Abstract
Although alternative DNA secondary structures (non-B DNA) can induce genomic rearrangements, their associated mutational spectra remain largely unknown. The helicase activity of WRN, which is absent in the human progeroid Werner syndrome, is thought to counteract this genomic instability. We determined non-B DNA-induced mutation frequencies and spectra in human U2OS osteosarcoma cells and assessed the role of WRN in isogenic knockdown (WRN-KD) cells using a supF gene mutation reporter system flanked by triplex- or Z-DNA-forming sequences. Although both non-B DNA and WRN-KD served to increase the mutation frequency, the increase afforded by WRN-KD was independent of DNA structure despite the fact that purified WRN helicase was found to resolve these structures in vitro. In U2OS cells, ∼70% of mutations comprised single-base substitutions, mostly at G·C base-pairs, with the remaining ∼30% being microdeletions. The number of mutations at G·C base-pairs in the context of NGNN/NNCN sequences correlated well with predicted free energies of base stacking and ionization potentials, suggesting a possible origin via oxidation reactions involving electron loss and subsequent electron transfer (hole migration) between neighboring bases. A set of ∼40,000 somatic mutations at G·C base pairs identified in a lung cancer genome exhibited similar correlations, implying that hole migration may also be involved. We conclude that alternative DNA conformations, WRN deficiency and lung tumorigenesis may all serve to increase the mutation rate by promoting, through diverse pathways, oxidation reactions that perturb the electron orbitals of neighboring bases. It follows that such “hole migration” is likely to play a much more widespread role in mutagenesis than previously anticipated.
Keywords: DNA Helicase, DNA Structure, Gene Silencing, Mutagenesis Mechanisms, Oxidative Stress, Charge Transfer, DNA Secondary Structure, Mutation Frequency, U2OS WRN Knockdown, WRN Helicase Activity
Introduction
The application of high resolution array comparative genomic hybridization techniques has revealed the frequent occurrence of DNA sequence motifs capable of forming alternative (non-B) DNA conformations (e.g. triplex, quadruplex, Z-DNA, cruciforms, slipped structures) at the breakpoint junctions of chromosomal alterations (gross deletions and duplications) associated with human genetic disease, including cystic fibrosis, mental retardation, and multiple congenital anomalies (1). These observations have served to extend the generality of previous work that aimed to elucidate the molecular mechanisms underlying recurrent translocations (2–6) and the genetic instability observed in many model systems (7–16), both of which were suggestive of a direct mutagenic role for non-B DNA. In the same vein, analyses of DNA sequence motifs flanking human gross deletion breakpoints (9), genomic inversions that distinguish the human from the chimpanzee genome (17), and DNA sequence tracts involved in pathological gene conversion events (18) have provided evidence for a wide ranging role for DNA secondary structure in promoting gross genomic rearrangements. Despite these recent advances, few studies (8, 11) have attempted to address systematically the extent of the influence of non-B DNA-forming sequences in modulating mutational spectra. It therefore remains unclear whether other types of mutation, such as single base substitutions, might also be induced by the presence of non-B DNA.
Considerable work in vitro has documented the ability of helicases, such as BLM, WRN, DHX9, and FANCJ, to unwind non-B DNA conformations (19–23), consistent with their postulated role in maintaining genome integrity (reviewed in Ref. 24) (25, 26). The heritable deficiency of WRN, Werner syndrome (WS),2 is a rare recessive disorder characterized by the early onset of an aged appearance and age-related disorders including bilateral cataracts, skin changes, short stature, graying and loss of hair, osteoporosis, diabetes, and premature death (21, 27, 28). WS patients invariably carry inactivating mutations in the WRN gene encoding WRN (27, 29, 30), an evolutionarily conserved member of the RecQ helicase family that possesses both 3′ → 5′ helicase and exonuclease activities (31).
Fibroblast cultures from WS patients display a variety of chromosomal abnormalities, including reciprocal translocations, deletions, and inversions (reviewed in Ref. 31), elevated single base pair mutation frequencies (32–34), and modified bases (reviewed in Ref. 35). Fibroblasts from both WS patients and siRNA-mediated WRN knockdown exhibit a marked reduction in proliferation in culture, which is exacerbated by DNA damaging agents (36). An increase in DNA damage response nuclear foci and other defects thought to be associated with the resolution of either arrested DNA replication forks (36, 37) or mitotic recombination intermediates (38) are also evident in the absence of WRN.
WRN deficiency has also been shown to be associated with a profound reprogramming of genome-wide gene expression profiles (39–42) and increased intracellular levels of reactive oxygen species, suggesting that cellular senescence may result from the pleiotropic effect of several genes in association with redox imbalance (43–46).
To explore the complex interrelationship between non-B DNA structures, WRN deficiency, and mutagenesis, we used a plasmid system containing triplex- and Z-DNA-forming sequences flanking a reporter gene in transfection experiments performed in WT and WRN knockdown (WRN-KD) U2OS human osteosarcoma cell lines. Our results revealed that both non-B DNA and WRN deficiency serve to induce mutations. Further, mutational spectra were characterized by single base changes, mostly at G·C pairs. These mutations were found to be strongly dependent upon base stacking with neighboring bases, suggesting a role for oxidative damage through electron loss to the local environment leading to redistribution of the outer electron cloud between adjacent DNA bases (hole migration). Further analysis of a large number of predominantly G·C mutations from a lung cancer genome confirmed the dependence of such lesions on sequence context as would be predicted by a hole migration model. Hence, electron loss and redistribution within the DNA molecule appear to underlie mutations arising from such diverse sources as alternative DNA conformations, WRN deficiency, and the process of tumorigenesis.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids pCEX, pMEXy, pMEXr, pGG32y, and pSCG14 are derivatives of plasmid pSP189 in which inserts (Fig. 1A) were cloned between the EcoRI and XhoI restriction sites. Triplex-forming plasmids (Fig. 1B) formed concatenated multimer species during bacterial cell growth. Because plasmid dimerization limits the detection of single base substitutions and could alter deletion/rearrangement frequencies by providing a substrate for homologous recombination, we first determined the most suitable Escherichia coli genetic background to obtain monomer plasmids. Of the DH5α, HB101, DL795, DL733, and DL1649 strains tested, the highest yields of monomer DNA were obtained in CaCl2-competent E. coli strain HB101. The fraction of monomer DNA was additionally increased by transforming closed circular, relaxed, monomer plasmid prepared by linearization with PvuI followed by ligation with T4 DNA ligase. The ratios of monomer to concatenated multimer species ranged from >95% for pCEX to ∼70% for pGG32y as assessed by agarose gel electrophoresis (data not shown). Structural transitions from B-DNA to non-B DNA require negative supercoiling. Therefore, to separate closed circular DNA from functionally ineffective open circular DNA, large scale plasmid DNA preparations were purified by equilibrium centrifugation in CsCl-ethidium bromide continuous gradients (47) on a Beckman Optima LE-80K ultracentrifuge and resequenced.
