Abstract
CaV1.2 calcium channels play roles in diverse cellular processes such as gene regulation, muscle contraction, and membrane excitation and are diversified in their activity through extensive alternative splicing of the CaV1.2 mRNA. The mutually exclusive exons 8a and 8 encode alternate forms of transmembrane segment 6 (IS6) in channel domain 1. The human genetic disorder Timothy syndrome is caused by mutations in either of these two CaV1.2 exons, resulting in disrupted Ca2+ homeostasis and severe pleiotropic disease phenotypes. The tissue-specific pattern of exon 8/8a splicing leads to differences in symptoms between patients with exon 8 or 8a mutations. Elucidating the mechanisms controlling the exon 8/8a splicing choice will be important in understanding the spectrum of defects associated with the disease. We found that the polypyrimidine tract-binding protein (PTB) mediates a switch from exon 8 to 8a splicing. PTB and its neuronal homolog, nPTB, are widely studied splicing regulators controlling large sets of alternative exons. During neuronal development, PTB expression is down-regulated with a concurrent increase in nPTB expression. Exon 8a is largely repressed in embryonic mouse brain but is progressively induced during neuronal differentiation as PTB is depleted. This splicing repression is mediated by the direct binding of PTB to sequence elements upstream of exon 8a. The nPTB protein is a weaker repressor of exon 8a, resulting in a shift in exon choice when nPTB replaces PTB in cells. These results provide mechanistic understanding of how these two exons, important for human disease, are controlled.
Keywords: Calcium Channels, Gene Regulation, Ion Channels, Neurodevelopment, RNA Splicing, hnRNP I, PTB, Timothy Syndrome, Alternative Splicing, nPTB
Introduction
Calcium ions play important roles in cellular signaling, and maintaining appropriate intracellular Ca2+ concentrations is critical in the control of many essential biological processes (1). CaV1.2 voltage-gated L-type calcium channels open upon membrane depolarization, allowing a rapid Ca2+ flux that initiates cellular events such as cardiac and smooth muscle contraction, hormone and neurotransmitter release, Ca2+-induced gene expression, cell motility, and membrane excitation (2–7). Recent genome-wide association studies have linked variation in the CACNA1C gene, which encodes the CaV1.2 channel, with bipolar disorder, schizophrenia, and autism spectrum disorders (8–10). Elucidating how CaV1.2 currents are modulated will be important for understanding the control of multiple cellular processes and how these processes misfunction in neurological and psychiatric disease.
The functional properties of ion channels are often highly diversified through alternative splicing of their mRNAs. The CACNA1C gene transcript has multiple alternatively spliced segments that affect different portions of the channel protein (11–15). Exons 8a and 8 are spliced in a mutually exclusive pattern and encode different versions of the IS6 segment in the protein, which forms a portion of the CaV1.2 pore region (11, 16–18). (Note that two nomenclatures for exons 8a and 8 are found in the literature. We follow the most common nomenclature from the original description of the human locus, where exon 8a is placed upstream of exon 8 in the genomic sequence (GenBankTM accession number Z26263) (16, 19).) In other studies, the upstream exon is named 8 and the downstream exon 8A (20, 21). Functional studies comparing exon 8 and 8a CaV1.2 variants suggest that they have similar electrophysiological properties but different sensitivities to dihydropyridine drugs (18, 22). Exon 8a and 8 variants are expressed in different tissues, and the ratio of exon 8 to 8a inclusion has been found to change in hypertrophied heart of spontaneously hypertensive rats (18, 20, 21, 23, 24). Alternative exons 8a and 8 are conserved in CACNA1C genes across vertebrate species, and a similar pair of exons is present in the related CACNA1D gene encoding the CaV1.3 L-type calcium channel (16, 25, 26). These exons clearly serve a conserved and important function. However, the molecular mechanisms controlling their splicing remain unknown.
In Timothy syndrome, single nucleotide missense mutations in either of the two mutually exclusive exons (8a or 8) of the CACNA1C gene alter the function of the channel in controlling cellular calcium concentration and lead to devastating disease phenotypes (20, 21). Patients with Timothy syndrome display dysfunctions in multiple organ systems, including congenital heart disease, syndactyly, immune deficiency, and autism. Interestingly, the spectrum of symptoms in Timothy syndrome differs depending on the exon (8 or 8a) that carries the mutation, presumably reflecting the different patterns of exon use between different cells and tissues (20, 27, 28). However, the mechanisms controlling this tissue-specific switching of exon 8a and 8 are not understood.
The factors that determine splicing choices in CaV1.2 have not been well studied. The most common mechanisms for affecting splice site choice involve the binding of specialized regulatory proteins to the pre-mRNA to alter splice site recognition by the spliceosome either positively or negatively. One such factor, the polypyrimidine tract-binding protein (PTB),2 is a widely expressed splicing regulator whose down-regulation during muscle and neuronal development allows induction of a variety of neuron- and muscle-specific splicing events (29–32). PTB carries four RNA recognition motifs and binds CU-rich regulatory elements adjacent to regulated exons (33–36). In this study, we found that the choice of splicing exon 8a or 8 in CaV1.2 is strongly regulated by PTB and, to a lesser extent, by its neuronal homolog, nPTB. PTB protein represses the splicing of exon 8a and thereby shifts the splicing choice to exon 8. These data establish the PTB protein as an important regulator of CaV1.2 splicing and provide the first insights into how these important disease exons are controlled. They also implicate CaV1.2 signaling as one component of the PTB splicing program that is induced during brain development.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
N2a (Neuro-2a) cells were grown according to the American Type Culture Collection-recommended protocols. Transfections were done with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Plasmids
The Dup E8a-8 and Dup E8a reporter minigenes were constructed by amplifying mouse genomic DNA containing exons 8a and 8 with flanking intron sequences using Pfu DNA polymerase and primers designed based on the mouse genome from UCSC Genome Browser. The PCR fragments were cloned into the template plasmid pDup4-1 (37, 38) between the ApaI and BglII sites, which were engineered into 5′- and 3′-ends of the amplicons. The same upper primer was used for the both minigenes (5′-GATCGATCAAGGGCCCGACCCACCATCCTAGACAAGT-3′), and the lower primers were 5′-GATCGATCAAAGATCTGCCTGCAGATGATAGCTGT-3′ for the Dup E8a-8 minigene and 5′-GATCGATCAAAGATCTTCAGGCACATACAGCAACCGAG-3′ for the Dup E8a minigene. Mutations in the minigenes were generated using the QuikChange® site-directed mutagenesis kit (Stratagene), followed by subcloning into pDup4-1 using the ApaI and BglII sites. All constructs were confirmed by sequencing. The PTB and nPTB cDNA expression plasmids were constructed in pcDNA3.1 (Invitrogen) as described previously (33, 39). Each carried an N-terminal FLAG tag.
