Abstract
Metacaspases (MCPs) are members of a new family of cysteine proteases found in plants, fungi, and protozoa that are structurally related to metazoan caspases. Recent studies showed that plant MCPs are arginine/lysine-specific cysteine proteases with caspase-like processing activities in vitro and in vivo, and some of the plant type II MCPs exhibit Ca2+ dependence for their endopeptidase activity in vitro. However, the mechanisms and biological relevance of Ca2+ dependence and self-processing of plant MCPs remains unclear. Here we show that recombinant AtMCP2d, the most abundantly expressed member of Arabidopsis type II MCPs at the transcriptional level, exhibits a strict Ca2+ dependence for its catalytic activation that is apparently mediated by intramolecular self-cleavage mechanism. However, rapid inactivation of AtMCP2d activity concomitant with Ca2+-induced self-processing at multiple internal sites was observed. Because active AtMCP2d can cleave its inactive form, intermolecular cleavage (autolysis) of AtMCP2d could also occur under our assay conditions. Ca2+-induced self-processing of recombinant AtMCP2d was found to correlate with the sequential appearance of at least six intermediates, including self-cleaved forms, during the proenzyme purification process. Six of these peptides were characterized, and the cleavage sites were mapped through N-terminal protein sequencing. Mutation analysis of AtMCP2d revealed that cleavage after Lys-225, which is a highly conserved residue among the six Arabidopsis type II MCPs, is critical for the catalytic activation by Ca2+, and we demonstrate that this residue is essential for AtMCP2d activation of H2O2-induced cell death in yeast. Together, our results provide clues to understand the mode of regulation for this class of proteases.
Keywords: Arabidopsis, Calcium, Caspase, Cell Death, Cysteine Protease, Activation, Metacaspase
Introduction
Proteolytic enzymes, such as cysteine proteases, play important roles in programmed cell death (PCD)2 of eukaryotes. In animals, one conserved group of cysteine proteases called caspases, which cleaves target proteins after an aspartic acid residue, is indispensable for the regulation of apoptosis, the most well characterized form of PCD (1). Two groups of caspase-related cysteine proteases designated as paracaspase (found in metazoans and slime mode) and metacaspase (found in fungi, protozoa, and plants) were identified by iterative, structure-based homology searches (2). Metacaspases (MCPs) and paracaspases possess the catalytic dyad of histidine and cysteine residues that comprise the active site of caspases, whereas their overall sequence similarity is limited but significant. Thus, they have been proposed to be potential ancestors of animal caspases and may be similarly involved in the regulation of PCD in diverse organisms (3–5).
In plants, MCPs can be classified into type I and type II subclasses, based on their sequence similarity and domain structure (2). In the Arabidopsis thaliana genome, nine MCPs were annotated (3, 6). Of these, three genes belong to the type I class (AtMCP1a to -1c/AtMCA1 to -3), containing an N-terminal prodomain consisting of a zinc finger domain followed by a glutamine- and proline-rich (AtMCP1a/AtMC3) or proline-rich region (AtMCP1b/AtMC2 and AtMCP1c/AtMC1). Six type II MCPs (AtMCP2a to -2f/AtMC4 to -9) lack any apparent prodomain and only contain two domains predicted to relate to the p20 and p10 domains of animal caspases. However, more recent biochemical studies with recombinant MCPs from plants, fungi, and protozoa revealed that MCPs are cysteine proteases that prefer arginine or lysine residue at the P1 position of substrate target sites and do not cleave any synthetic caspase substrates in vitro (6–12). Recent studies have revealed that mammalian paracaspases possess proteolytic target specificity similar to that of MCPs (13, 14), thus indicating that MCPs and paracaspases are not true caspase orthologs. However, accumulating evidence supports the idea that plant and fungal MCPs could participate in the regulation of PCD observed under stress conditions (12, 15–18) or during aging (7, 15) and embryogenesis (8).
Compared with type I MCPs and paracaspases, several enzymatic properties of type II MCPs have been better characterized. Recombinant AtMCP2b/AtMC5, AtMCP2d/AtMC4, and AtMCP2e/AtMC8 from Arabidopsis and mcII-Pa from Norway spruce show pH optima at neutral to weakly basic pH (pH 7–8) and Ca2+ dependence for their catalytic activity in vitro as well as autocatalytic processing upon expression in Escherichia coli or Saccharomyces cerevisiae (6–8, 12). In contrast to these type II MCPs, the enzymatic properties of AtMC9/AtMCP2f are apparently different. The activity of AtMC9/AtMCP2f is controlled via its autocatalytic processing, which can be modulated by a reducing agent, dithiothreitol (DTT), and weakly acidic pH conditions (pH 5–6), S-nitrosylation, and a serine protease inhibitor, AtSerpin-1 (6, 19, 20). Vercammen et al. (6) reported that autocatalytic processing of proAtMC9 in the presence of DTT is a prerequisite for its endopeptidase activity in vitro, whereas AtMC9R/A, a zymogen form that has a point mutation in the processing site Arg-183, does not exhibit any peptidase activity. Given their structural and biochemical characteristics, AtMC9/AtMCP2f and other type II members (AtMC4 to -8/AtMCP2a to -2e) can be classified into two distinct subgroups from the type II AtMCP members. Overall, evidence indicates that multiple factors, such as Ca2+, pH, redox status, S-nitrosylation, and specific protease inhibitors, may be used to control the physiological activities of plant type II MCPs in vivo.
Although more is known about how the activity of AtMC9/AtMCP2f may be regulated via multiple mechanisms, the modes of regulation for other type II MCPs, such as AtMCP2d, remain to be clarified. Phylogenetically, it is interesting to note that AtMC9/AtMCP2f is clearly distinct from the other five type II MCPs, with only less than 40% identity at the protein sequence level, and contains a very short linker region between the p20- and p10-like domains (see supplemental Fig. S1). In A. thaliana, AtMCP2d/AtMC4 is the most abundantly expressed member in the type II subfamily at the transcriptional level and shows Ca2+ dependence for its endopeptidase activity (6, 7). However, there has been no further study on the mechanisms of its Ca2+ dependence and self-processing. Regulation of MCP activity by Ca2+ is of particular interest because alterations of its concentration in a cell could trigger programmed cell death (21–23), which may be mediated by MCPs.
Here we present a detailed investigation of how and to what extent Ca2+ can influence the activity of recombinant AtMCP2d/AtMC4, including the characterization of Ca2+-dependent activation and self-processing of the enzyme in vitro. For simplicity, we refer to Arabidopsis metacaspases as AtMCPs throughout this paper (3). Our results demonstrate that self-processing at Lys-225 is critical for Ca2+-induced self-processing of recombinant AtMCP2d (rAtMCP2d) concomitant with the activation of its catalytic activity in vitro. Interestingly, a rapid irreversible inactivation of rAtMCP2d was observed to follow this zymogen activation step. We also provide evidence that its specific processing at Lys-225 is required for H2O2-induced yeast cell death. Taken together, our results suggest the functional importance of cleavage-dependent activation of plant type II MCPs and uncover a novel self-inactivation mechanism that may be involved in the desensitization of signaling pathways mediated through MCPs.