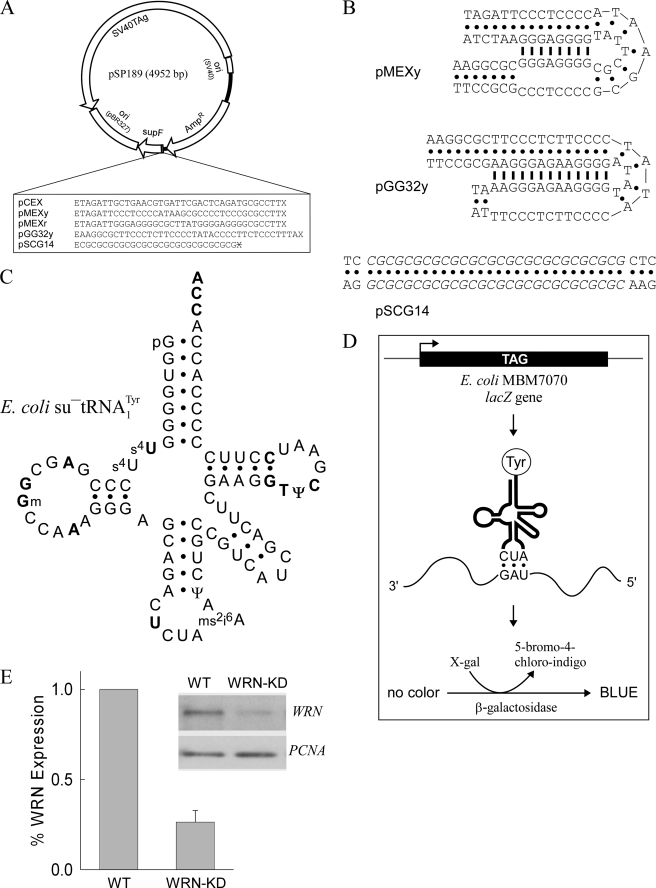
FIGURE 1.
Overview of the experimental strategy. A, map of the parental pSP189 shuttle vector. The DNA sequences cloned upstream of the supF gene between the EcoRI (E) and XhoI (X) restriction sites are shown. X with a line through it denotes an inactivated XhoI site introduced following cloning. B, representative triplex and Z-DNA structures formed by the cloned inserts in pMEXy, pGG32y, and pSCG14. Only one of four possible isomers is shown, viz. the 3′-YRR conformer, in which the 3′ purine-rich strand of the purine·pyrimidine tract with mirror repeat symmetry engages in the triplex structure. The potential triplex structures formed by pMEXr are the same as for pMEXy. Bullets, Watson-Crick base-pairs; thick lines, Hoogsteen base-pairs; italics, Z-DNA region. C, schematic of the cloverleaf structure and sequence composition, including commonly modified bases, of the su−tRNATyr, the product of the supF gene. Bold type, invariably conserved residues in tRNAs; p, phosphate group at the 5′ end. D, scheme of the chromosomal lacZ gene in the indicator E. coli strain MBM7070, showing the presence (not to scale) of the relevant TAG amber stop codon (top). Upon transformation of MBM7070 cells with the supF-containing plasmids (A), initiation of lacZ gene transcription (arrow), activated by exogenous IPTG, yields fully translated mRNA products in which amber stop codons are read by su−tRNATyr molecules (middle). The encoded β-galactosidase (bottom) hydrolyzes the culture media supplemented with 5-bromo-4-chloro-indoyl-β-galactoside (X-gal), which upon condensation yields 5-bromo-4-chloro-indigo, conferring a blue color upon bacterial colonies. E, representative Western blot and quantitation of the WRN and PCNA proteins expressed in U2OS whole cell extracts. WT, cells harboring the integrated pshSCR plasmid, containing a scrambled shRNA insert; WRN-KD, WRN knockdown cells harboring the integrated pshWRN plasmid containing a WRN-targeting shRNA insert. The graph shows the fraction of WRN protein expressed in WRN-KD relative to WT cells normalized to the amount of PCNA (means ± S.D. from four independent experiments).
Cell Types and Relevant Genotypes
E. coli strains DH5α (supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 gyrA96 thi-1 relA1) and HB101 (F− supE44 hsdsB20(rB−mB−) recA13 ara-14 leuB6 Δ(gpt-proA)62 Δ(mcrC-mrr) thi-1 lacY1 gaIK2 rpsL20 xyl1-5 mtl-1 λ−) were obtained from the ATCC (Manassas, VA). The E. coli K12 SH28-derivative strains DL795 (deoA Δ(mrr hsdRMS mcrBC)2 mcrA (e14°) sbcC201 supE recA:CmR), DL733 (JM83 sbcC202 phoR:Tn10), and DL1649 (MG1655 ΔlacZ (ΔsbcDC)PsbcDC-lacZaph) were a kind gift from David R. Leach (University of Edinburgh, Edinburgh, UK). Control and WRN knockdown gene expression were obtained by stable transfection of the human U2OS osteosarcoma cell line with pSilencer 3.1-H1 hygro (AM5766) in which either a scrambled shRNA sequence (WT) with no homology with known genes or the WRN-targeting shRNA (WRN-KD) insert GGATCCCGTGAAGAGCAAGTTACTTGCTTCAAGAGAGCAAGTAACTTGCTCTTCATTTTTTGGAAAAGCTT was cloned between the BamHI and HindIII restriction sites (48) to yield pshSCR and pshWRN, respectively.
Mutation-Reporter Plasmid Transfection in WT and WRN-KD Cells
WT and WRN-KD U2OS human osteosarcoma cells were maintained in h-DMEM (DMEM containing glucose/glutamine/sodium pyruvate supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA), 1× streptomycin/penicillin (Invitrogen) solution, and 200 μg/ml hygromycin B (Calbiochem, La Jolla CA). For transfections, ∼80% confluent cells were treated with 1× trypsin (Invitrogen), and ∼1.2 million cells were mixed with 110 μl of premixed cell line nucleofector kit V (Lonza), 1.8 μl of plasmid DNA at 2 μg/μl and electroporated. Transfected cells were allowed to recover for 24 h in McCoy's 5A medium (Invitrogen) supplemented with 1× l-glutamine and 10% fetal bovine serum and then grown for an additional 48-h period in h-DMEM. Plasmid DNA was isolated using QIAprep spin miniprep kits (Qiagen); cleaved with DpnI, AatII, and Tth111I (to remove plasmids that did not replicate in mammalian cells as well as any episomal pshWRN that was occasionally released upon transfection) in 100-μl reaction volumes; extracted with an equal volume of phenol/chloroform containing 0.1% hydroxyquinoline and 0.2% β-mercaptoethanol; ethanol-precipitated in 300 mm sodium acetate (pH 5.2), 70% ethanol, and 50 μg/ml glycogen for 16 h at −20 °C, and dissolved in 10 μl of H2O.
Mutation Screening
To determine supF reporter-gene mutation frequencies and spectra, plasmids were extracted from WT and WRN-KD cells and transformed in the indicator E. coli strain MBM7070. The supF gene is transcribed in bacteria into su−tRNATyr (49) (Fig. 1C), which enables E. coli strain MBM7070 to read through amber stop codons in the chromosomal lacZ gene (Fig. 1D); hence, supF mutants are found as white colonies among blue wild-type colonies on X-gal/IPTG/ampicillin-containing agar plates. For screening, 50 and 100 μl (of 1 ml) of transfected E. coli strain MBM7070 cells (47) were plated on X-gal/IPTG/ampicillin-containing plates and grown for ∼16 h at 37 °C. When replating was required to optimize the number of colonies on the plates, E. coli solutions, which were kept at 4 °C overnight, were prewarmed to 37 °C. This heating step was necessary to overcome extensive cell death observed with pGG32y-containing cells, a feature of E. coli harboring triplex-containing plasmids (7). All of the white colonies were picked and streaked on X-gal/IPTG/ampicillin-containing plates to confirm their phenotype. Mutation frequencies were determined by dividing the number of white (mutant) colonies by the total number of colonies (blue + white). The experiments were repeated 3–6 times.