Short Hairpin RNAs
The short hairpin RNAs psiRNA-sh-PTB and psiRNA-sh-nPTB were designed to target the 3′-untranslated region of the mouse PTB and nPTB genes. The template vector was psiRNA-W[H1.4], and the design of the short hairpin RNA was similar to that described previously (30). The oligonucleotides used were as follows: for PTB, 5′-TCCCGCAGTTGGAGTGACCTTACTTCAAGAGAGTAAGGTCACTTCAGCTGC-3′ (forward) and 5′-AAAAGCAGTTGGAGTGACCTTACTCTCTTGAATAAGGTCACTTCAGCTGC-3′ (reverse); and for nPTB, 5′-TCCCATGTCAGTACTGAGTAATGTCTTCAAGAGAGACATTACTTAGTGCTGACAT-3′ (forward) and 5′-AAAAATGTCAGTACTGAGTAATGTCTCTCTTGAAGACATTACTTAGTGCTGACAT-3′ (reverse). The short hairpin RNA expression plasmids were transfected into N2a cells grown in 6-well plates using Lipofectamine 2000. The efficiency of knockdown was verified by Western blotting.
RT-PCR
Cytoplasmic RNA was isolated from cells or mouse embryonic cortexes with an RNeasy mini kit (Qiagen) and reverse-transcribed with random hexamers and Superscript III (Invitrogen) according to the manufacturer's instructions. Splice products were PCR-amplified (25 cycles for N2a cells and 17 cycles for minigene splice products) using a 32P-labeled primer in the 3′-constitutive exon and an unlabeled primer in the 5′-constitutive exon. The primers used for the endogenous CaV1.2 transcripts in N2a cells or mouse embryonic cortex were ′-GGCATCACCAACTTCGACAA-3′ (forward) and 5′-TGATCCAGTCCAGGTAGCCTT-3′ (reverse). The primers used for transcripts derived from the Dup E8a-8 and Dup E8a minigenes were the Dup-8 forward primer (5′-GACACCATGCATGGTGCAC-3′) and the Dup-3 reverse primer (5′-AACAGCATCAGGAGTGGACAGATCCC-3′). The 32P-labeled PCR products were digested by BamHI at 37 °C for 3 h and separated on 7.5 M urea-8% polyacrylamide denaturing gels. Gels were dried, visualized by a Typhoon phosphorimaging system (Amersham Biosciences), and quantified using ImageQuant TL software (Amersham Biosciences).
Immunoblots
Tissue culture cell lysates for immunoblotting were isolated in radioimmune precipitation assay buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mm Tris, pH 8.0) as described previously (29). Whole cell lysates were resolved on 4–20% Tris/glycine gels (Invitrogen) in Tris/glycine/SDS running buffer. The proteins were transferred onto Protran® nitrocellulose membranes (Whatman) or Immobilon-FL PVDF membranes (Millipore). Membranes were probed under standard conditions with antibody raised against the PTB N terminus (1:3000) (39), the nPTB IS2 segment (1:500) (40), or GAPDH (1:10,000; Ambion). Blots were then probed with horseradish peroxidase-conjugated secondary antibodies (1:10,000; GE Healthcare) and developed using Femto SuperSignal reagents (Pierce) and Kodak Bio-Max XAR film. For blotting with fluorophore-conjugated secondary antibodies, the PVDF membranes were probed with ECL Plex Cy3-conjugated goat anti-mouse (1:5000; GE Healthcare) and Cy5-conjugated goat anti-rabbit (1:2500; GE Healthcare) secondary antibodies, dried, and scanned on a Typhoon phosphorimaging system. Quantification of fluorescent signals was performed using ImageQuant TL software.
Electrophoretic Mobility Shift Assay
The in vitro transcription of [α-32P]UTP-radiolabeled and unlabeled RNA probes and the purification of His-PTB were both performed as described previously (40, 41). 25 nm [α-32P]UTP-radiolabeled RNA was equilibrated in HeLa nuclear extracts or with various concentrations of purified His-PTB (10–1000 nm) in binding buffer (20 mm HEPES, pH 7.6, 20% glycerol, 80 mm potassium glutamate, 100 ng/μl tRNA, 0.02% Nonidet P-40, 6 mm MgCl2, and 1 mm DTT). All reaction components except the RNA probe were mixed and incubated for 5 min at 30 °C; the RNA probe was then added and incubated for another 25 min at 30 °C. In the supershift assay experiment, after the incubation of radiolabeled RNA probe with HeLa nuclear extract for 30 min, anti-PTB or anti-nPTB antibody was added to the reactions and incubated at room temperature for an additional 30 min. Each reaction mixture was loaded onto a prerun native polyacrylamide gel (6% (w/v) 19:1 acrylamide/bisacrylamide and 1× Tris borate/EDTA). Gels were resolved at 200 V for 90 min before they were dried, exposed to a PhosphorImager screen, and visualized as described above.
RESULTS
PTB Protein Expression Stimulates CaV1.2 Exon 8 Splicing over Exon 8a Splicing
The control of the splicing choice between exons 8a and 8 in CaV1.2 is of great interest for understanding both the regulation of calcium channel function and the role of these exons in Timothy syndrome. In examining these exons, we found that multiple sequence blocks surrounding the two exons are conserved across mammalian species. Most notably, a conserved sequence encompassing the putative branch point of exon 8a is enriched in C and U nucleotides, which is a hallmark of exons that are repressed by the PTB protein (Fig. 1A) (34). We assayed the splicing of exons 8a and 8 in N2a mouse neuroblastoma cells by RT-PCR. These exons are identical in length and highly similar in sequence. To distinguish the different spliced forms, we used primers within exons 7 and 9 to amplify the region containing exons 8 and 8a, followed by digestion of the amplified products with BamHI, which cleaves exon 8a but not exon 8. This produced DNA fragments of diagnostic sizes for single exon 8 or 8a insertion as well as for the products of double exon skipping (without exons 8a and 8, ΔExon) and double exon insertion (Exon (8a+8)) (Fig. 1B). These latter two products were detectable in N2a cells, but the single exon insertions predominated as expected for a pair of mutually exclusive exons. In untreated N2a cells, the exon 8-included mRNA was the most abundant transcript, making up 82% of the CaV1.2 mRNA containing one exon. Notably, the knockdown of PTB protein in N2a cells using a short hairpin RNA dramatically changed the splicing pattern, with the inclusion level of exon 8 being reduced from 82 to 7–10% and the splicing of exon 8a proportionally increased (Fig. 1C, lanes 1–3). Western blot analysis confirmed that PTB protein was successfully depleted. As observed previously, nPTB protein expression was strongly induced by PTB knockdown (30, 32, 42). The neuronally expressed homolog of PTB, nPTB, has both common and different targets compared with PTB. The strong shift from exon 8 to 8a splicing seen with the loss of PTB and gain of nPTB indicates that this switch was affected primarily by PTB protein and not nPTB. It is possible that PTB could act negatively on one exon while nPTB acted positively on the other. However, the depletion of the low amount of nPTB in N2a cells did not affect the exon 8/8a ratio at all, and the double PTB/nPTB knockdown had the same effect as the single PTB knockdown (Fig. 1C, lanes 4–6).