EXPERIMENTAL PROCEDURES
Gene Constructs
To create recombinant AtMCP2d constructs, clone 183F14 (expressed sequence tag clone of AtMCP2d; accession number H37084) was used as a template for PCR. All oligonucleotide primers used in this study are listed in supplemental Table S1. The coding sequence of AtMCP2d was amplified by PCR using the following set of oligonucleotide primers (AtCAS2F and AtCAS2R), and then the PCR products were directly cloned into a pCR2.1-TOPO (Invitrogen). An NheI-EcoRI fragment of AtMCP2d was then subcloned into the NheI and EcoRI sites of the expression vector pET28a (Novagen) to generate His-AtMCP2d/pET28a. For generation of N-terminally T7 epitope-tagged and C-terminally hexahistidine-tagged AtMCP2d, the AtMCP2d-encoding sequence amplified from clone 183F14 using the oligonucleotide primers EL1457 and EL1458 was cloned into pCR2.1-TOPO, and then a BamHI-SalI fragment of AtMCP2d was subcloned into the BamHI and XhoI sites of the expression vector pET23a (Novagen) to generate pNW88 (T7-AtMCP2d-His/pET23a).
Three mutant forms of AtMCP2d (C139A, R190G, and R190G/K271G) were constructed using the QuikChange XL site-directed mutagenesis kit (Stratagene) and the following set of mutagenesis primers: EL2506 and EL2507, generating plasmid pNW246 (His-AtMCP2dC139A/pET28a) or pNW251 (T7-AtMCP2dC139A-His/pET23a); EL3228 and EL3229, generating plasmid pNW247 (His-AtMCP2dR190G/pET28a); EL3645 and EL3646, generating plasmid pNW248 (His-AtMCP2dR190G/pET28a) or pNW259 (His-AtMCP2dR190G,K271G/pET28a). A K225G mutant of AtMCP2d was created in pENTR/D-TOPO (Invitrogen), using the overlapping PCR method with the following primer set: EL4244 and EL4225, generating plasmid NW270. For the His-AtMCP2dK225G construct, a BamHI-SalI fragment of AtMCP2d was subcloned into the BamHI and XhoI sites of the pET28a (Novagen) vector to generate pNW273 (His6-AtMCP2dK225G/pET28a).
N-terminally or C-terminally truncated mutants (AtMCP2d(1-190), AtMCP2d(1–225), AtMCP2d(1–271), and AtMCP2d(226–418)) were created by PCR using the following set of primers: EL3447 and EL3448 for AtMCP2d(1–190), EL3447 and EL4500 for AtMCP2d(1–225), EL3447 and EL3449 for AtMCP2d(1–271), and EL4501 and EL3450 for AtMCP2d(226–418). Then the PCR products were directly cloned into a pENTR/D-TOPO. A BamHI-SalI fragment of each of these AtMCP2d mutants was subcloned into the BamHI and XhoI sites of the pET23a vector to generate pNW256 (T7-AtMCP2d(1–190)-His6/pET23a), pNW255 (T7-AtMCP2d(1–271)-His6/pET23a), pNW272 (T7-AtMCP2d(1–225)-His6/pET23a), or pNW273 (T7-AtMCP2d(226–418)-His6/pET23a).
For generation of C-terminally V5 epitope- and hexahistidine-tagged AtMCP2d (AtMCP2d-V5-His6), the coding sequence of AtMCP2d was amplified by PCR and directly cloned into a yeast expression vector pYES2.1/V5-His-TOPO (Invitrogen). Mutant forms of AtMCP2d (C139A or K225G) were also created by site-directed mutagenesis as described above and given pNW197m (AtMCP2dC139A-V5-His) or pNW274 (AtMCP2dK225G-V5-His), respectively.
To express AtMCP2d-V5-His and its mutant forms (C139A or K225G) in plant cells, the BamHI-XbaI fragment of the coding sequence from each yeast expression construct (pNW197, pNW197m, or pNW274) were subcloned into the plant binary vector EL103, generating pNW210 (35S:AtMCP2d-V5-His), pNW211 (35S:AtMCP2dC139A-V5-His), or pNW275 (35S:AtMCP2dK225G-V5-His). All DNA clones were identified by restriction digestion and then confirmed by DNA sequencing.
Bacterial Expression and Purification of Recombinant Proteins
The bacterial expression constructs were transformed into BL21(DE3)pLysS (Novagen), and then transformants were used for production and purification of recombinant AtMCP2d proteins (rAtMCP2d). rAtMCP2d or its mutant forms were induced by adding 0.4 mm isopropyl-1-thio-β-d-galactopyranoside to the cell culture with an A600 of 0.4–0.6. Then the cell culture was shaken (180 rpm) at 22 °C for 3-6 h. Cells were harvested, washed with distilled water, and resuspended in extraction buffer (50 mm potassium phosphate, pH 7.4, 500 mm NaCl, 10 mm 2-mercaptoethanol, 10% (w/v) glycerol, 5 μg/ml aprotinin). Cells were then broken by sonication, and insoluble materials were removed by centrifugation (20,000 × g, 20 min, 4 °C). The soluble extracts from the rAtMCP2d-expressing strain, supplemented with 10 mm imidazole, were mixed with Ni2+-nitrilotriacetic acid-agarose (Qiagen) equilibrated with 10 mm imidazole in extraction buffer and incubated for 1 h at 4 °C. After washing with extraction buffer supplemented with 20 mm imidazole, bound rAtMCP2d was eluted with 150 mm imidazole in extraction buffer. The purified samples were further passed through a PD-10 column (GE Healthcare) equilibrated with sample buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10 mm DTT, 1 mm EDTA, and 20% (w/v) glycerol) and then stored at −80 °C until use. Protein concentration was determined with a protein assay kit (Bio-Rad) using bovine serum albumin (BSA) as the standard.
Yeast Expression
Yeast cells were grown at 28 °C, and liquid cultures were shaken at 260 rpm as described previously (7). In brief, yeast cultures harboring expression vectors were grown overnight in 3 ml of noninducing selective medium (SC glucose), diluted in 10 ml of fresh SC glucose medium (adjusted to an A600 of ∼0.05) and then cultured until the A600 reached 0.4–0.6. The cells collected by brief centrifugation were resuspended in inducing medium (SC galactose) and grown for 6–24 h. For stimulation of yeast apoptosis, H2O2 (Sigma-Aldrich) was added to a final concentration of 1.2 mm into SC galactose media and then cells were grown for 24 h. Cell viability of yeast cell cultures was determined as described previously (7).
Plant Expression
Agrobacterium tumefaciens strain GV3101/pMP90 was transformed with pNW210, pNW211, or pNW275. A single colony of transformed A. tumefaciens was inoculated in 3 ml of Luria-Bertani broth containing 50 μg ml−1 gentamycin and 50 μg ml−1 kanamycin at 28 °C overnight. Bacteria pellets were resuspended in water to an A600 of about 1.0, and then the resuspended bacteria (1 ml) were infiltrated into the abaxial side of 1–2-month-old tobacco (Nicotiana tabacum cv. Samsun NN) leaves using a 1-ml syringe without a needle. At 2 days after agro-infiltration, crude extracts were prepared from the infiltrated zones using lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 10 mm DTT, and 5 μg/ml aprotinin) and then subjected to immunoblot analysis using an anti-V5 epitope antibody.