Plasmid DNA was isolated from randomly selected white colonies and supF mutations were determined by direct DNA sequencing using primers 189 (5′-CAAAAAAGGGAATAAGGGCG-3′) and 201 (5′-CGTTTCTGGGTGAGCAAAA-3′), which hybridize within the ampicillin resistance gene. The mutations induced by non-B DNA conformations were assessed by comparing the mutation frequencies and spectra obtained with sequences cloned upstream of the supF gene (Fig. 1A), which included: 1) a control sequence (pCEX) unable to form any known non-B DNA structure; 2) two sequences from the human c-MYC promoter (pMEXy and pMEXr) capable of adopting metastable triplex structures (Fig. 1B); and 3) two sequences (pGG32y and pSCG14) predicted to fold into stable triplex and Z-DNA conformations (Fig. 1B), respectively.
Helicase Assays in Vitro
For plasmid DNA, 1 μg of plasmid DNA was incubated with 60 nm purified WRN protein (50) at 35 °C for 30 min. Fifty units of mung bean nuclease were added for 5 min, and the reactions were stopped by chilling on ice. The products were separated by 0.8% agarose gel electrophoresis, and the percentage of unwinding by WRN was determined from the total amount of supercoiled plasmid remaining after the reactions. For synthetic triplex DNA, PAGE-purified 41R 5′-AAACAACACTGGGGGAGGGGGACGGTGAAGGCCAAGTTCCC-3′, 41Y 5′-GGGAACTTGGCCTTCACCGTCCCCCTCCCCCAGTGTTGTTT-3′, and 31R (3′-over-hang) 5′-GGAAGCGGTAGGGGGAGGGGGCAGTCGAGCG-3′(triplex region underlined) oligonucleotides were 5′-end-labeled and annealed, and 5 nm labeled triplex DNA was incubated with purified WRN (19). The reaction products were resolved by 12% PAGE and quantified (Molecular Dynamics, Sunnyvale, CA).
8-OxodG Analysis
The concentration of 8-oxodG (8-oxodG/dG) was determined by HPLC (51) on genomic DNA isolated from wild-type and WRN-KD U2OS cells using the QIAamp DNA mini kit (Qiagen). DNA was digested and dephosphorylated by nuclease P1 and alkaline phosphatase and analyzed by HPLC using a reverse phase analytical column coupled with a photodiode array detector (SPD-M10A; Shimadzu, Columbia, MD) followed by electrochemical detection (CoulArray; ESA, Chelmsford, MA).
Databases and Statistical Analyses
A data set of 54,422 germline missense and nonsense mutations was retrieved from the Human Gene Mutation Database (52). These mutations, as well as those mutations identified in the WT and WRN-KD cells, were recategorized according to their occurrence in the 10 possible alternative dinucleotide pairs. To allow the spectrum of mutations found in the WT and WRN-KD cells to be compared directly with that of the inherited disease mutations recorded in the Human Gene Mutation Database, each type of mutation at a particular position was counted only once. A Chi square test was then performed to assess the significance of the disproportionate occurrence of certain types of mutation found in the WT and WRN-KD cells within a specific dinucleotide pair as compared with those mutations recorded in the Human Gene Mutation Database. All of the results were corrected for multiple testing.
Approximately 50,000 somatic single nucleotide substitutions identified in a non-small cell lung cancer genome (53) were used to determine the number of mutations that occurred at G·C base-pairs (39,615 of 50,675 or 78%) in the context of 64 possible NGNN and their complementary NNCN sequences (using in-house Perl scripts) during the process of tumorigenesis. Mutations were mapped onto the reference human genome assembly hg19; the numbers of each tetranucleotide sequence (i.e. AGAA, TGAC, etc.) mutated in the lung cancer genome were divided by the number of tetranucleotide sequences present either within 1 kb of each mutation site (Fkb) or genome-wide (Fgw).
A one-way analysis of variance Tukey test was used to determine the statistical significance of the mutation frequencies. Differences in the numbers of transversions and transitions generated were assessed by means of McNemar's test between two correlated proportions.
RESULTS
The ability of repetitive DNA sequences to adopt non-B DNA conformations and to induce genomic instability is well documented (2–6, 8–11, 17, 18). However, few studies have reported on the mutational spectra induced by such elements (8, 10, 11). In addition, the potential role of those proteins whose function is to preserve genome stability, such as WRN, in counteracting non-B DNA-induced mutagenesis has not yet been explored in vivo. To this end, we measured mutation frequencies and spectra in a plasmid reporter system (Fig. 1A) in which the supF gene was preceded by sequences capable of forming either triplex structures of increasing thermal stability or a Z-DNA conformation (Fig. 1B). These plasmids were transfected into human osteosarcoma U2OS cells containing an integrated plasmid encoding either a control shRNA (WT) or a targeting shRNA designed to stably knockdown WRN (WRN-KD) (48). Under standard cell culture conditions, the average level of WRN protein in WRN-KD cells relative to its WT counterpart was found to be 0.26 ± 0.06 (mean ± S.D.), as determined by immunoblot analysis (Fig. 1E and supplemental text).
Non-B DNA-forming Sequences and WRN-KD Both Increase Mutation Frequencies
We postulated that if one function of the WRN helicase were to resolve triplex and Z-DNA structures, as observed in vitro (8, 11, 23), then mutation frequencies might increase in WRN-KD cells as compared with WT cells because both the number and stability of such structures would be significantly greater in the former cell type. We verified by native PAGE that purified WRN protein (50) was able to unwind the third, purine-rich, strand of a synthetic triplex in vitro and to partially protect from mung bean nuclease cleavage the Z-DNA-forming repeat in supercoiled pSCG14 (data not shown).
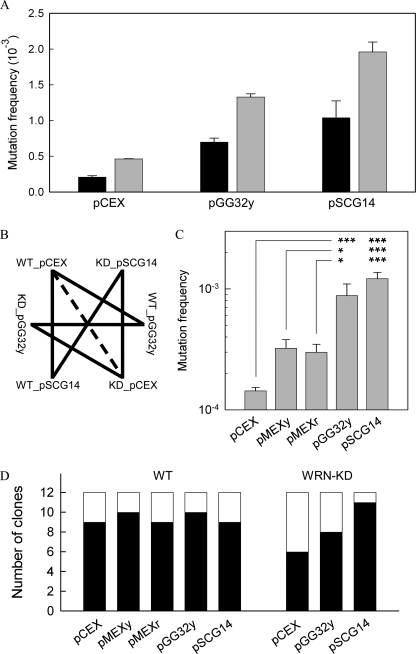
To determine the spontaneous and non-B DNA-induced mutation frequencies in WT and WRN-KD cells, 3–6 separate experiments were performed in both cell lines in parallel using plasmids pCEX, pGG32y, and pSCG14 (Fig. 1). In WT cells, from a total of 232,938 colonies, 129 were confirmed to be mutant (white), with relative mutation frequencies of 2.1 × 10−4 ± 2.0 × 10−5 (mean, S.E.) for pCEX, 7.0 × 10−4 ± 5.8 × 10−5 for pGG32y, and 1.0 × 10−3 ± 2.4 × 10−4 for pSCG14 (Fig. 2A). For the WRN-KD cells, a total of 144,832 colonies were counted, and of these, 190 were mutants, yielding mutation frequencies of 4.6 × 10−4 ± 5.8 × 10−6 for pCEX, 1.3 × 10−3 ± 4.5 × 10−5 for pGG32y, and 2.0 × 10−3 ± 1.4 × 10−4 for pSCG14 (Fig. 2A). Thus, the mutation frequencies generated in the WRN-KD cells were invariably ∼2-fold (1.9–2.2) greater than in WT cells, irrespective of the relative ability of the various plasmids to adopt non-B DNA conformations.