FIGURE 1.
Depletion of PTB switches splicing from exon 8 to 8a in N2a cells. A, upper panel, diagram of the CaV1.2 genomic sequence from exons 7 to 9 showing the mutually exclusive splicing pattern of exons 8a and 8. Lower panel, PhastCons plot of placental mammal conservation (UCSC Genome Browser) showing the conservation of CT-rich elements (highlighted in gray) surrounding the potential branch point (indicated by the asterisk). The RNA probe used in Fig. 4 is also indicated. B, digestion maps of the amplified exon 8a/8 splice variants. Exons 7, 8a, 8, and 9 are shown as boxes, and introns are shown as lines. The 104-nt exons 8a and 8 comprise a pair of mutually exclusive exons encoding alternate forms of the CaV1.2 channel IS6 pore region (11, 18). The PCR-amplified region for exons 8a and 8 encompasses exons 7–9, with the upper primer in exon 7 and the 32P-labeled lower primer in exon 9. Exons 8a and 8 are not strictly mutually exclusive, and a low level of CaV1.2 transcripts can be observed where both exons are included or both excluded. The four mRNA products are indicated as Exon 8 or Exon 8a for either exon inclusion, Exon (8a+8) for double exon insertion, and ΔExon for double exon skipping. Exon 8a contains a BamHI cleavage site not present in exon 8. After PCR amplification, the products were digested by BamHI to distinguish all four species. The 32P-labeled 3′-fragment of the four PCR products are underlined, and their molecular sizes indicated. C, depletion of PTB promotes exon 8a splicing. Upper panel, RT-PCR assay of exon 8a and 8 splicing in N2a cells expressing the vector control (ctrl) or the PTB (sh-PTB)- and/or nPTB (sh-nPTB)-targeted shRNAs. The labeled PCR products were cleaved with BamHI and separated on denaturing gels. The molecular identities and sizes of the products are indicated on the left. The percent of single exon 8 inclusion relative to the total single exon inclusion products (exons 8 and 8a) is shown below. Lower panel, immunoblot of the PTB and nPTB proteins in cells treated with control vector (lane 1) and in cells treated with short hairpin vectors targeting PTB (lanes 2 and 3), nPTB (lanes 4 and 5), or both (lane 6). GAPDH expression was monitored as a loading control. D, overexpression of PTB and nPTB proteins promotes exon 8 splicing. Upper panel, RT-PCR assay of exon 8a and 8 splicing in N2a cells transfected with the pcDNA3.1 control vector or with PTB and nPTB expression plasmids. Lower panel, immunoblot of PTB and nPTB proteins in N2a cells transfected with PTB and nPTB expression plasmids. GAPDH expression served as a loading control.
To confirm the role of PTB in the regulation of exons 8a and 8, we examined the effects of PTB and nPTB overexpression on the splicing (Fig. 1D). Although the basal inclusion level of exon 8 was relatively high (82%), 2-fold overexpression of PTB protein further increased exon 8 splicing to 94% (lanes 1 and 2). In contrast, >10-fold overexpression of nPTB increased the exon 8 inclusion to 91% (lane 3). These results suggest that PTB and nPTB have the same effect of repressing exon 8a and/or enhancing exon 8 but that PTB has much stronger activity than nPTB.
PTB Regulates Exon 8a and 8 Splicing in Heterologous Transcripts Expressed from a Splicing Reporter Gene
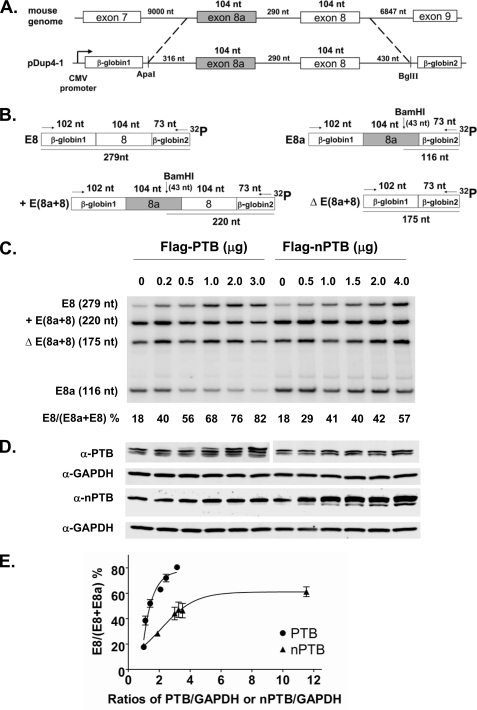
To examine the effect of PTB and nPTB on CaV1.2 splicing more carefully, we constructed the Dup E8a-8 minigene, which contains exons 8a and 8 and portions of their adjacent introns inserted between β-globin exons 1 and 2 and is driven by the CMV promoter (Fig. 2A). When expressed from this plasmid in N2a cells, exons 8 and 8a showed a somewhat different pattern of use than the endogenous CaV1.2 transcripts, with higher levels of double exon inclusion and double exon skipping. Presumably, as seen in other minigenes, the flanking exons and the large flanking introns that are deleted in the minigene are likely to include binding sites for additional regulators and thus affect the efficiency of splice product formation. Alternatively, differences in the transcriptional regulation of the transfected and endogenous genes could affect their exon inclusion rates.
FIGURE 2.
PTB switches splicing from exon 8a to 8 in transcripts expressed from a transfected minigene. A, diagram of the mouse genomic sequence encompassing exons 8a and 8 cloned into the pDup4-1 minigene reporter (37). The resulting Dup E8a-8 minigene contains 1244 nt of mouse genomic sequence, including 316 nt of upstream intron, the 104-nt exon 8a, the 104-nt exon 8, the complete 290-nt intron between exons 8a and 8, and 430 nt of downstream intron. B, digestion maps of exon 8a/8 splice products derived from the Dup E8a-8 minigene. Similar to native CaV1.2 transcripts, exons 8a and 8 in the Dup E8a-8 minigene are not strictly mutually exclusive, and transcripts in which both exons are included or both excluded are observed. These products are labeled as described in the legend of Fig. 1. C, RT-PCR assay of exon 8a and 8 splicing in N2a cells cotransfected with the Dup E8a-8 minigene and increasing amounts of FLAG-PTB and FLAG-nPTB plasmids. Lanes and products are labeled as described in the legend of Fig. 1. D, immunoblot of PTB and nPTB expression in transfected N2a cells. GAPDH expression was monitored as a loading control. E, dose-response curves for PTB and nPTB. The amount of expressed PTB or nPTB relative to GAPDH is plotted versus the percent exon 8 splicing. Note that exon 8 was more sensitive to small changes in PTB expression compared with nPTB, and its maximum level of splicing was much higher at high PTB concentrations compared with nPTB.