Enzyme Assay
MCP activity was measured by incubation of purified rAtMCP2d for 10–20 min at 30 °C with a 100 μm concentration of the fluorogenic substrate Boc-Gly-Arg-Arg-MCA (Boc-GRR-MCA; Bachem Bioscience) in a reaction buffer (100 μl) containing 25 mm HEPES-KOH (pH 7.5), 150 mm NaCl, 0–50 mm CaCl2, 0.1 mm EDTA, 1 mm DTT, 0.011–0.174 μm purified recombinant protein. The release of 7-amino-4-methyl-coumarin (AMC) was continuously monitored for 10–20 min with a microtiter plate reader (Synergy HT, Multidetection, BIO-TEK) at an excitation wavelength of 360 nm and an emission wavelength of 460 nm. Data are expressed as increases in relative fluorescence as a function of time. The specific enzyme activity to hydrolyze the peptidyl substrate was calculated as nmol of substrate hydrolyzed/mg of protein/min using a standard curve of AMC in the enzyme reaction buffer. Inhibitory effects of protease inhibitors, such as biotin-X-d-Phe-Pro-Arg-chloromethylketone (biotin-FPR-cmk; Calbiochem), d-Phe-Pro-Arg-chloromethylketone (FPR-cmk; Calbiochem), benzyloxycarbonyl-Phe-Ala-chloromethylketone (Z-FA-cmk; Bachem), and benzyloxycarbonyl-Phe-Lys-2,4,6-trimethylbenzoyloxymethyl ketone (Z-FK-tbmk; Bachem) were assayed under the same reaction conditions, using concentrations as indicated. Initial rates for the hydrolysis of Boc-GRR-MCA were calculated and estimated for determination of the Michaelis-Menten parameters Km and Vmax, The first order rate constant (Ko) value was determined from V0/[E], where V0 and [E] represent initial velocity and enzyme concentration, respectively.
To assay for rAtMCP2d activity in crude cell extracts of galactose-induced yeast cultures (10 ml), the cells were suspended in lysis buffer (100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 10 mm DTT, 1% CHAPS, and 5 μg/ml aprotinin (Sigma-Aldrich). Following cell lysis by vortexing with glass beads, the cell lysates were centrifuged (20,000 × g, 20 min, 4 °C), and the resultant supernatant was used for measurement of in vitro metacaspase activity as described above.
Antibody Production and Purification
The purified rAtMCP2d was used to immunize rabbits, and total IgG (H+L) fraction was purified from the antiserum by affinity chromatography with Protein A-Sepharose (Millipore) according to the manufacturer's instructions.
SDS-PAGE and Immunoblotting
Proteins were separated by 12% (v/v) SDS-PAGE. Separated proteins were electrophoretically transferred to a PVDF membrane (0.45 μm, Immobilon-P, Millipore) with a semidry blotter (Bio-Rad). Nonspecific binding was blocked with 5% (w/v) nonfat milk in TBST buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% (w/v) Tween 20) for 1 h at room temperature. The membrane was probed with anti-T7 epitope mAb (1:5,000 dilution; Novagen), anti-V5 epitope mAb (1:5,000 dilution; Invitrogen), or polyclonal anti-AtMCP2d antibody (1:20,000 dilution) at room temperature for 1 h. Immunoreactive polypeptides were detected using the appropriate secondary antibodies (anti-mouse IgG or anti-rabbit IgG) conjugated with horseradish peroxidase or alkaline phosphatase.
Determination of N-terminal Amino Acid Sequences of rAtMCP2d and Its Processed Forms
10 μg of purified rAtMCP2d was incubated for 10 min in a total volume of 500 μl of reaction mixture containing 25 mm HEPES-KOH (pH 7.5), 150 mm NaCl, 0.1 mm EDTA, 1 mm DTT, 5% (w/v) glycerol, and 0 or 10 mm CaCl2. The reaction was stopped by adding 125 μl of 100% trichloroacetic acid, incubated on ice for 30 min, and then precipitated by centrifugation (at 13,000 × g, 10 min, 4 °C), followed by washing the pellets with 80% cold acetone. Precipitants were dissolved in 30 μl of 1× SDS sample buffer (50 mm Tris-HCl, pH 6.8, 1% SDS, 10% glycerol, 1% 2-mercaptoethanol, 12.5 mm EDTA, and 0.02% bromphenol blue), separated on 12% SDS-PAGE, and then transferred to an Immobilon-PSQ membrane (0.2 μm pore size; Millipore) with a semidry blotter. After staining the membrane with 0.1% Coomassie Brilliant Blue in 50% methanol, followed by washing with 50% methanol and nanopure water, excised polypeptide bands from the membrane were submitted to the Protein Facility (Iowa State University) for N-terminal amino acid sequence determination performed by automated N-terminal Edman degradation using a 494 Procise protein/peptide sequencer with 140C PTH amino acid analyzer (Applied Biosystems).
Affinity Labeling with Biotin-FPR-cmk and Detection
Labeling of proteins with biotin-FPR-cmk was performed by incubating purified rAtMCP2d with 1–100 μm biotin-FPR-cmk in the enzyme assay mixture for 0–10 min at room temperature. Proteins were separated by SDS-PAGE and transferred to a PVDF membrane with a semidry blotter. Nonspecific binding was blocked with 5% (w/v) nonfat milk in TBST buffer for 1 h at room temperature. The membrane was washed with TBST buffer and then incubated with streptavidin-conjugated HRP (1:2,000 dilution; GE Healthcare) for 3 h at room temperature. The affinity-labeled proteins were visualized by Western Lightning-ECL enhanced chemiluminescence reagent (PerkinElmer Life Sciences) exposed to an autoradiography film (HyBlot CL, Denville Scientific).
RESULTS
Reversible Activation and Rapid Inactivation of Recombinant AtMCP2d by Calcium
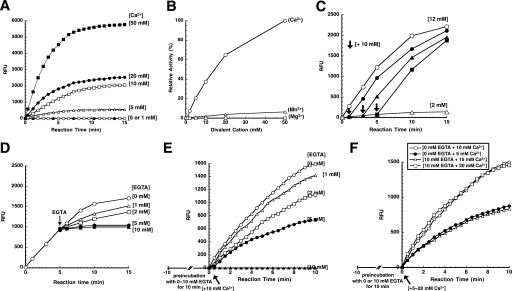
Heterologous expression of the MCPs is required to obtain a sufficient amount of starting materials for their detailed biochemical characterization. Although in vitro endopeptidase activity of recombinant type II MCPs has been detected in millimolar levels of Ca2+ (6–8, 12), little is known about the mechanism through which Ca2+ stimulates their activities. We first analyzed the effects of Ca2+ concentration on Boc-GRR-MCA-hydrolyzing activity (GRRase activity) with hexahistidine-tagged rAtMCP2d in vitro. The reaction was started by adding both Ca2+ and the peptidyl substrate Boc-GRR-MCA simultaneously. In the absence of Ca2+ or when a low concentration of Ca2+ (1 mm) was added, no significant activity was detected after the start of the reaction (Fig. 1A). However, a strong acceleration of the rate of Boc-GRR-MCA hydrolysis was observed in Ca2+ concentration ranges over 5 mm at the start of the reaction (Fig. 1A). This acceleration was essentially instantaneous with the addition of Ca2+ over 5 mm, and concurrently, the rate of Boc-GRR-MCA hydrolysis decreased with reaction times ranging from 5 to 10 min onward (Fig. 1A). On the other hand, a specific mutant form of rAtMCP2d having a mutation at its catalytic center, Cys-139 (rAtMCP2dC139A), did not show any obvious GRRase activity in the presence of Ca2+ (not shown). We also tested the effects of other divalent cations, such as Mg2+ and Mn2+, on the GRRase activity of rAtMCP2d. Mn2+ slightly activated the latent GRRase activity, with the reaction showing ∼20-fold less stimulated activity as compared with Ca2+, whereas Mg2+ showed no significant effect (Fig. 1B). These data indicate a specific requirement for Ca2+ to stimulate the latent rAtMCP2d activity in vitro, albeit a relatively high concentration of Ca2+ is needed.
FIGURE 1.