FIGURE 2.
Mutation frequencies and spectra in WT and WRN-KD cells. A, fractions of white (mutant) colonies to the total number of colonies for the indicated plasmids (x axis) isolated from WT cells (black) and WRN-KD cells (gray). B, statistical z-test pair-wise comparisons of the data from A. Solid lines, p < 0.001; dashed line, 0.01 < p < 0.05. C, as in A for WT cells; 374 mutant (white) colonies were obtained from a total of 915,386 colonies. The mutation frequencies were 1.4 × 10−4 ± 1.0 × 10−5 (mean, S.E.) for pCEX, 3.2 × 10−4 ± 5.8 × 10−5 for pMEXy, 3.0 × 10−4 ± 5.0 × 10−5 for pMEXr, 8.8 × 10−4 ± 2.2 × 10−4 for pGG32y, and 1.2 × 10−3 ± 1.6 × 10−4 for pSCG14; ***, p < 0.001; *, p = <0.001- 0.023; pair-wise comparisons between pGG32y (or pSCG14) and pCEX (top line), pMEXy (middle line), and pMEXr (bottom line) are shown. D, mutation spectra of selected mutant colonies. Black, single nucleotide changes in the supF gene; white, deletions disrupting the supF gene.
Pair-wise statistical (z-test) comparisons within the data set indicated that in both WT and WRN-KD cells, the non-B DNA-forming plasmid inserts increased the mutation frequencies over background levels (p < 0.001) (Fig. 2B). The increases in the number of mutant colonies observed in WRN-KD cells relative to WT cells for each of the plasmids were also significant (p = 9.8 × 10−5 to 1.5 × 10−2) (Fig. 2B). Additional experiments performed in WT cells, which also included the less stable triplex-forming plasmids pMEXr and pMEXy (Fig. 2C), further indicated that the mutation frequency was directly proportional to the stability of the non-B DNA structure (Fig. 1B) predicted to form in the respective plasmids.
Mutational Spectra in Human Cells
To characterize the mutational spectra, we sequenced the supF mutation-reporter gene in 12 clones, randomly chosen, for each of the plasmids studied. For the control pCEX plasmid in WT cells, nine clones (75%) revealed the presence of one or more base changes within the supF gene, whereas in the remaining three clones, the entire supF gene was deleted (Fig. 2D). The prevalence of single base mutations remained unchanged when the non-B DNA-forming plasmids were used. Indeed, the ratio of single base changes to deletions did not appear to be affected by the type of plasmid employed. In WRN-KD cells, single base substitutions were present in 6, 8, and 11 cases in pCEX, pGG32y, and pSCG14, respectively (Fig. 2D), and hence were more abundant than deletions, as found with WT cells. This conclusion was further supported by results obtained from 24 additional clones with pCEX, pMEXr, pMEXy, pSCG14, and pGG32y, 22 of which contained single base changes in the supF gene (not shown).
To determine the relative contribution of mutations arising in bacterial cells, pCEX, pGG32y, and pSCG14 were transformed directly into E. coli MBM7070 cells, which were plated on X-gal/IPTG/ampicillin-containing plates. From a total of 287,398 colonies in two separate experiments, 13 mutant colonies (4.5 × 10−5) were confirmed, without significant differences (z-test) in frequency between plasmids (3.8 × 10−5 for pCEX, 6.6 × 10−5 for pGG32y, and 3.3 × 10−5 for pSCG14). Thus, mutations in bacteria did not contribute significantly to the mutation frequencies observed in the human cells.
Plasmid DNA was also extracted from 26 mutant colonies (of 154,541 screened) derived from WT human cells transfected with pCEX and retransformed in fresh competent MBM7070 cells. Of these, six yielded blue (i.e. wild-type) colonies, indicating that mutations were present in the lac operon of the recipient bacterial cells, rather than in the plasmid supF gene, at a frequency of ∼3.9 × 10−5 (6/154,541).
In summary, the following conclusions could be drawn: 1) the mutational spectra of the supF gene in human cells comprised predominantly single base changes; 2) the presence of non-B DNA conformations or WRN-KD increased mutation frequencies but did not alter the underlying mutational spectra; 3) the presence of non-B DNA structures increased mutation frequencies to an extent that was dependent upon their relative thermal stability; and 4) most mutations (from 68% in pCEX to 96% in pSCG14) in the supF gene arose in the human cells. Thus, additivity (rather than synergy) with respect to mutation frequencies was evident between the presence of non-B DNA conformations and WRN helicase deficiency, allowing one to conclude that WRN helicase activity does not provide protection against non-B DNA-directed mutation.
GA·TC Dinucleotides Represent Mutation Hot Spots
Of the 120 clones sequenced (Fig. 3), 94 (78%) were found to harbor base changes within the supF gene (47 for WT cells and 47 for WRN-KD cells), whereas only 26 clones (22%) carried deletions that abolished supF gene function (supplemental text and supplemental Fig. S1). Of the 94 clones with base changes, 92 contained single base substitutions, and 1 clone lacked a nucleotide, whereas the other displayed a double base substitution (Fig. 3). Hence, no insertions were detected. Overall, 123 base changes occurred (63 in WT cells and 60 in WRN-KD cells), or 1.3 base changes per clone on average, with up to 5 base changes per clone. A survey of the nucleotides most commonly substituted revealed three hot spots at nucleotide positions 41, 67, and 76 (Fig. 3), which together accounted for more than a third (37%) of all the base substitutions. The second and third hot spots shared a DNA sequence motif (TTCGAA) that possesses dyadic symmetry, in which TTC is complementary to GAA. Similarly, the first hot spot occurred within the anticodon loop of the su−tRNATyr molecule (Fig. 1C), which also corresponds to a region of inverted repeat symmetry. Inspection of the nucleotides that underwent sequence changes suggested a bias toward G·C base pairs when followed by A·T pairs. Indeed, in 103 of 121 (85%) cases, G·C base pairs were changed within a GA·TC dinucleotide context with a significantly higher prevalence of the G → C (49%) type of transversion over the alternative G → A (28%, p = 5.2 × 10−12), G → T (23%, p = 9.2 × 10−14), or combined G → A + G → T (p = 3.7 × 10−8) types of mutation. On the other hand, no difference was observed in the frequency of transversions versus transitions when either the GA·TC (p = 0.257) dinucleotide or all sequences (p = 0.252) were analyzed. In summary, 1) G → C transversions within GA·TC dinucleotides were the preferred type of mutation, and 2) triplex or Z-DNA-forming sequences and WRN-KD did not alter the propensity for G → C transversions to occur within a GA·TC context.
FIGURE 3.
SupF-inactivating single base changes. Sequence composition of the supF gene (below) and base changes (above) found in all sequenced mutant plasmids isolated from WT and WRN-KD U2OS cells. Δ, nucleotide deleted; underlining, double-base substitution; double underlining, GA·TC dinucleotides.