As seen for the endogenous gene, the splicing of transcripts from the transfected minigene is regulated by PTB. For the single exon inclusion products, 18% contain exon 8, and 82% contain exon 8a. Coexpression of PTB protein strongly shifted the splicing in a dose-dependent manner to favor exon 8, reaching 82% exon 8 splicing at the highest titration point (Fig. 2, C and D). Coexpression of nPTB also increased exon 8 inclusion, but not as strongly and reaching a maximum of 57% exon 8 splicing. The immunoblots for PTB and nPTB used different antibodies whose signals cannot be directly compared. However, plotting exon 8 splicing relative to the ratios of PTB and nPTB to GAPDH expression revealed clear differences in the dose-response curves for the two splicing regulators (Fig. 2E). PTB could drive exon 8 splicing to a much higher level than nPTB. Exon 8 was also sensitive to smaller changes in PTB concentration than seen for nPTB. For example, it required 4 μg of transfected nPTB plasmid to shift the splicing to the same degree as 0.5 μg of PTB plasmid. These data confirm that PTB is a significantly stronger regulator of the exon 8/8a splicing choice compared with nPTB.
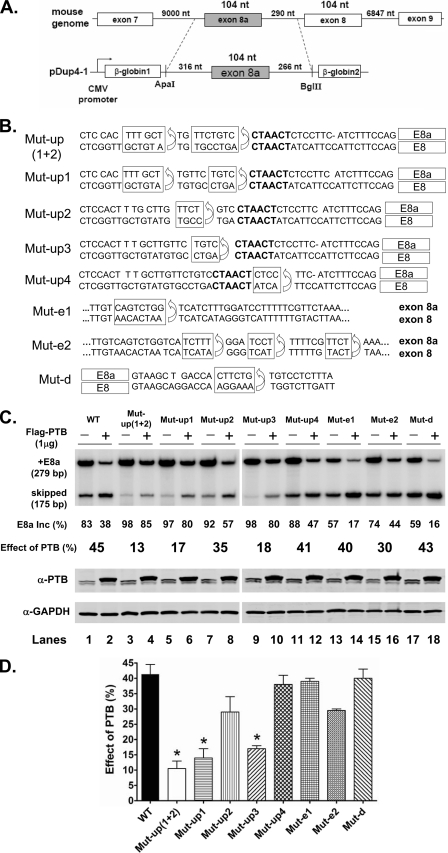
Intron Sequences Upstream of Exon 8a Mediate Splicing Repression by PTB
The expression of PTB shifts splicing of CaV1.2 from exon 8a inclusion to exon 8. This could result from PTB acting as a repressor of exon 8a, an enhancer of exon 8, or both. The presence of pyrimidine elements upstream of exon 8a, as often seen for PTB-repressed exons, led us to favor the first model. To measure the effect of PTB on exon 8a specifically, we constructed a single exon 8a splicing reporter containing the pyrimidine elements in the upstream intron (Fig. 3A). We aligned the exon 8 and 8a sequences and made mutations in the exon 8a clone that converted a series of different pyrimidine elements upstream, downstream, or within exon 8a to the sequence found in exon 8 (Fig. 3B). The wild-type and mutant 8a reporters were transiently expressed in N2a cells and tested for their response to coexpressed PTB. In the absence of the PTB expression clone, the wild-type reporter exhibited 83% inclusion of exon 8a. Coexpressed PTB reduced exon 8a inclusion to 38% (Fig. 3, C and D), indicating that exon 8a is indeed highly repressed by the protein. Mutation of several of the upstream pyrimidine elements both increased the basal level of exon 8a splicing and greatly reduced its repression by PTB (Mut-up1 and Mut-up3; 97% basal inclusion reduced to 80% by PTB). Several elements had little effect on the splicing of exon 8a, including those altered by Mut-up2, Mut-up4, and Mut-e2 (Fig. 3, C and D; see also supplemental Fig. 1). Combining mutations Mut-up2 and Mut-up1 gave a somewhat stronger effect than Mut-up1 alone. Mut-e1 and Mut-d, within the exon and downstream intron, reduced the basal level of inclusion, but still exhibited strong repression by PTB. These elements may alter positive-acting elements affecting the exon. These results support the model that PTB represses exon 8a through binding to pyrimidine elements in the upstream intron.
FIGURE 3.
Efficient repression of exon 8a by PTB requires upstream intron sequences. A, diagram of the Dup E8a minigene containing 686 nt of mouse genomic sequence, including 316 nt of upstream intron, the 104-nt exon 8a, and 266 nt of downstream intron. This Dup-E8a minigene without mutations is labeled WT in C and D. B, mutations in the Dup-E8a minigene. The genomic sequences encompassing exons 8a (upper lines) and 8 (lower lines) are aligned. Sequence elements upstream of exon 8a (Mut-up1 to Mut-up4), within the exon (Mut-e1 and Mut-e2), and downstream of exon 8a (Mut-d) were replaced with the corresponding sequence within or adjacent to exon 8 to generate a series of mutant Dup-E8a minigenes. The boxed exon 8a sequence element was replaced in each mutant with the element in exon 8 below. Mut-up(1 + 2) combines the mutations in Mut-up1 and Mut-up2. The putative branch points (CTAACT) of exons 8a and 8 are shown in boldface. C, upper panel, minigene reporters were cotransfected with 1 μg of empty vector pcDNA3.1+ (−) or FLAG-PTB expression vector (+), and the resulting splicing was assayed by RT-PCR. The transfected minigene is indicated above, and the percent exon 8a inclusion is indicated below, along with the percent change in exon 8a inclusion (Inc) with PTB cotransfection. Lower panel, immunoblot assay of PTB expression in the transfected N2a cells using anti-PTB antibody. GAPDH expression was monitored as a loading control. D, bar graph quantifying the total percent change in splicing with PTB coexpression for the Dup E8a minigene and each mutant. *, p < 0.01 when the experimental values from duplicate experiments were compared with the wild type by one-way analysis of variance and Tukey's tests.