Divalent cation-dependent activation of GRRase activity by rAtMCP2d. Reactions were carried out at room temperature for 15 min in a total volume of 100 μl containing 25 mm HEPES-KOH (pH 7.5), 150 mm NaCl, 0.1 mm EDTA, 1 mm DTT, 5% (w/v) glycerol, 22 nm purified recombinant protein, and various concentrations of Ca2+, Mn2+, or Mg2+ (0–50 mm). Reactions were started by adding reaction mixture (90 μl) to the enzyme solution (10 μl), and then the fluorescence of the released AMC was measured as described under “Experimental Procedures.” GRRase activity is expressed as relative fluorescence units (RFU). A, Ca2+-dependent activation profile of GRRase activity. B, comparison of dose-dependent activation of GRRase activity by three divalent cations (Ca2+, Mg2+, and Mn2+). The initial rate of Boc-GRR-MCA hydrolysis was measured, and specific activity of the enzyme is expressed as relative values when compared with 50 mm Ca2+-dependent AtMCP2d activity set as 100%. C, rapid activation of GRRase activity by Ca2+. Reactions were started by adding 2 mm Ca2+, and GRRase activity was further monitored after the addition of 10 mm Ca2+ at the indicated times (indicated by arrows) at 1, 3, and 5 min after the start of the reaction. Ca2+-dependent activation of GRRase activity when the reaction was started by adding 12 mm Ca2+ is also shown as a control. D, reversible inhibition of GRRase activity by EGTA. Reactions were started by adding 10 μl of 100 mm CaCl2 to the enzyme solution (90 μl), and then various concentrations of EGTA (final concentrations 0, 1, 2, 5, and 10 mm) were added to the enzyme mixture at 5 min after the start of the reaction (indicated by an arrow). In C and D, the reaction was stopped by adding 100 μl of 100 mm EGTA after incubation for various periods, and then the fluorescence of the released AMC was measured as described under “Experimental Procedures.” E, effect of EGTA during preincubation on Ca2+-dependent GRRase activity. Preincubation was carried out for 10 min in the presence of 0–10 mm EGTA in the reaction buffer except for 10 mm Ca2+. After the preincubation period, the reaction was started by the addition of 10 mm Ca2+ and continued for 10 min. F, the reaction was carried out under the same conditions as for E except that different Ca2+ concentrations (5–20 mm) were added to start the reaction.
We subsequently investigated the kinetic parameters (the values for the apparent Vmax and Km) for GRRase activity of rAtMCP2d in the presence of 2, 10, or 50 mm Ca2+. Our data indicate that the apparent Km is very similar at each of the Ca2+ concentrations tested, whereas the apparent Vmax value increases with increasing Ca2+ (supplemental Table S2). These data indicate that activation of rAtMCP2d activity by Ca2+ is mediated by an acceleration of the rate of the enzyme activity of rAtMCP2d without significant changes in its substrate affinity.
We next tested whether Ca2+-dependent activation of rAtMCP2d is reversible. A standard peptidase assay was initiated, and the enzyme was added, followed several min later by the addition of Ca2+ and/or a strong Ca2+-specific chelating agent, EGTA. When the reaction was first stimulated with 2 mm Ca2+, the rate of Boc-GRR-MCA hydrolysis was markedly enhanced by the addition of 10 mm Ca2+ at 1, 3, and 5 min after the start of the reaction, whose kinetic curves were very similar to that in 12 mm Ca2+ alone (Fig. 1C). These results suggest that activation of rAtMCP2d by Ca2+ is reversible at the early reaction phase (<5 min). On the other hand, when EGTA was added at 5 min after the start of the reaction with 10 mm Ca2+, the rate of Boc-GRR-MCA hydrolysis concomitantly decreased with increasing EGTA concentrations (Fig. 1D). Similar results were obtained when EGTA was added at 1 or 3 min after the start of the reaction (data not shown). However, preincubation of rAtMCP2d with EGTA did not affect Ca2+-dependent activation of GRRase activity, although EGTA decreased the capacity to induce GRRase activity, depending on the initial dose of both EGTA and Ca2+ concentration added in the reaction mixtures (Fig. 1, E and F). Together, these results strongly suggest that activation of GRRase activity by Ca2+ and inhibition of GRRase activity by EGTA are reversible in the early period of the reaction.
We further tested whether the decrease of GRRase activity at the later reaction period (10 min) is due to substrate depletion, feedback inhibition of cleaved substrate, or inactivation of the catalytic activity. When rAtMCP2d was preincubated with 10 mm Ca2+ for 10 min in the absence of substrate or when rAtMCP2d reacted for 10 min in the presence of substrate was subjected to a new enzyme reaction with the addition of fresh substrates, no GRRase activity was detected in subsequent reaction periods of up to 10 min (data not shown). This finding strongly suggests that the termination of GRRase activity in the later reaction period is due to a Ca2+-dependent inactivation of rAtMCP2d activity. Thus, activation and concomitant inactivation of rAtMCP2d activity occurred under our assay conditions upon the addition of Ca2+, and this complication is reflected in the kinetics of Ca2+-dependent rAtMCP2d activity that shows a rapid cessation of activity after 5–10 min.
Calcium-dependent Autocatalytic Processing of Recombinant AtMCP2d
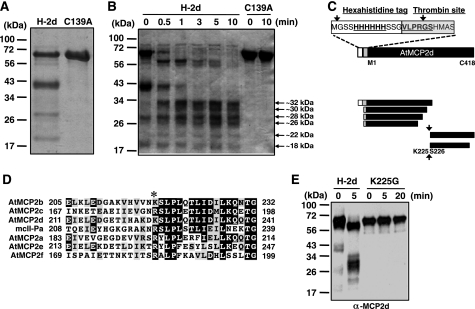
Proteases of the caspase family are known to require activation by proteolytic processing of their inactive zymogen, and similarly, it has been suggested that plant MCPs may require such a mechanism for catalytic regulation (6–8, 12). We therefore addressed the question of whether activation and inactivation of rAtMCP2d by Ca2+ are mediated by an autocatalytic mechanism. SDS-PAGE analysis of purified rAtMCP2d revealed that it underwent processing at multiple internal sites during its production in bacterial cells and/or purification process, whereas a variant (C139A) with a specific mutation at the catalytic site cysteine showed few detectable cleavage products (Fig. 2A). This is consistent with earlier results demonstrating that recombinant proteins of plant type II MCPs, including AtMCP2b, AtMCP2d, AtMCP2e, and AtMCP2f, can be autocatalytically processed in bacterial cells (6–8, 12).
FIGURE 2.
Analysis of self-processing of purified rAtMCP2d and its C139A mutant form. A, ∼5 μg of purified rAtMCP2d (H-2d) and its C139A mutant form (C139A) were separated on 12% SDS-PAGE, and bands were visualized by Coomassie Blue staining. B, ∼5 μg of purified rAtMCP2d (H-2d) and the C139A form were incubated in 100 μl of reaction mixture containing 10 mm Ca2+ for 0–10 min at room temperature and then subjected to SDS-PAGE. Self-processing of pro-rAtMCP2d and processed forms indicated with arrowhead (∼18–32 kDa) was visualized by Coomassie Blue staining. The positions of molecular mass markers are shown on the left. C, schematic representation of self-processing sites in the rAtMCP2d proteins determined by N-terminal protein sequencing. The size and structure of each fragment at the C termini are tentatively shown. D, comparison of the regions around the cleavage sites of AtMCP2d with other Arabidopsis type II MCPs and mcII-Pa. Positions for possible conserved cleavage sites are indicated by an asterisk. E, mutation of rAtMCP2d at Lys-225 (Lys → Gly; K225G) resulted in loss of the Ca2+-induced self-processing property. The reaction was carried out in the reaction mixture, including 22 nm rAtMCP2d (H-2d) or K225G and 10 mm Ca2+, for 0–20 min at room temperature and then subjected to immunoblot analysis using an anti-MCP2d antibody.