The 8-OxodG Level Is Increased in WRN-KD Cells
To explore potential mechanisms underlying the observed base changes, we first assessed whether mutations at the GA·TC dinucleotide occurred more frequently than expected by chance alone. A comparison of the relative number of unique mutations (e.g. the 13 G → C transversions in dinucleotide GA at position 41 (Fig. 3) were counted as one mutation) within each type of dinucleotide sequence context between the supF gene (Fig. 3) and the Human Gene Mutation Database (52) indicated a very significant level of enrichment (p = 5.4 × 10−11) for mutations in the GA·TC dinucleotide in the supF gene (Table 1). Hence, a very specific mechanism must have led to the high prevalence of GA·TC mutations.
TABLE 1.
Number of unique mutations found in the supF gene and the number of disease-associated missense/nonsense mutations recorded in the HGMD (irrespective of the host gene) for each dinucleotide pair
The p values (shown on a logarithmic scale) indicate over-representation (−log10p > 0) or under-representation (−log10p < 0) of a given type of mutation in the WT and WRN-KD cells as compared with the corresponding mutation type recorded in the Human Gene Mutation Database (HGMD).
| Dinucleotide pair | Number of mutations |
− log10pa | |
|---|---|---|---|
| In HGMD | In supF | ||
| AA·TT | 6,396 | 0 | 1 |
| AC·GT | 11,501 | 4 | 1 |
| AG·CT | 9,684 | 12 | −1 |
| AT·AT | 5,130 | 1 | 0 |
| CA·TG | 16,826 | 15 | −1 |
| CC·GG | 20,188 | 21 | −1 |
| CG·CG | 14,165 | 20 | 0 |
| GA·TC | 13,155 | 38 | 10 |
| GC·GC | 9,159 | 3 | 0 |
| TA·TA | 2,636 | 0 | 0 |
| Total | 54,422 | 50 | |
a All probabilities were corrected so as to allow for multiple testing.
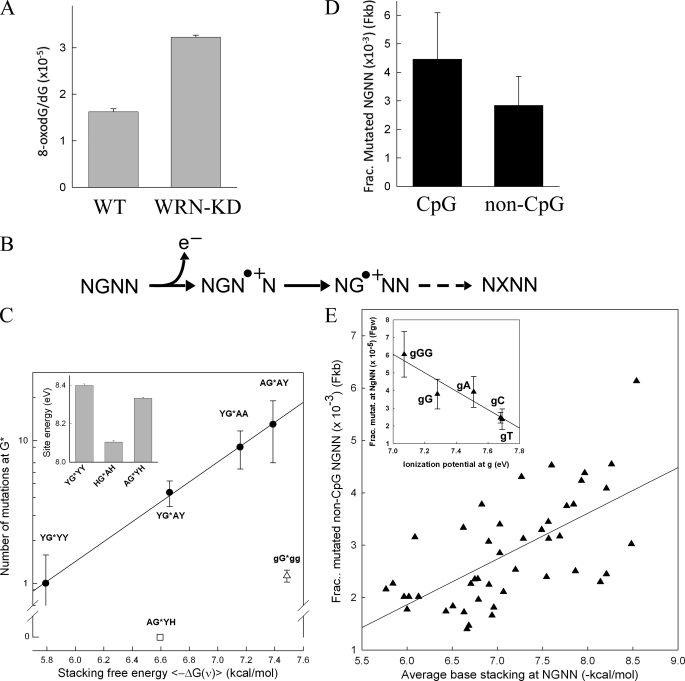
With an estimated frequency of ∼1/40,000 guanines, 8-oxodG is the most common oxidative lesion in vertebrate genomic DNA (54). To assess whether WRN deficiency led to an increase in the basal level of oxidative damage in U2OS cells, 8-oxodG levels were determined in genomic DNA. A significant (p = 1.4 × 10−5) 2-fold increase, from 1.6 ± 0.06 to 3.2 ± 0.04 × 10−5, was observed in the fraction of 8-oxodG in WRN-KD cells as compared with their wild-type counterparts (Fig. 4A). Hence, a 2-fold increase in the frequency of mutations in WRN-KD cells in plasmid DNA was paralleled by a 2-fold increase in oxidative damage in genomic DNA, supporting the view that WRN deficiency led to a hyperoxidative cellular environment, which in turn contributed to an elevation in the mutation rate.
FIGURE 4.
Oxidative damage and single base changes. A, concentration of 8-oxodG divided by the concentration of dG in genomic DNA of WT and WRN-KD cells (means ± S.D. from three DNA purifications). B, schematic diagram for hole migration showing the abstraction of an electron (e−) (step 1) by a radical species in the solvent (not shown) from a base downstream of the target G in an NGNN sequence, forming a radical cation (hole). The hole migrates to the upstream guanine (step 2) through the reorganization of the outer electron cloud involving base stacking between the interacting bases. A drop in the ionization potential traps the hole at the oxidized guanine, which through multiple steps (step 3) (including DNA replication and repair) eventually gives rise to a mutation. C, correlation between single base changes in the supF gene and base stacking at NGNN sequences. The y axis shows the number of mutations (mean ± S.E.) at G residues (from Fig. 3; C residues were also counted with 3′ → 5′ flanking sequences) in NGNN/NNCN sequences. The x axis shows the average free energy contribution [−ΔG(ν)] to nearest neighbor base stacking in single-stranded DNA for the NG, GN, and NN dinucleotides within NGNN (from Table 4 of Ref. 61) with ϵi = 2]; Y, T or C; H, T, C or A; G, a G residue at either the first, third, or fourth position within NGNN. Inset, site energy (eV) for the nucleobase G in 5′-NGN-3′ (from Table 2 of Ref. 55). HGAH = YGAY+YGAA+AGAY. D, the fractions of single nucleotide variants from a lung cancer genome (53) at each of the 64 possible NGNN/NNCN tetranucleotide sequences (compared with the reference human genome assembly hg19) to the number of NGNN/NNCN sequences found within 1 kb (Fkb) of each mutation site were calculated and averaged for the 19 CpG-containing sequences (4.46 ± 1.63 × 10−3) and the 45 non-CpG-containing sequence combinations (2.84 ± 1.02 × 10−3) (p = 0.00051). For the data normalized to the genome-wide number of NGNN/NNCN sequences (Fgw), the respective fractions were 5.23 ± 2.03 × 10−5 (CpG) and 3.36 ± 1.25 × 10−5 (non-CpG); p = 0.00032). E, the Fkb data (D) for the 45 non-CpG-containing sequences (y axis) were plotted against the average base stacking energies for the NGNN sequences (x axis), as determined from Ref. 61 (see C); r = 0.64; p < 0.001. A significant correlation was also obtained when the Fgw fractions were analyzed. Inset, the Fgw data for the combined NGGG/CCCN, NGGN/NCCN, NGAN/NTCN, NGCN/NGCN, and NGTN/NACN were plotted against the ionization potential values of the upstream G (Ref. 54). r = −0.94; p = 0.018.