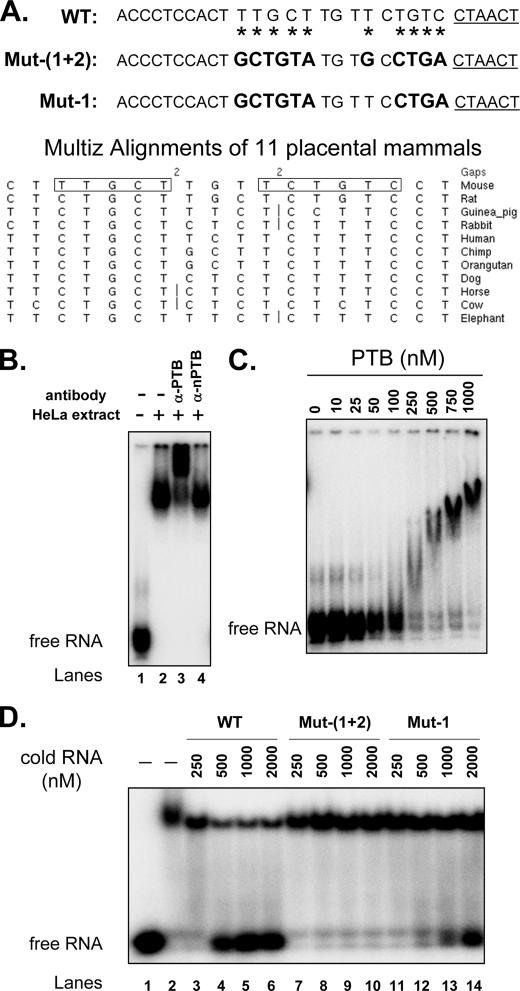
To confirm that PTB directly bound to the splicing repressor elements upstream of the exon 8a branch point, we performed EMSAs. We synthesized a 30-nucleotide (nt) RNA probe (WT) containing the putative exon 8a branch point and the upstream pyrimidine elements whose mutation affected PTB regulation (Figs. 1A and 4A, upper panel). These elements are largely conserved across mammalian species with some substitutions that are not likely to affect PTB binding (Fig. 4A, lower panel). This probe was incubated in HeLa nuclear extract and analyzed on native electrophoretic gels. This wild-type probe was quantitatively assembled in the extract into a slowly migrating molecular complex (Fig. 4B, lanes 1 and 2). This complex was supershifted in the gel by anti-PTB antibody (Fig. 4B, lane 3), indicating that the RNA was indeed bound by PTB. Antibody to nPTB, which is not present in HeLa extract (39), served as a negative control (Fig. 4B, lane 4). PTB binding was further confirmed by native gel assay using purified PTB, which showed a clear dose-dependent assembly of wild-type RNA into a PTB complex (Fig. 4C). To measure the sequence specificity of the PTB-RNA interaction, we performed competition experiments. We measured PTB binding to the radioactive wild-type probe in the presence of increasing concentrations of unlabeled wild-type RNA or RNA containing Mut-up1 and Mut-up(1+2) that strongly reduced PTB-dependent repression of splicing. As expected, wild-type RNA strongly competed for PTB binding (Fig. 4D, lanes 3–6). In contrast, Mut-up1 and Mut-up(1+2) eliminated the ability of an RNA to compete for PTB binding against wild-type RNA (Fig. 4D, lanes 7–14). Thus, PTB specifically binds to the upstream sequence that mediates PTB-dependent repression of exon 8a. Given the additional blocks of conserved sequences surrounding these exons, it is likely that PTB also binds to sites more distal from the exon.
FIGURE 4.
PTB binds specifically to sequences upstream of exon 8a. A, upper panel, RNA probes containing the wild-type sequence upstream of the putative exon 8a branch point (underlined) or mutant sequences as indicated. Mutated nucleotides are indicated by asterisks on the wild-type probe and are highlighted in boldface on the mutant RNA probes. Lower panel, alignment (UCSC Genome Browser) of placental mammal intron sequence affecting the regulation of exon 8a by PTB. The boxed sequences are those mutated in the upper panel. B, EMSA of PTB binding to the wild-type probe in HeLa extract. In vitro transcribed labeled RNA probe was incubated in binding buffer (lane 1) or HeLa nuclear extract (lane 2) or in HeLa nuclear extract followed by the addition of anti-PTB (lane 3) or anti-nPTB (lane 4) antibody. C, EMSA of purified PTB binding to the sequence upstream of exon 8a. The wild-type probe was incubated in binding buffer containing increasing concentrations of His-PTB protein, as indicated. D, the pyrimidine-rich elements upstream of exon 8a specifically compete for PTB binding. Lane 1, EMSA of labeled wild-type RNA (25 nm) not incubated with PTB; lanes 2–14, EMSA of labeled wild-type RNA (25 nm) preincubated with purified PTB (1000 nm) and unlabeled wild-type or mutant RNA probes at the concentrations indicated. Note that mutation of the upstream elements drastically reduced the ability of the RNA probe to compete with the wild-type probe for PTB binding.
CaV1.2 Splicing Switches from Exon 8 to 8a Concurrent with the Decline in PTB Expression in Developing Cortex
During the differentiation of neuronal progenitor cells into neurons, PTB expression is progressively reduced, altering the splicing of a set of PTB target exons (30, 32, 43). Expression of nPTB is initially induced during neuronal differentiation but later decreases as neurons mature after birth (data not shown). If the PTB protein is essential to proper regulation of CaV1.2 splicing in the brain as seen in tissue culture, there should be a shift from exon 8 to 8a splicing during brain development. To examine this, we measured the splicing of the two exons in developing mouse cortex from embryonic days 12 to 18. We found that exon 8 was used in ∼60% of the CaV1.2 mRNA at embryonic day 12 but was progressively repressed during development, reaching 10% inclusion by embryonic day 18 (Fig. 5A). As expected, exon 8a showed a coordinate increase in splicing during this time period. Immunoblot assay of the PTB proteins from embryonic cortex confirmed that PTB protein concentration sharply decreased from embryonic days 12 to 18 (Fig. 5B). In contrast, nPTB expression was moderately increased. Thus, this segment of the CaV1.2 channel is modified during brain development, and the change in exons 8 and 8a correlates well with the loss of PTB during neuronal differentiation.
FIGURE 5.
CaV1.2 exons 8a and 8 shift their splicing during development of embryonic mouse cortex. A, exon 8 progressively decreased in inclusion as exon 8a increased from embryonic days 12 (E12) to 18 (E18). The PCR products and sizes are indicated on the left. Note that transcripts excluding both exons 8a and 8 (ΔExon) were also observed in embryonic cortex. From 7 to 10 embryonic cortices were pooled for RNA and protein extraction at each time point. B, immunoblot of proteins in developing cortex shows decreasing PTB protein expression between embryonic days 12 and 18, whereas nPTB expression moderately increased, as normalized to GAPDH expression.
DISCUSSION
PTB Proteins Control the Exon 8/8a Splicing Choice in the CaV1.2 Calcium Channel
We found that the splicing factor PTB strongly repressed exon 8a of the CaV1.2 calcium channel transcript and thereby shifted splicing to the exon 8 isoform. Depletion of PTB from cells stimulated exon 8a, and overexpression of PTB repressed exon 8a splicing. These effects were also observed on transcripts from a transfected splicing reporter containing exon 8a but not exon 8. PTB bound directly to conserved sequence elements upstream of the putative exon 8a branch point, and mutation of these elements greatly reduced PTB-mediated splicing repression. Thus, PTB is a key factor determining the choice between exons 8 and 8a in CaV1.2 expression.