To analyze its Ca2+-dependent activation and inactivation, purified rAtMCP2d was incubated in an enzyme reaction mixture with or without 10 mm Ca2+ for 0–10 min, and aliquots were withdrawn at intervals and then analyzed by SDS-PAGE. The band pattern in terms of peptide composition of purified rAtMCP2d was not obviously changed in the absence of Ca2+ or in the presence of 1 mm Ca2+ over the course of our assay (data not shown). However, the spectrum of polypeptides was dramatically changed in the presence of 10 mm Ca2+, leading to the appearance of smaller polypeptides ranging from 18 to 32 kDa as well as a reduction in the amount of both the proenzyme and its cleaved form of ∼40 kDa (Fig. 2B). On the other hand, rAtMCP2dC139A did not show any obvious self-processing, indicating that activation of latent GRRase activity as well as self-processing of rAtMCP2d that is activated by Ca2+ depend on its catalytic center Cys-139 residue (Fig. 2B). In addition, we confirmed by SDS-PAGE that EGTA directly inhibits Ca2+-dependent activation of GRRase activity by chelating Ca2+, and this showed clear dependence on the ratio between [Ca2+] and [EGTA] added in the reaction mixtures (see Fig. 1, D–F). At 5 min after the reaction was started, most of the addition of 5 or 10 mm EGTA at 5 min after the start of the reaction prevents disappearance of ∼58-kDa fragment and two fragments of ∼40 kDa during the additional incubation period, whereas other fragments were largely unaffected (supplemental Fig. S2A). This indicates that chelation of calcium ion by EGTA directly blocks further self-processing of calcium-activated rAtMCP2d. In addition, EGTA did not cause further cleavage or disappearance (autolysis) of smaller self-processed forms (smaller than 34 kDa) of rAtMCP2d induced by Ca2+; nor did it increase any degradation of rAtMCP2dC139A (supplemental Fig. S2B). Therefore, our results demonstrate that the proteolytic cleavage of rAtMCP2d observed upon the addition of Ca2+ under our conditions depends on its catalytic activity and is not due to the presence of any Ca2+-dependent protease co-purified from the host bacteria.
In order to map the self-processing sites of rAtMCP2d, we determined the N-terminal sequences for the major intermediates by a protein sequencer. We confirmed that four major fragments of rAtMCP2d as purified by Ni2+-conjugated nitrilotriacetic acid-agarose column chromatography contained N-terminal fusion tag sequences, which can be confirmed by immunoblot analysis using anti-His antibody (data not shown), although the first Met residue was truncated (Fig. 2C). Immunoblot analysis of self-processed forms using anti-His antibody confirmed that three processed forms with ∼32, ∼40, or ∼56 kDa have an N-terminal hexahistidine tag, whereas the other smaller forms do not (data not shown). N-terminal protein sequencing of self-processed forms revealed the N-terminal sequences for six polypeptides: ∼32 kDa (no first Met residue in the N-terminal tag part); ∼30, ∼28, and ∼26 kDa (all cleaved after the thrombin recognition sequence with an artificial cleavage site due to the trypsin-like activity of AtMCP2d); and ∼22 and ∼18 kDa (both cleaved after Lys-225 of AtMCP2d). The presence of multiple intermediates suggests that the enzyme recognizes several different internal cleavage sites, especially between the N-terminal (p20) and C-terminal (p10) regions, during its autocatalytic processing (Fig. 2C). Importantly, sequence comparison of plant type II MCPs revealed that Lys-225 of AtMCP2d and the following C-terminal sequences are relatively conserved among plant type II MCPs (Fig. 2D), and it was shown that cleavage at the corresponding Arg-183 residue of AtMCP2f is critical for its self-processing and enzymatic activity under reducing conditions (6). Thus, we created a mutant form of rAtMCP2d where Lys-225 is replaced by a Gly (K225G) and compared its self-processing property with the non-mutated form. Fig. 2E shows that the purified K225G form did not contain significant cleaved or self-processed forms and remained intact even after prolonged incubation with 10 mm Ca2+. Our results show that the initial self-processing of rAtMCP2d occurs at Lys-225, and this is required for further cleavage at other internal sites.
Calcium-dependent Autolysis of rAtMCP2d Mutants
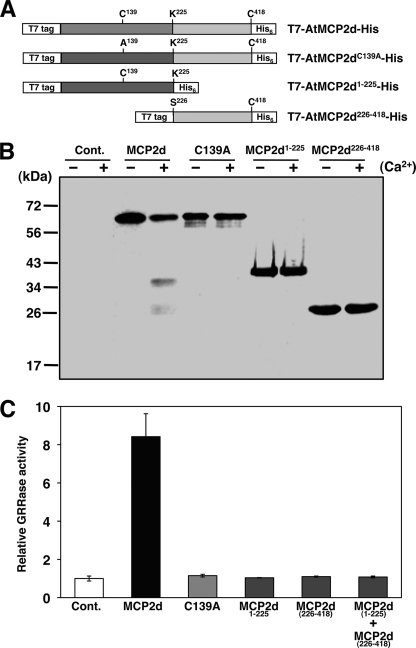
Although Ca2+-dependent activation of rAtMCP2d depends on the specific cleavage at Lys-225, the presence of multiple processed forms, including intermediates, suggests that the activated enzyme recognizes several different internal cleavage sites during its activation, maturation, and autoprocessing (Fig. 2). In order to analyze if each of the N-terminal and C-terminal regions of rAtMCP2d cleaved at Lys-225 are active, we created the corresponding truncation constructs in an inducible expression vector for E. coli (Fig. 3A) and compared the GRRase activity and autocatalytic processing using extracts from individual transformants. The data showed that similar to the catalytic inactive variant (C139A) or rAtMCP2d, neither rAtMCP2d(1–225) nor rAtMCP2d(226–418) is active, and these mutant variants did not show any obvious Ca2+-induced processing patterns on SDS-PAGE (Fig. 3, B and C). In addition, attempted reconstitution of active forms of rAtMCP2d by mixing lysates containing each of the two recombinant forms (rAtMCP2d(1–225) plus rAtMCP2d(226–418)) was unsuccessful under our assay conditions (Fig. 3C). This latter observation suggested that structural features prior to separation of the two domains could be important determinants for the catalytic regulation of AtMCP2d.
FIGURE 3.
Calcium-dependent autolysis of rAtMCP2d mutants in vitro. A, constructs for biochemical analysis of rAtMCP2d used in this study are shown. These four constructs have the T7-epitope and hexahistidine sequences at N termini and C termini, respectively. B, extracts from BL21(DE3)pLysS cells transformed with various rAtMCP2d constructs were incubated in the reaction mixture with or without 10 mm Ca2+ for 10 min at room temperature and then subjected to immunoblot analysis using anti-MCP2d antibody. C, relative GRRase activity in extracts from each transformants. For MCP2d-N + MCP2d-C, equal amounts of extracts from each transformant were mixed and preincubated on ice for 10 min, and then the reaction was started. Cont., empty vector (pET23a); MCP2d, T7-AtMCP2d-His; C139A, T7-AtMCP2d(C139A)-His; MCP2d1–225, T7-AtMCP2d(1–225)-His; MCP2d226–418, T7-AtMCP2d(226–418)-His. Error bars, S.E.