A Role for Hole Migration
The one-electron oxidation type of reaction, in which a nucleobase loses an electron to a radical species in the solvent, is believed to represent an efficient mechanism of mutation at guanine residues because of their low ionization potential (54). In addition, the migration of a charge from a distal base toward G (X•+ + G → X + G•+) (hole migration) through π-stacking (55–59) has been proposed to contribute to mutagenesis (60). Hole migration behavior (55–59) would predict that the number of mutations at any given G in the context of NGNN should increase with increasing local base stacking and with a decrease in ionization potentials afforded by 3′ base stacking (54) (Fig. 4B). Hence, we determined the number of mutations at G (Fig. 3) in NGNN sequences (and their NNCN complements) with respect to known values of sequence-dependent base stacking (61) and ionization potentials (55). The number of mutations was found to increase in the order YGYY < YGAY < YGAA < AGAY (Fig. 4C) and was directly proportional to the average stacking energy within these tetranucleotide sequences. By contrast, no mutations were observed for the four AGYX (where X is C, T, or A) sequences despite favorable stacking, whereas a small number of mutations were detected when at least one other G (gGgg) was present adjacent to G. The estimated site energy (eV) at G for the YGYY and AGYH sequences, with an average of ≤1 mutations, was also higher than for the group of sequences (YGAY + YGAA + AGAY) with an average of more than four mutations (Fig. 4C, inset). Thus, despite a relatively small number of observations, these data were strongly suggestive of a role for hole migration in oxidation-induced mutations.
To further verify this conclusion, we analyzed ∼50,000 somatic single nucleotide variants reported in a lung cancer genome (53). In passing, we note that the lung cancer genome was determined from tumor cells taken from a heavy smoker; smoking is known to cause oxidative damage and has been claimed to induce cellular senescence through WRN down-regulation (46). Analysis of the 39,615 mutations (78%) that occurred at G·C base-pairs in the context of all 64 permutations of NGNN/NNCN sequences revealed that for the 19 types of tetranucleotide containing a CpG step, the fraction of mutated sequences (both Fkb and Fgw; see “Experimental Procedures”) were significantly overrepresented (Fig. 4D). Hence, deamination of 5-methylcytosine at methylated CpG dinucleotides was likely the mutational mechanism involved (62). For the remaining 45 types of non-CpG-containing sequences, a positive correlation was observed between the fractions (both Fkb and Fgw) of mutated sequences and their average base stacking (Fig. 4E). In addition, a significant correlation was evident between the fractions of mutated sequences and the ionization potential values at the target G as a function of its 3′ next neighbor(s) (inset). We therefore conclude that the mechanism of hole migration was common to mutations induced by both non-B DNA-containing plasmids and WRN deficiency in cultured cells, as well as in a lung cancer genome.
DISCUSSION
The central conclusion emerging from this study was the unanticipated role of charge transfer (hole migration) in targeting guanine residues for oxidation, leading to mutation, in contexts as diverse as the formation of non-B conformations by specific DNA sequences, partial WRN deficiency, and lung tumorigenesis. Although these three contexts are structurally and functionally very different, they all ultimately appear to converge on the abstraction of an electron from a nucleobase and the subsequent reorganization of electron orbitals by base-base interactions along the DNA chain. Thus, a purely physical property of DNA (hole migration) appears to play a critical role in mutagenesis in a wide variety of contexts.
In addition, we observed that in the human osteosarcoma U2OS cell line, DNA sequences with the capacity to adopt either triplex or Z-DNA structures served to increase spontaneous mutation frequencies in cis in a manner that was directly proportional to their predicted thermal stability. These non-B DNA-forming sequences increased mutation frequencies without altering the basic composition of the mutational spectra. Thus, sequences capable of forming non-B DNA increase the rate of mutagenesis such that all types of mutation are increased proportionately, microlesions as well as large deletions. To our knowledge, no other type of DNA sequence has so far been reported to be capable of inducing different types of mutational events, both small and large.
We have also shown that chronic deficiency of the WRN protein led to a ∼2-fold increase in mutation frequency (both single base substitutions and deletions) irrespective of the presence or absence of non-B DNA-forming sequences. Given the ability of WRN to resolve non-B DNA (Refs. 21, 23, and 63 and this study) conformations in vitro, a synergistic effect on mutation frequency might reasonably have been expected between WRN-KD and non-B DNA-forming sequences. Because no synergy was observed, a role for WRN in reducing mutation frequencies via a mechanism(s) that is dependent upon its cellular helicase activity appears unlikely. Moreover, the higher level of 8-oxodG in WRN-KD as compared with wild-type U2OS cells suggests that the observed 2-fold increase in mutations was probably caused by an overall hyperoxidative state in the WRN-deficient cells (43).
This conclusion concurs with the view that the senescent phenotype induced by WRN deficiency is associated with oxidative stress and consequent DNA damage (43, 44). Indeed, the increase in life-span of WS patients treated with pioglitazone, a potent PPARγ agonist that mediates the up-regulation of antioxidant enzymes, such as catalase and copper/zinc superoxide dismutase, has been interpreted in terms of a reduction in oxidative stress (28). In a different type of study (46), cigarette smoke, which contains numerous reactive oxygen species and cytotoxic by-products of oxidation reactions, has been found to induce DNA damage, WRN down-regulation, and cellular senescence in lung fibroblasts. Either WRN overexpression or treatment with the antioxidant N-acetylcysteine served to attenuate WRN down-regulation, senescence-associated-β-galactoside activity, and the senescent phenotype in human pulmonary fibroblasts in culture exposed to H2O2 or cigarette smoke extract (46). Hence, redox imbalance appears to play a significant role in WRN deficiency-associated senescence.
The G•+ radical cation is known to be converted to several species, including 8-oxodG, 2-amino-5-[(2-deoxy-β-d-erythro-pentofuranosyl)amino]-4H-imidazol-4-one (Iz), its hydrolysis product 2,2-diamino-4-[(2-deoxy-β-d-erythro-pentofuranosyl)amino]-5(2H)-oxazolone, and others (54). 8-oxodG and Iz have been shown to base pair with A and G, respectively, giving rise to T·A and C·G transversions (64–66). In our study, mutations at GA·TC dinucleotides yielded mostly transversions with the ranking: G·C → C·G > T·A. These base changes are consistent with Iz·G and 8-oxodG·A mispairing intermediates, respectively (54, 64–66). Iz has also been shown to arise from both direct deprotonation of G•+ in single-stranded DNA and from 8-oxodG through long range hole migration (65). Thus, the high G·C → C·G transition rates observed herein are consistent with hole migration activity.
8-oxo-purines have been noted to accumulate with age in normal individuals, as well as in cells from prematurely aged WS patients and WRN-KD cells in culture (reviewed in Ref. 35). Base adducts, such as 8-oxo-purines and Fapy lesions, associate stably with the WRN protein, and the subsequent complexes are believed to trigger/initiate base excision repair (24, 35, 39, 67). In the absence (or reduction) of WRN, base lesions may accumulate, thereby increasing the mutation frequency.
The increase in mutations in the supF gene by the upstream triplex and Z-DNA forming repeats is unambiguous. However, the underlying mutational mechanism remains to be determined. If pausing at the non-B DNA-forming motifs occurred in vivo, either during DNA replication or transcription (68–70), this might have increased exposure of the downstream supF sequences to the solvent and hence to oxidative damage. Consistent with this interpretation, the genome-wide increase in mutation rates observed in late-replicating regions are believed to arise from the accumulation of single-stranded DNA regions and their exposure to endogenous oxidative DNA damage (71).