PTB is expressed in many cell types but is repressed during neuronal and muscle cell differentiation (29, 30, 32). We found that in the embryonic cortex, exon 8 predominated early in development at embryonic day 12, but splicing was strongly shifted to exon 8a by day 18, coincident with the loss of PTB expression. During this period, the neuronally expressed homolog of PTB, nPTB, increased in expression before being reduced after birth. We found that exon 8a was significantly more sensitive to PTB expression than nPTB, in agreement with its developmental pattern of splicing. PTB and nPTB are highly similar in peptide sequence and have similar RNA-binding properties, and yet we have found a group of exons, including exon 8a, that are differentially affected by the two proteins. Other exons seem to be equally sensitive to either PTB or nPTB (30, 42). These results present interesting mechanistic questions regarding how a differential or equivalent response to the two PTB proteins can be encoded in target exon sequences.
CaV1.2 exons 8 and 8a are members of a class of exons whose alternative splicing is conserved across vertebrate species and that show blocks of conserved sequence in their adjacent intron sequences (44–47). Exons of this class are generally regulated by multiple factors, and the PTB-binding sites adjacent to exon 8a comprise only a portion of the conserved nucleotides presenting potential regulatory elements for these exons. It is thus likely that other factors are affecting these exons in addition to PTB. Indeed, in cardiomyocytes, PTB proteins are expressed, but exon 8a is spliced into the CaV1.2 mRNA. In these cells, there must be additional mechanisms for countering PTB action (18, 23, 24). We have found that two other alternative exons in CaV1.2, exons 9* and 33, are strongly regulated by proteins in the Fox family of splicing factors (48). Coexpression and knockdown of Fox1 and Fox2 were found to have only weak effects on exons 8 and 8a (data not shown). More work is needed to identify the larger set of regulatory factors and sequence elements controlling these exons.
A number of mechanisms have been described that enforce the mutually exclusive choice between two exons (49). CaV1.2 exons 8a and 8 are separated by a several hundred-nucleotide intron that has compatible major class splice sites. Moreover, the downstream exon 8 does not apparently have a distal branch point that would interfere with recognition of exon 8a. Thus, the mutually exclusive splicing pattern is not likely due to the steric interference or splice site incompatibility mechanisms described for tropomyosin and Jnk1 exons (50, 51). It is possible that factors combine to positively affect one exon and negatively affect the other, as seen with mutually exclusive exons in the FGFR2 transcript (52, 53). We did not see a significant positive effect of PTB on the splicing of exon 8 expressed from a minigene (data not shown), but other factors could play such a role. The strong splicing of exon 8 seen in N2a cells that is converted to almost complete exon 8a splicing by PTB depletion indicates that either exon can be efficiently spliced in N2a cells. Notably, the splicing of exons 8a and 8 is not strictly mutually exclusive, as both the double exon-included and double exon-skipped products can be observed in cells. Exons 8 and 8a are both 104 nt long. Thus, both the double exon-included and double exon-skipped transcripts will cause a frameshift in the mRNA and are unlikely to produce a functional protein product. At this position in the mRNA, upstream from many exon-exon junctions, these transcripts are predicted to be targets of the nonsense-mediated RNA decay pathway (54, 55). The mutually exclusive choice in splicing exons 8 and 8a is not apparently rigidly enforced, but the incorrect products are degraded, as proposed for the FGFR2 transcript (54).
We recently demonstrated that, similar to the regulation of exons 8 and 8a by PTB, the Fox proteins regulate the splicing of CaV1.2 alternative exons 9* and 33 during cortical development (48). Thus, these channels undergo extensive functional modification at the level of splicing during brain development. It will be interesting to test whether PTB proteins also control the splicing of similar exons in the CaV1.3 channel during development (16, 25, 26).
Alternative Splicing of CaV1.2 in Timothy Syndrome
The regulation of the CaV1.2 exon 8/8a switch is also relevant to its role in neurological disease, most notably Timothy syndrome. There is great interest in splicing-targeted therapies for a number of human disorders, including spinal muscular atrophy, frontotemporal dementia with parkinsonism on chromosome 17, and familial dysautonomia (56–59). Although such therapies still need significant development, there have been promising results using complementary oligonucleotides to cause exon skipping in dystrophin transcripts in the treatment of Duchenne muscular dystrophy (60–63). Conversely, oligonucleotides that block a repressor element of exon 7 in the SMN2 transcript can stimulate its splicing, providing a promising strategy for the treatment of spinal muscular atrophy (64–67).
Timothy syndrome is an interesting setting for considering therapeutic alteration of splicing. One characterized Timothy syndrome mutation (G1216A in DNA or G406R in protein) greatly slows the voltage-dependent inactivation of the CaV1.2 channel (21, 68, 69). The prolonged Ca2+ current from this channel is presumably harmful to cells and may account for the pleiotropic effects seen in the disease (27). Although they clearly produce aberrant proteins, it is possible that the nucleotide substitutions also alter the splicing of the exons. This should be examined in the patient RNA. If this is not the case, then the Timothy syndrome mutations are likely to be deleterious only in cells where the exon containing the mutation is spliced into the mRNA. The different spectrum of symptoms produced by mutations in exon 8 or 8a is presumably due to the different patterns of tissue-specific exon use (20, 21, 28, 70). The presence of the other functional exon in these mutated genes creates an unusual situation for splicing-targeted therapy, as one can envision shifting the exon choice in tissues where the mutated exon is predominant. Although exons 8 and 8a will not be exactly equivalent, this exon switch may still produce a more functional channel than that containing the mutation.
In analogy to the results on SMN2, blocking the PTB-binding site upstream of exon 8a with an antisense oligonucleotide would be predicted to activate exon 8a splicing and shift the choice away from exon 8. This could be beneficial to patients carrying mutations in the downstream exon 8. Drugs that interfere with PTB function would also be predicted to activate exon 8a. However, PTB has many targets, and such a treatment would have multiple other effects. In patients with exon 8a mutations, switching the splicing to exon 8 might be accomplished by blocking one of the exon 8a splice sites. This is known to induce exon skipping in dystrophin and other transcripts (62, 71–73). In addition to the brain, CaV1.2 channels also play important roles in cardiac and vascular smooth muscles. CACNA1C splice variants contribute to important differences in channel activity between these tissues. It will be important to identify the other factors controlling CaV1.2 splicing, if we are to fully understand the intricate control of CaV1.2 function in normal tissue and in disease.
Acknowledgments
We thank Peter Stoilov for the sh-PTB and sh-nPTB plasmid constructs and Diane Lipscombe for discussions and critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM49662 (to D. L. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- PTB
- polypyrimidine tract-binding protein
- nPTB
- neuronal PTB
- nt
- nucleotide(s).