It was reported that recombinant mcII-Pa from Norway spruce was automatically cleaved at two internal sites after Arg-188 and Lys-269 when produced in bacteria, although an inactive form of mcII-Pa (with a C139A mutation) was not (8). However, the dependence of mcII-Pa processing and activation by Ca2+ was not investigated, and the importance of Arg-188 and Lys-269 in enzyme activity is undefined (8). Sequence alignment between AtMCP2d and mcII-Pa revealed that these two MCPs are the most similar at their predicted amino acid sequence as compared with other type II members of AtMCP (supplemental Fig. S1, A-C), and the two self-processing sites of mcII-Pa are conserved in AtMCP2d as well (supplemental Fig. S3A). Bozhkov et al. (8) proposed that self-processing at the Arg-188 and Lys-269 sites of mcII-Pa can produce three domains: (i) a caspase-like catalytic domain (p20); (ii) a p10-like domain of canonical caspases; and (iii) a linker region between p20- and p10-like domains specific for plant type II MCPs.
To address the question of whether cleavage of rAtMCP2d at the two putative processing sites (Arg-190 and Lys-271), as has been reported in mcII-Pa, is required for catalytic activation, we constructed several mutant versions of rAtMCP2d that contain either one or both of these residues mutated to Gly. In addition, we also created C-terminal truncations to remove either the predicted p10-like domain or the p10 plus linker domains together. These various constructs are summarized in supplemental Fig. S3B. To compare the biochemical activity for the various AtMCP2d variants, we produced the recombinant proteins in bacteria, and extracts from individual transformants were subjected to an in vitro enzyme assay followed by immunoblot analysis. The data showed that a mutation either at Arg-190 or mutations at both Arg-190 and Lys-271 retained their ability to undergo Ca2+-dependent autolysis with different patterns, depending on the particular mutation (supplemental Fig. S4A), and these mutants are still catalytically active (supplemental Fig. S4B). In addition, it appeared that there may be multiple self-processing sites in addition to Arg-190 and Lys-271 in rAtMCP2d that can be targeted (supplemental Fig. S4A). On the other hand, it was evident that C-terminally truncated rAtMCP2d mutants (AtMCP2d(1–190) and AtMCP2d(1–271)) were catalytically inactive and did not show any obvious autolysis in the presence or absence of Ca2+ (supplemental Fig. S4, C and D). Together, our results suggest that self-processing at the two processing sites of Arg-190 or Lys-271 is not required for rAtMCP2d activation, but these sites could be secondary cleavage sites after activation of AtMCP2d cleaved at Lys-225 in vitro.
Mechanism of Ca2+-induced Autocatalytic Processing of rAtMCP2d
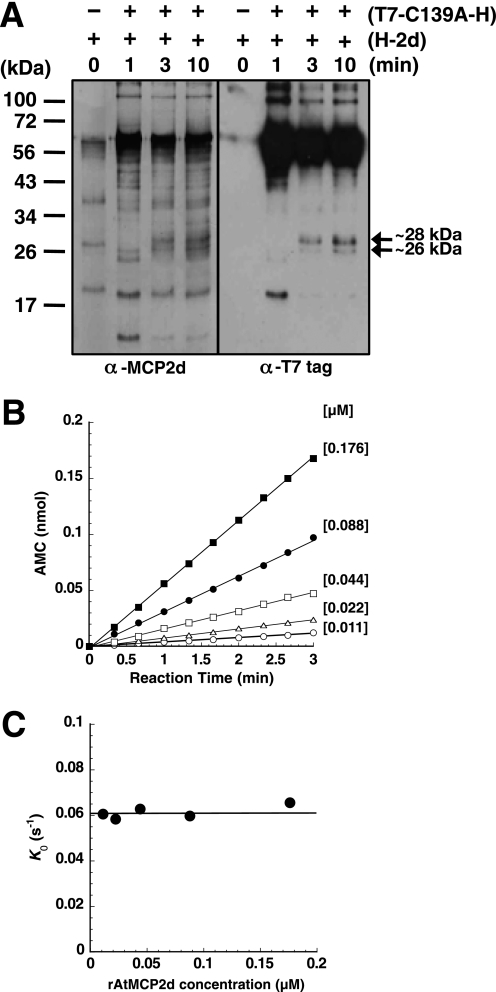
In considering the mechanism of Ca2+-induced rAtMCP2d activation and autolysis, it is possible that a given molecule cleaves itself (intramolecular processing) because this enzyme has multiple internal residues as potential cleavage sites. Alternatively, intermolecular processing that involves one rAtMCP2d molecule cleaving another may occur. To address these possibilities, we examined if an enzymatically inactive form of rAtMCP2d (T7-AtMCP2dC139A-His) is cleavable by an active form of rAtMCP2d (His-AtMCP2d). Co-incubation of T7-AtMCP2dC139A-His with His-AtMCP2d resulted in the appearance of two processed polypeptides detectable with an anti-T7 antibody (∼26 and ∼28 kDa) in a time-dependent manner. Production of other polypeptides ranging from 24 to 36 kDa are also detectable with an anti-MCP2d antibody (Fig. 4A). However, T7-AtMCP2dC139A-His was not processed very well by His-AtMCP2d. This may be due to the low enzyme concentration (11 nm) used in this assay, where active enzyme may be bound to the plastic surface of our assay tubes. We thus tested for effects of detergents or BSA as a potential protectant of the rAtMCP2d activity. As shown in supplemental Fig. S5A, all four of the non-ionic detergents we tested caused partial inhibition of the GRRase activity, whereas BSA did not show any significant effect. Immunoblot analysis of self-processing patterns of rAtMCP2d in the presence or absence of 0.05% Tween 20 indicated that Tween 20 does not significantly alter the observed Ca2+-stimulated self-processing pattern of rAtMCP2d (supplemental Fig. S5B). Similar results were obtained with 0.05% Triton X-100 (data not shown). These results support the idea that poor intermolecular cleavage of T7-AtMCP2dC139A-His by rAtMCP2d is not due to low enzyme concentration. Therefore, it seems likely that activated rAtMCP2d can cleave its mutant form in trans but also concomitantly undergoes autocleavage and inactivation, thereby leading to no further accumulation of processed T7-AtMCP2dC139A-His. Autolysis of rAtMCP2d appears to be more efficient than trans-cleavage between rAtMCP2d molecules under our conditions. The more rapid inactivation of rAtMCP2d activity observed in the presence of non-ionic detergents suggests that subunit interactions in the activated enzyme could be important.
FIGURE 4.
Intermolecular and intramolecular processing of rAtMCP2d in the presence of Ca2+. A, intermolecular cleavage of the catalytic inactive form of rAtMCP2d (C139A) by active rAtMCP2d. 1.0 μg (22 pmol) of purified T7-AtMCP2dC139A-His (T7-C139A-H) was co-incubated with 0.05 μg (1.1 pmol) of purified rAtMCP2d (H-2d) in a 100 μl of the reaction mixture for 0, 3, and 10 min at room temperature and then subjected to SDS-PAGE. Intermolecular processing of T7-AtMCP2dC139A-His by rAtMCP2d was detected by immunoblotting using anti-MCP2d antibody or anti-T7 epitope antibody. The positions of molecular mass markers are shown on the left. B, enzyme concentration-dependent profiles of GRRase activity in the presence of 10 mm Ca2+. Reactions were carried out under the same conditions as for Fig. 1 except that different concentrations of rAtMCP2d (0.011–0.176 μm) were used. Relative fluorescent units (RFU) were counted for 3 min. Relative fluorescent units were converted into the relative AMC amount compared with a known AMC standard, and non-linear regression of the data is shown. Concentrations of rAtMCP2d indicated are in μm. C, replot of the first order rates (K0) of the reaction (obtained from B) as a function of rAtMCP2d concentration.