Oxidative DNA damage has been proposed as a major source of mutation both in somatic and germline cells (72). Our data support this view and point to a critical role for the early steps of the reaction, including hole migration. Those genomic regions with the propensity to fold transiently into non-B DNA secondary structures may become hot spots for mutagenesis, particularly under conditions, such as WS and cancer, that are characterized by an elevated cellular basal oxidative state.
Acknowledgments
We thank Dr. Herng-Hsiang Lo (ICMB/CRED Protein and Metabolite Analysis Facility, University of Texas, College of Pharmacy, Austin, TX) for the measurement of 8-oxodG and other nucleosides. We thank Patricia L. Opresko for the U2OS cells, Luis Coletta for DNA sequencing, the members of the K. Vasquez and R. Wood labs for comments and suggestions, and Sarah Henninger for manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants CA093729 (to K. M. V.) and HHSN261200800001E (to A. B.). This work was also supported by National Institute of Environmental Health Sciences Center Grant P30ES007784, by the Intramural Program of the National Institute on Aging, National Institutes of Heath, and by the National Institutes of Health through M. D. Anderson Cancer Center Support Grant CA016672.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, and Fig. S1.
- WS
- Werner syndrome
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- IPTG
- isopropyl β-d-thiogalactopyranoside
- 8-oxodG
- 7,8-dihydro-8-oxo-2′-deoxyguanosine
- Iz
- 2-amino-5-[(2-deoxy-b-d-erythro-pentofuranosyl)amino]-4H-imidazol-4-one.
REFERENCES
- 1. Quemener S., Chen J. M., Chuzhanova N., Bénech C., Casals T., Macek M., Jr., Bienvenu T., McDevitt T., Farrell P. M., Loumi O., Messaoud T., Cuppens H., Cutting G. R., Stenson P. D., Giteau K., Audrézet M. P., Cooper D. N., Férec C. (2010) Hum. Mutat. 31, 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Inagaki H., Ohye T., Kogo H., Kato T., Bolor H., Taniguchi M., Shaikh T. H., Emanuel B. S., Kurahashi H. (2009) Genome Res. 19, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kato T., Inagaki H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B. S., Kurahashi H. (2006) Science 311, 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gotter A. L., Nimmakayalu M. A., Jalali G. R., Hacker A. M., Vorstman J., Conforto Duffy D., Medne L., Emanuel B. S. (2007) Genome Res. 17, 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gotter A. L., Shaikh T. H., Budarf M. L., Rhodes C. H., Emanuel B. S. (2004) Hum. Mol. Genet. 13, 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gajecka M., Gentles A. J., Tsai A., Chitayat D., Mackay K. L., Glotzbach C. D., Lieber M. R., Shaffer L. G. (2008) Genome. Res. 18, 1733–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bacolla A., Jaworski A., Connors T. D., Wells R. D. (2001) J. Biol. Chem. 276, 18597–18604 [DOI] [PubMed] [Google Scholar]
- 8. Wang G., Vasquez K. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13448–13453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bacolla A., Jaworski A., Larson J. E., Jakupciak J. P., Chuzhanova N., Abeysinghe S. S., O'Connell C. D., Cooper D. N., Wells R. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wojciechowska M., Napierala M., Larson J. E., Wells R. D. (2006) J. Biol. Chem. 281, 24531–24543 [DOI] [PubMed] [Google Scholar]
- 11. Wang G., Christensen L. A., Vasquez K. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2677–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wells R. D., Dere R., Hebert M. L., Napierala M., Son L. S. (2005) Nucleic Acids Res. 33, 3785–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bacolla A., Wells R. D. (2004) J. Biol. Chem. 279, 47411–47414 [DOI] [PubMed] [Google Scholar]
- 14. Wang G., Carbajal S., Vijg J., DiGiovanni J., Vasquez K. M. (2008) J. Natl. Cancer Inst. 100, 1815–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mirkin E. V., Mirkin S. M. (2007) Microbiol. Mol. Biol. Rev. 71, 13–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang G., Vasquez K. M. (2009) Mol. Carcinog. 48, 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kolb J., Chuzhanova N. A., Högel J., Vasquez K. M., Cooper D. N., Bacolla A., Kehrer-Sawatzki H. (2009) Chromosome Res. 17, 469–483 [DOI] [PubMed] [Google Scholar]
- 18. Chuzhanova N., Chen J. M., Bacolla A., Patrinos G. P., Férec C., Wells R. D., Cooper D. N. (2009) Hum. Mutat. 30, 1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jain A., Bacolla A., Chakraborty P., Grosse F., Vasquez K. M. (2010) Biochemistry 49, 6992–6999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. London T. B., Barber L. J., Mosedale G., Kelly G. P., Balasubramanian S., Hickson I. D., Boulton S. J., Hiom K. (2008) J. Biol. Chem. 283, 36132–36139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fry M., Loeb L. A. (1999) J. Biol. Chem. 274, 12797–12802 [DOI] [PubMed] [Google Scholar]
- 22. Popuri V., Bachrati C. Z., Muzzolini L., Mosedale G., Costantini S., Giacomini E., Hickson I. D., Vindigni A. (2008) J. Biol. Chem. 283, 17766–17776 [DOI] [PubMed] [Google Scholar]
- 23. Brosh R. M., Jr., Majumdar A., Desai S., Hickson I. D., Bohr V. A., Seidman M. M. (2001) J. Biol. Chem. 276, 3024–3030 [DOI] [PubMed] [Google Scholar]
- 24. Bohr V. A. (2008) Trends Biochem. Sci. 33, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chakraborty P., Grosse F. (2010) Nucleic Acids Res. 38, 4722–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Compton S. A., Tolun G., Kamath-Loeb A. S., Loeb L. A., Griffith J. D. (2008) J. Biol. Chem. 283, 24478–24483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muftuoglu M., Oshima J., von Kobbe C., Cheng W. H., Leistritz D. F., Bohr V. A. (2008) Hum. Genet. 124, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yokote K., Saito Y. (2008) J. Am. Geriatr. Soc. 56, 1770–1771 [DOI] [PubMed] [Google Scholar]
- 29. Huang S., Lee L., Hanson N. B., Lenaerts C., Hoehn H., Poot M., Rubin C. D., Chen D. F., Yang C. C., Juch H., Dorn T., Spiegel R., Oral E. A., Abid M., Battisti C., Lucci-Cordisco E., Neri G., Steed E. H., Kidd A., Isley W., Showalter D., Vittone J. L., Konstantinow A., Ring J., Meyer P., Wenger S. L., von Herbay A., Wollina U., Schuelke M., Huizenga C. R., Leistritz D. F., Martin G. M., Mian I. S., Oshima J. (2006) Hum. Mutat. 27, 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Friedrich K., Lee L., Leistritz D. F., Nürnberg G., Saha B., Hisama F. M., Eyman D. K., Lessel D., Nürnberg P., Li C., Garcia-F-Villalta M. J., Kets C. M., Schmidtke J., Cruz V. T., Van den Akker P. C., Boak J., Peter D., Compoginis G., Cefle K., Ozturk S., López N., Wessel T., Poot M., Ippel P. F., Groff-Kellermann B., Hoehn H., Martin G. M., Kubisch C., Oshima J. (2010) Hum. Genet. 128, 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ozgenc A., Loeb L. A. (2005) Mutat. Res. 577, 237–251 [DOI] [PubMed] [Google Scholar]