REFERENCES
- 1. Carafoli E., Klee C. B. (eds) (1999) Calcium as a Cellular Regulator, Oxford University Press, New York [Google Scholar]
- 2. Catterall W. A. (2000) Annu. Rev. Cell Dev. Biol. 16, 521–555 [DOI] [PubMed] [Google Scholar]
- 3. Dolmetsch R. E., Pajvani U., Fife K., Spotts J. M., Greenberg M. E. (2001) Science 294, 333–339 [DOI] [PubMed] [Google Scholar]
- 4. Stea A., Soong T. W., Snutch T. P. (1995) in Handbook of Receptors and Channels: Ligand- and Voltage-gated Ion Channels (North R. A. ed) pp. 113–152, CRC Press, Boca Raton, FL [Google Scholar]
- 5. Striessnig J. (1999) Cell Physiol. Biochem. 9, 242–269 [DOI] [PubMed] [Google Scholar]
- 6. Tsien R. W., Wheeler D. B. (1999) in Calcium as a Cellular Regulator (Carafoli E., Klee C. eds) pp. 171–199, Oxford University Press, New York [Google Scholar]
- 7. Zühlke R. D., Pitt G. S., Deisseroth K., Tsien R. W., Reuter H. (1999) Nature 399, 159–162 [DOI] [PubMed] [Google Scholar]
- 8. Ferreira M. A., O'Donovan M. C., Meng Y. A., Jones I. R., Ruderfer D. M., Jones L., Fan J., Kirov G., Perlis R. H., Green E. K., Smoller J. W., Grozeva D., Stone J., Nikolov I., Chambert K., Hamshere M. L., Nimgaonkar V. L., Moskvina V., Thase M. E., Caesar S., Sachs G. S., Franklin J., Gordon-Smith K., Ardlie K. G., Gabriel S. B., Fraser C., Blumenstiel B., Defelice M., Breen G., Gill M., Morris D. W., Elkin A., Muir W. J., McGhee K. A., Williamson R., MacIntyre D. J., MacLean A. W., St C. D., Robinson M., Van Beck M., Pereira A. C., Kandaswamy R., McQuillin A., Collier D. A., Bass N. J., Young A. H., Lawrence J., Ferrier I. N., Anjorin A., Farmer A., Curtis D., Scolnick E. M., McGuffin P., Daly M. J., Corvin A. P., Holmans P. A., Blackwood D. H., Gurling H. M., Owen M. J., Purcell S. M., Sklar P., Craddock N. (2008) Nat. Genet. 40, 1056–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nyegaard M., Demontis D., Foldager L., Hedemand A., Flint T. J., Sørensen K. M., Andersen P. S., Nordentoft M., Werge T., Pedersen C. B., Hougaard D. M., Mortensen P. B., Mors O., Børglum A. D. (2010) Mol. Psychiatry 15, 119–121 [DOI] [PubMed] [Google Scholar]
- 10. Wang K., Zhang H., Ma D., Bucan M., Glessner J. T., Abrahams B. S., Salyakina D., Imielinski M., Bradfield J. P., Sleiman P. M., Kim C. E., Hou C., Frackelton E., Chiavacci R., Takahashi N., Sakurai T., Rappaport E., Lajonchere C. M., Munson J., Estes A., Korvatska O., Piven J., Sonnenblick L. I., Alvarez Retuerto A. I., Herman E. I., Dong H., Hutman T., Sigman M., Ozonoff S., Klin A., Owley T., Sweeney J. A., Brune C. W., Cantor R. M., Bernier R., Gilbert J. R., Cuccaro M. L., McMahon W. M., Miller J., State M. W., Wassink T. H., Coon H., Levy S. E., Schultz R. T., Nurnberger J. I., Haines J. L., Sutcliffe J. S., Cook E. H., Minshew N. J., Buxbaum J. D., Dawson G., Grant S. F., Geschwind D. H., Pericak-Vance M. A., Schellenberg G. D., Hakonarson H. (2009) Nature 459, 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abernethy D. R., Soldatov N. M. (2002) J. Pharmacol. Exp. Ther. 300, 724–728 [DOI] [PubMed] [Google Scholar]
- 12. Cheng X., Pachuau J., Blaskova E., Asuncion-Chin M., Liu J., Dopico A. M., Jaggar J. H. (2009) Am. J. Physiol. Heart Circ. Physiol. 297, H680–H688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liao P., Yong T. F., Liang M. C., Yue D. T., Soong T. W. (2005) Cardiovasc. Res. 68, 197–203 [DOI] [PubMed] [Google Scholar]
- 14. Lipscombe D., Pan J. Q., Gray A. C. (2002) Mol. Neurobiol. 26, 21–44 [DOI] [PubMed] [Google Scholar]
- 15. Soldatov N. M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4628–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lipscombe D., Castiglioni A. J. (2004) in Calcium Channel Pharmacology (McDonough S. I. ed) pp. 369–409, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 17. Tang Z. Z., Liang M. C., Lu S., Yu D., Yu C. Y., Yue D. T., Soong T. W. (2004) J. Biol. Chem. 279, 44335–44343 [DOI] [PubMed] [Google Scholar]
- 18. Welling A., Ludwig A., Zimmer S., Klugbauer N., Flockerzi V., Hofmann F. (1997) Circ. Res. 81, 526–532 [DOI] [PubMed] [Google Scholar]
- 19. Soldatov N. M. (1994) Genomics 22, 77–87 [DOI] [PubMed] [Google Scholar]
- 20. Splawski I., Timothy K. W., Decher N., Kumar P., Sachse F. B., Beggs A. H., Sanguinetti M. C., Keating M. T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8089–8096; Discussion 8086–8088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Splawski I., Timothy K. W., Sharpe L. M., Decher N., Kumar P., Bloise R., Napolitano C., Schwartz P. J., Joseph R. M., Condouris K., Tager-Flusberg H., Priori S. G., Sanguinetti M. C., Keating M. T. (2004) Cell 119, 19–31 [DOI] [PubMed] [Google Scholar]
- 22. Zühlke R. D., Bouron A., Soldatov N. M., Reuter H. (1998) FEBS Lett. 427, 220–224 [DOI] [PubMed] [Google Scholar]
- 23. Tang Z. Z., Hong X., Wang J., Soong T. W. (2007) Cell Calcium 41, 417–428 [DOI] [PubMed] [Google Scholar]
- 24. Tang Z. Z., Liao P., Li G., Jiang F. L., Yu D., Hong X., Yong T. F., Tan G., Lu S., Wang J., Soong T. W. (2008) Biochim. Biophys. Acta 1783, 118–130 [DOI] [PubMed] [Google Scholar]
- 25. Jurkat-Rott K., Lehmann-Horn F. (2004) J. Physiol. 554, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koschak A., Reimer D., Huber I., Grabner M., Glossmann H., Engel J., Striessnig J. (2001) J. Biol. Chem. 276, 22100–22106 [DOI] [PubMed] [Google Scholar]
- 27. Liao P., Soong T. W. (2010) Pflugers Arch. 460, 353–359 [DOI] [PubMed] [Google Scholar]
- 28. Lipscombe D. (2005) Curr. Opin. Neurobiol. 15, 358–363 [DOI] [PubMed] [Google Scholar]
