 2010;6:784–793. doi: 10.4161/auto.6.6.12510. [DOI] [PubMed] [Google Scholar]
- 189.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]