- 32. Fukuchi K., Martin G. M., Monnat R. J., Jr. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5893–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kyoizumi S., Kusunoki Y., Seyama T., Hatamochi A., Goto M. (1998) Hum. Genet. 103, 405–410 [DOI] [PubMed] [Google Scholar]
- 34. Moser M. J., Bigbee W. L., Grant S. G., Emond M. J., Langlois R. G., Jensen R. H., Oshima J., Monnat R. J., Jr. (2000) Cancer Res. 60, 2492–2496 [PubMed] [Google Scholar]
- 35. Das A., Boldogh I., Lee J. W., Harrigan J. A., Hegde M. L., Piotrowski J., de Souza Pinto N., Ramos W., Greenberg M. M., Hazra T. K., Mitra S., Bohr V. A. (2007) J. Biol. Chem. 282, 26591–26602 [DOI] [PubMed] [Google Scholar]
- 36. Dhillon K. K., Sidorova J., Saintigny Y., Poot M., Gollahon K., Rabinovitch P. S., Monnat R. J., Jr. (2007) Aging Cell 6, 53–61 [DOI] [PubMed] [Google Scholar]
- 37. Sidorova J. M. (2008) DNA Repair 7, 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prince P. R., Emond M. J., Monnat R. J., Jr. (2001) Genes Dev. 15, 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deschênes F., Massip L., Garand C., Lebel M. (2005) Hum. Mol. Genet. 14, 3293–3308 [DOI] [PubMed] [Google Scholar]
- 40. Turaga R. V., Paquet E. R., Sild M., Vignard J., Garand C., Johnson F. B., Masson J. Y., Lebel M. (2009) Cell Cycle 8, 2080–2092 [DOI] [PubMed] [Google Scholar]
- 41. Johnson J. E., Cao K., Ryvkin P., Wang L. S., Johnson F. B. (2010) Nucleic Acids Res. 38, 1114–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kyng K. J., May A., Kølvraa S., Bohr V. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12259–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Massip L., Garand C., Paquet E. R., Cogger V. C., O'Reilly J. N., Tworek L., Hatherell A., Taylor C. G., Thorin E., Zahradka P., Le Couteur D. G., Lebel M. (2010) FASEB J. 24, 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Szekely A. M., Bleichert F., Nümann A., Van Komen S., Manasanch E., Ben Nasr A., Canaan A., Weissman S. M. (2005) Mol. Cell. Biol. 25, 10492–10506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ungvari Z., Krasnikov B. F., Csiszar A., Labinskyy N., Mukhopadhyay P., Pacher P., Cooper A. J., Podlutskaya N., Austad S. N., Podlutsky A. (2008) Age 30, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nyunoya T., Monick M. M., Klingelhutz A. L., Glaser H., Cagley J. R., Brown C. O., Matsumoto E., Aykin-Burns N., Spitz D. R., Oshima J., Hunninghake G. W. (2009) Am. J. Respir. Crit. Care Med. 179, 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 48. Harrigan J. A., Wilson D. M., 3rd, Prasad R., Opresko P. L., Beck G., May A., Wilson S. H., Bohr V. A. (2006) Nucleic Acids Res. 34, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kraemer K. H., Seidman M. M. (1989) Mutat. Res. 220, 61–72 [DOI] [PubMed] [Google Scholar]
- 50. Brosh R. M., Jr., Opresko P. L., Bohr V. A. (2006) Methods Enzymol. 409, 52–85 [DOI] [PubMed] [Google Scholar]
- 51. Lau S. S., Peters M. M., Kleiner H. E., Canales P. L., Monks T. J. (1996) Adv. Exp. Med. Biol. 387, 267–273 [DOI] [PubMed] [Google Scholar]
- 52. Stenson P. D., Mort M., Ball E. V., Howells K., Phillips A. D., Thomas N. S., Cooper D. N. (2009) Genome Med. 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee W., Jiang Z., Liu J., Haverty P. M., Guan Y., Stinson J., Yue P., Zhang Y., Pant K. P., Bhatt D., Ha C., Johnson S., Kennemer M. I., Mohan S., Nazarenko I., Watanabe C., Sparks A. B., Shames D. S., Gentleman R., de Sauvage F. J., Stern H., Pandita A., Ballinger D. G., Drmanac R., Modrusan Z., Seshagiri S., Zhang Z. (2010) Nature 465, 473–477 [DOI] [PubMed] [Google Scholar]
- 54. Burrows C. J., Muller J. G. (1998) Chem. Rev. 98, 1109–1152 [DOI] [PubMed] [Google Scholar]
- 55. Senthilkumar K., Grozema F. C., Guerra C. F., Bickelhaupt F. M., Lewis F. D., Berlin Y. A., Ratner M. A., Siebbeles L. D. (2005) J. Am. Chem. Soc. 127, 14894–14903 [DOI] [PubMed] [Google Scholar]
- 56. Lewis F. D., Daublain P., Cohen B., Vura-Weis J., Wasielewski M. R. (2008) Angew. Chemie-Int. Ed. 47, 3798–3800 [DOI] [PubMed] [Google Scholar]
- 57. Candeias L. P., Steenken S. (1993) J. Am. Chem. Soc. 115, 2437–2440 [Google Scholar]
- 58. Shao F., Augustyn K., Barton J. K. (2005) J. Am. Chem. Soc. 127, 17445–17452 [DOI] [PubMed] [Google Scholar]
- 59. Sugiyama H., Saito I. (1996) J. Am. Chem. Soc. 118, 7063–7068 [Google Scholar]
- 60. Bergeron F., Auvré F., Radicella J. P., Ravanat J. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 5528–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Friedman R. A., Honig B. (1995) Biophys. J. 69, 1528–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cooper D. N., Mort M., Stenson P. D., Ball E. V., Chuzhanova N. A. (2010) Hum. Genomics 4, 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Constantinou A., Tarsounas M., Karow J. K., Brosh R. M., Bohr V. A., Hickson I. D., West S. C. (2000) EMBO Rep. 1, 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kino K., Sugiyama H. (2000) Nucleic Acids Symp. Ser. 44, 139–140 [DOI] [PubMed] [Google Scholar]
- 65. Kino K., Sugiyama H. (2001) Chem. Biol. 8, 369–378 [DOI] [PubMed] [Google Scholar]
- 66. Kino K., Sugiyama H. (2005) Mutat. Res. 571, 33–42 [DOI] [PubMed] [Google Scholar]
- 67. Bukowy Z., Harrigan J. A., Ramsden D. A., Tudek B., Bohr V. A., Stevnsner T. (2008) Nucleic Acids Res. 36, 4975–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Belotserkovskii B. P., De Silva E., Tornaletti S., Wang G., Vasquez K. M., Hanawalt P. C. (2007) J. Biol. Chem. 282, 32433–32441 [DOI] [PubMed] [Google Scholar]
- 69. Ditlevson J. V., Tornaletti S., Belotserkovskii B. P., Teijeiro V., Wang G., Vasquez K. M., Hanawalt P. C. (2008) Nucleic Acids Res. 36, 3163–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Christensen L. A., Finch R. A., Booker A. J., Vasquez K. M. (2006) Cancer Res. 66, 4089–4094 [DOI] [PubMed] [Google Scholar]
- 71. Stamatoyannopoulos J. A., Adzhubei I., Thurman R. E., Kryukov G. V., Mirkin S. M., Sunyaev S. R. (2009) Nat. Genet. 41, 393–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stoltzfus A. (2008) Mutat. Res. 644, 71–73 [DOI] [PubMed] [Google Scholar]