- 29. Boutz P. L., Chawla G., Stoilov P., Black D. L. (2007) Genes Dev. 21, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boutz P. L., Stoilov P., Li Q., Lin C. H., Chawla G., Ostrow K., Shiue L., Ares M., Jr., Black D. L. (2007) Genes Dev. 21, 1636–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fairbrother W. G., Fairbrother W., Lipscombe D. (2008) BioEssays 30, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T. (2007) Mol. Cell 27, 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amir-Ahmady B., Boutz P. L., Markovtsov V., Phillips M. L., Black D. L. (2005) RNA 11, 699–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu H., Zhang W., Reed R. B., Liu W., Grabowski P. J. (2002) RNA 8, 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oberstrass F. C., Auweter S. D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D. L., Allain F. H. (2005) Science 309, 2054–2057 [DOI] [PubMed] [Google Scholar]
- 36. Spellman R., Smith C. W. (2006) Trends Biochem. Sci. 31, 73–76 [DOI] [PubMed] [Google Scholar]
- 37. Modafferi E. F., Black D. L. (1997) Mol. Cell. Biol. 17, 6537–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Modafferi E. F., Black D. L. (1999) RNA 5, 687–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Markovtsov V., Nikolic J. M., Goldman J. A., Turck C. W., Chou M. Y., Black D. L. (2000) Mol. Cell. Biol. 20, 7463–7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sharma S., Falick A. M., Black D. L. (2005) Mol. Cell 19, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Black D. L. (1992) Cell 69, 795–807 [DOI] [PubMed] [Google Scholar]
- 42. Spellman R., Llorian M., Smith C. W. (2007) Mol. Cell 27, 420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Q., Lee J. A., Black D. L. (2007) Nat. Rev. Neurosci. 8, 819–831 [DOI] [PubMed] [Google Scholar]
- 44. Modrek B., Lee C. J. (2003) Nat. Genet. 34, 177–180 [DOI] [PubMed] [Google Scholar]
- 45. Sorek R., Ast G. (2003) Genome Res. 13, 1631–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sugnet C. W., Srinivasan K., Clark T. A., O'Brien G., Cline M. S., Wang H., Williams A., Kulp D., Blume J. E., Haussler D., Ares M., Jr. (2006) PLoS Comput. Biol. 2, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yeo G. W., Van Nostrand E., Holste D., Poggio T., Burge C. B. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2850–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tang Z. Z., Zheng S., Nikolic J., Black D. L. (2009) Mol. Cell. Biol. 29, 4757–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith C. W. (2005) Cell 123, 1–3 [DOI] [PubMed] [Google Scholar]
- 50. Letunic I., Copley R. R., Bork P. (2002) Hum. Mol. Genet. 11, 1561–1567 [DOI] [PubMed] [Google Scholar]
- 51. Smith C. W., Nadal-Ginard B. (1989) Cell 56, 749–758 [DOI] [PubMed] [Google Scholar]
- 52. Carstens R. P., Wagner E. J., Garcia-Blanco M. A. (2000) Mol. Cell. Biol. 20, 7388–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Del Gatto F., Breathnach R. (1995) Mol. Cell. Biol. 15, 4825–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jones R. B., Wang F., Luo Y., Yu C., Jin C., Suzuki T., Kan M., McKeehan W. L. (2001) J. Biol. Chem. 276, 4158–4167 [DOI] [PubMed] [Google Scholar]
- 55. Lejeune F., Maquat L. E. (2005) Curr. Opin. Cell Biol. 17, 309–315 [DOI] [PubMed] [Google Scholar]
- 56. Cooper T. A., Wan L., Dreyfuss G. (2009) Cell 136, 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rubin B. Y., Anderson S. L. (2008) Neuromol. Med. 10, 148–156 [DOI] [PubMed] [Google Scholar]
- 58. Tazi J., Bakkour N., Stamm S. (2009) Biochim. Biophys. Acta 1792, 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang G. S., Cooper T. A. (2007) Nat. Rev. Genet. 8, 749–761 [DOI] [PubMed] [Google Scholar]
- 60. Aartsma-Rus A., Janson A. A., Heemskerk J. A., De Winter C. L., Van Ommen G. J., Van Deutekom J. C. (2006) Ann. N.Y. Acad. Sci. 1082, 74–76 [DOI] [PubMed] [Google Scholar]
- 61. Jearawiriyapaisarn N., Moulton H. M., Buckley B., Roberts J., Sazani P., Fucharoen S., Iversen P. L., Kole R. (2008) Mol. Ther. 16, 1624–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van Deutekom J. C., Janson A. A., Ginjaar I. B., Frankhuizen W. S., Aartsma-Rus A., Bremmer-Bout M., den Dunnen J. T., Koop K., van der Kooi A. J., Goemans N. M., de Kimpe S. J., Ekhart P. F., Venneker E. H., Platenburg G. J., Verschuuren J. J., van Ommen G. J. (2007) N. Engl. J. Med. 357, 2677–2686 [DOI] [PubMed] [Google Scholar]
- 63. Wood M. J., Gait M. J., Yin H. (2010) Brain 133, 957–972 [DOI] [PubMed] [Google Scholar]
- 64. Hua Y., Vickers T. A., Baker B. F., Bennett C. F., Krainer A. R. (2007) PLoS Biol. 5, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Khoo B., Krainer A. R. (2009) Curr. Opin. Mol. Ther. 11, 108–115 [PMC free article] [PubMed] [Google Scholar]
- 66. Singh N. N., Shishimorova M., Cao L. C., Gangwani L., Singh R. N. (2009) RNA Biol. 6, 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Williams J. H., Schray R. C., Patterson C. A., Ayitey S. O., Tallent M. K., Lutz G. J. (2009) J. Neurosci. 29, 7633–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barrett C. F., Tsien R. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2157–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thiel W. H., Chen B., Hund T. J., Koval O. M., Purohit A., Song L. S., Mohler P. J., Anderson M. E. (2008) Circulation 118, 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liao P., Zhang H. Y., Soong T. W. (2009) Pflugers Arch. 458, 481–487 [DOI] [PubMed] [Google Scholar]
- 71. Aartsma-Rus A., van Vliet L., Hirschi M., Janson A. A., Heemskerk H., de Winter C. L., de Kimpe S., van Deutekom J. C., 't Hoen P. A., van Ommen G. J. (2009) Mol. Ther. 17, 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Svasti S., Suwanmanee T., Fucharoen S., Moulton H. M., Nelson M. H., Maeda N., Smithies O., Kole R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wheeler T. M., Lueck J. D., Swanson M. S., Dirksen R. T., Thornton C. A. (2007) J. Clin. Invest. 117, 3952–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]