To obtain information on the nature of the steps involved in the autocatalytic processing of wild-type rAtMCP2d upon stimulation by Ca2+, we monitored the kinetics of GRRase activity at the early step of the reaction, where the rate of GRRase activity is linear. Typical progress curves obtained at varying concentrations of added rAtMCP2d are shown in Fig. 4B. The initial rate of the hydrolysis increased in a time-dependent and enzyme dose-dependent manner. The first order rate constant (k0) for Ca2+-activated GRRase activity can be plotted as a function of the pro-rAtMCP2d concentration in each measurement. As shown in Fig. 4C, k0 constants for each enzyme concentration are very similar, indicating that autocatalytic activation of rAtMCP2d does not depend on the initial enzyme concentration. Thus, it seems most likely that initial autocatalytic activation of rAtMCP2d by Ca2+ is mediated by an intramolecular mechanism.
Identification of the Active rAtMCP2d Forms by Affinity Labeling and SDS-PAGE
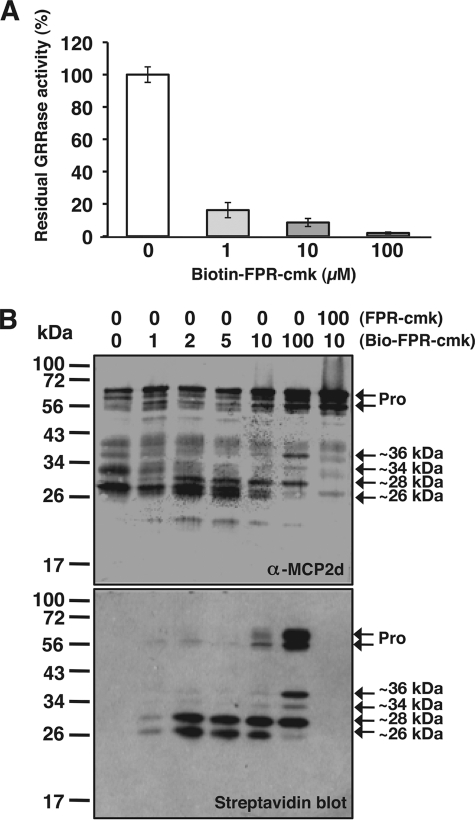
The above results strongly suggest that cleavage of pro-rAtMCP2d can occur via an intramolecular and intermolecular reaction that hydrolyzes GRR substrate and itself concomitantly, whereas this autolysis also leads to eventual inactivation (Fig. 4). However, one question that remained is whether the pro-rAtMCP2d itself can cleave other pro-rAtMCP2d molecules or whether, following an initial activation step, further cleavage only involved the newly formed mature enzymes. Having established the conditions for assaying the autolysis and catalytic activity of rAtMCP2d upon activation by Ca2+, we can determine whether proenzyme and/or processed forms of rAtMCP2d are catalytically active. To this end, the detailed inhibitory effects on pro-rAtMCP2d cleavage by the addition of a biotinylated peptide inhibitor (biotin-FPR-cmk) that can inhibit GRRase activity with an IC50 of <1 μm (Fig. 5A) were studied. Because biotin-FPR-cmk is an irreversible inhibitor, newly generated active forms of rAtMCP2d are immediately inhibited, and the protein can then accumulate during incubation with biotin-FPR-cmk. The biotinylated forms can subsequently be detected with a biotin-strepavidin detection system, and the results can be compared with the self-processing patterns of rAtMCP2d as detected by immunoblots with an anti-MCP2d antibody. In this way, the various forms of the pro-rAtMCP2d protein that is activated and can bind to a substrate would be revealed through active site tagging with the biotinylated inhibitor peptide. The fate of the activated enzyme can also be followed in a subsequent time course once it has been biotinylated.
FIGURE 5.
Inhibition of calcium-dependent autolysis of rAtMCP2d by biotin-FPR-cmk inhibitor. A, dose-dependent inhibition of rAtMCP2d activity by biotin-FPR-cmk. Reactions were carried out under the same conditions as for Fig. 1 except for initial Ca2+ concentration (10 mm) and enzyme concentration (0.11 μm). Residual GRRase activity is expressed as relative units when compared with control without inhibitor. B, affinity labeling of the active form of rAtMCP2d by biotin-FPR-cmk. The reaction was carried out in 100 μl of reaction mixture containing 0.11 μm rAtMCP2d, 10 mm Ca2+, 0–100 μm biotin-FPR-cmk for 5 min at room temperature and then subjected to SDS-PAGE. For binding specificity of biotin-FPR-cmk to active rAtMCP2d, 100 μm FPR-cmk was added as a competitor in the reaction mixture, including 10 μm biotin-FPR-cmk. Autolysis of rAtMCP2d and biotin-labeled rAtMCP2d were detected by immunoblotting using an anti-MCP2d antibody and by streptavidin blot, respectively. The positions of molecular mass markers are shown on the left. Error bars, S.E.
As shown in Fig. 5B, it is apparent that at the lower (1–5 μm) biotin-FPR-cmk concentrations, only two bands were specifically biotinylated, corresponding to peptides with 26 and 28 kDa apparent molecular mass. The amounts of these two biotinylated forms as determined by both biotin labeling (Fig. 5B, bottom) and protein immunolabeling (Fig. 5B, top) were apparently correlated. At higher (10 μm) biotin-FPR-cmk concentration, where both of the ∼26- and ∼28-kDa peptides were generated, proenzyme forms at 62 and 57 kDa were also weakly biotinylated. Because weak GRRase activity of rAtMCP2d (10–20% of GRRase activity compared with control) (Fig. 5A) could be detectable at lower concentrations of biotin-FPR-cmk (1–5 μm), biotinylated proenzymes appear to be processed by residual activated enzymes. In contrast, four major peptides, including proenzymes as well as peptides at 36, 34, and 28 kDa apparent molecular mass, were biotinylated much more intensely at the highest concentration of biotin-FPR-cmk (100 μm) used, where GRRase activity of rAtMCP2d is highly inhibited (Fig. 5A). At this concentration, a peptide of 36 kDa remarkably accumulated, whereas the biotinylated peptide of 26 kDa became underrepresented, as seen on our protein blot with anti-AtMCP2d antibody. These results suggest that the 36-kDa peptide may be an intermediate for the generation of the 26-kDa peptide cleaved by activated proenzymes. The specificity of biotinylation for the different forms of rAtMCP2d by biotin-FPR-cmk was demonstrated by a competition experiment with a 10-fold excess of FPR-cmk that completely prevents labeling by 10 μm biotin-FPR-cmk (Fig. 5B). In addition, we confirmed that another peptidyl inhibitor, Z-FK-tbmk, which inhibits type II MCP activity in vitro (6, 7), can also competitively prevent affinity labeling of rAtMCP2d by biotin-FPR-cmk, whereas the inactive analog Z-FA-cmk cannot (supplemental Fig. S6).
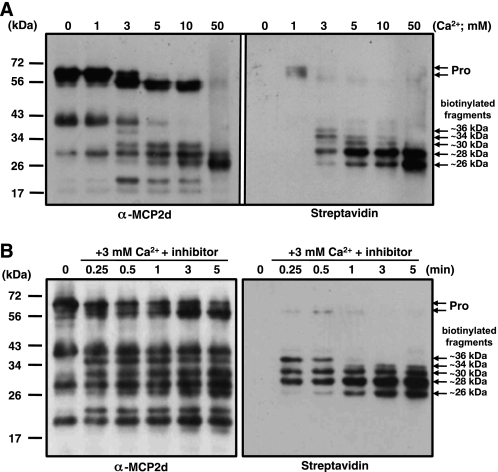
To determine the role of Ca2+ in the activation of rAtMCP2d, we performed affinity labeling with 10 μm biotin-FPR-cmk in the presence of different concentrations of this divalent cation. We found that no biotinylated peptides were detected when the reaction was carried out in the absence of Ca2+ (Fig. 6A), and this observation correlates with the fact that rAtMCP2d does not display any significant GRRase activity under this condition (Fig. 1A). At the lowest Ca2+ concentration tested (i.e. 1 mm), a very small percentage of the proenzymes appeared to be biotinylated in the presence of 10 μm biotin-FPR-cmk, although there is no obvious autolysis or protease activity at this concentration. This labeling could not be prevented in the presence of 100 μm Boc-GRR-MCA substrate (data not shown), confirming that 10 μm biotin-FPR-cmk effectively inhibits GRRase activity (Fig. 5A). Also, it seems that 1 mm Ca2+ can induce a conformational change in the precursor form of rAtMCP2d, allowing it to bind to appropriate substrate targets albeit at a lower apparent affinity. However, this concentration of Ca2+ cannot stimulate significant levels of latent GRRase activity (Fig. 1A). On the other hand, Ca2+-dependent autolysis of rAtMCP2d in the presence of the inhibitor was evident with increasing concentrations of Ca2+, corresponding to the generation of several processed forms ranging from 36 to 26 kDa in apparent molecular mass (Fig. 6B). At 50 mm Ca2+, the amounts of the biotinylated peptides at 28 and 26 kDa as determined by both biotin labeling and immunolabeling were highly correlated. However, proenzymes were not well biotinylated at 3–10 mm Ca2+, although relatively large amounts of proenzymes can be detectable by protein labeling. At 3 mm Ca2+, our time course analysis revealed that self-processing of rAtMCP2d was not completed by 5 min, but several intermediate forms of biotin inhibitor-labeled polypeptides ranging from 26 to 40 kDa and a precursor form of ∼56 kDa were apparent at the early time of reaction (∼1 min), and then three polypeptides of 26–33 kDa accumulated sequentially at 1–5 min (Fig. 6B). These results suggest that rAtMCP2d proenzymes can be biotinylated after Ca2+-dependent activation and that Ca2+-stimulated proenzymes can modify themselves as well as other biotinylated forms intermolecularly, which may then be converted to several inactive polypeptides that remain biotinylated at its active site.
FIGURE 6.
Calcium- and time-dependent autolysis of rAtMCP2d in the presence of biotin-FPR-cmk. A, effect of Ca2+ concentrations on autolysis of rAtMCP2d. The reaction was carried out in 100 μl of reaction mixture containing 0.22 μm rAtMCP2d, 10 μm biotin-FPR-cmk, and various concentrations of Ca2+ (0, 1, 3, 5, 10, and 50 mm) for 10 min at room temperature and then subjected to SDS-PAGE. B, time-dependent autolysis of rAtMCP2d in the presence of 3 mm Ca2+. The reaction was carried out as in A except for 3 mm Ca2+ and, with or without 10 μm biotin-FPR-cmk, for 5 min at room temperature and then subjected to SDS-PAGE. In A and B, autolysis of rAtMCP2d and biotin-labeled rAtMCP2d were detected by immunoblotting using anti-MCP2d antibody and by streptavidin blot, respectively. The positions of molecular mass markers are shown on the left.
Self-processing of AtMCP2d at Lys-225 Is Essential for Its Cell Death-inducing Activity in Yeast Cells
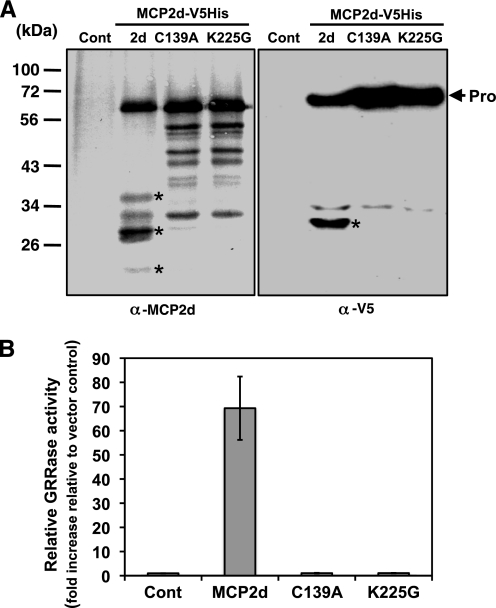
We and others recently reported that two Arabidopsis type II MCPs, AtMCP2b and AtMCP2e, can successfully be expressed as an active form and mediate cell death activation in the budding yeast S. cerevisiae (7, 12). These type II AtMCPs can functionally but partially complement yeast metacaspase-1 (YCA1) in yca1Δ cells when the cell death program can be triggered by H2O2 (7, 12). To determine whether the cleavage at Lys-225 is functionally relevant for AtMCP2d in vivo, we created three different constructs (wild-type, C139A, or K225G form of AtMCP2d) with a V5 epitope and hexahistidine fusion tag at their C termini, whose expressions are under the control of the inducible GAL1 promoter. Induction of each of these three AtMCP2d-derived constructs in the yca1Δ mutant was verified by immunoblotting using an anti-V5 epitope antibody or our anti-MCP2d antibody. As shown in Fig. 7A, specific self-processing of AtMCP2d required both its catalytic active site (Cys-139) and self-processing site (Lys-225) because each mutant form displayed similar nonspecific degradation patterns probably due to endogenous yeast protease activities. Cell lysates prepared from yeast expressing wild-type AtMCP2d showed an increase in GRRase activity, with >70-fold higher GRRase compared with extracts from induced yeast with either the empty vector or the mutant forms of AtMCP2d (Fig. 7B). This result demonstrates that specific processing of AtMCP2d at Lys-225 is essential for its catalytic activation in vivo when expressed in yca1Δ cells. Similar results were obtained when these constructs were expressed in the wild-type strain (BY4741) (data not shown).
FIGURE 7.
Expression of AtMCP2d-V5-His6 and its variants and detection of their GRRase activity in yeast lysates. A, immunoblot analysis of inducible expression of AtMCP2d-V5-His6 (2d) and its mutant variants (C139A and K225G). Total cell lysates were prepared from wild-type yeast cells (BY4741 strain) transformed with various rAtMCP2d constructs at 6 h post-galactose shift and then subjected to immunoblot analysis using anti-MCP2d antibody or anti-V5 epitope antibody. Bands corresponding to specific bands in the MCP2d sample, which are not detectable in both the C139A and K225G samples, are indicated with an asterisk on both blots. Antibody used and the positions of molecular mass markers are shown on the left. B, relative GRRase activity in extracts from each transformant. In the meantime, soluble fractions from each strain were prepared and then subjected to in vitro GRRase activity measurements. Each value represents the mean ± S.E. (error bars) of three independent samples per experiment. Cont., empty vector, pYES2.1; 2d, AtMCP2d-V5-His6; C139A, AtMCP2dC139A-V5-His6; K225G, AtMCP2dK225G-V5-His6.
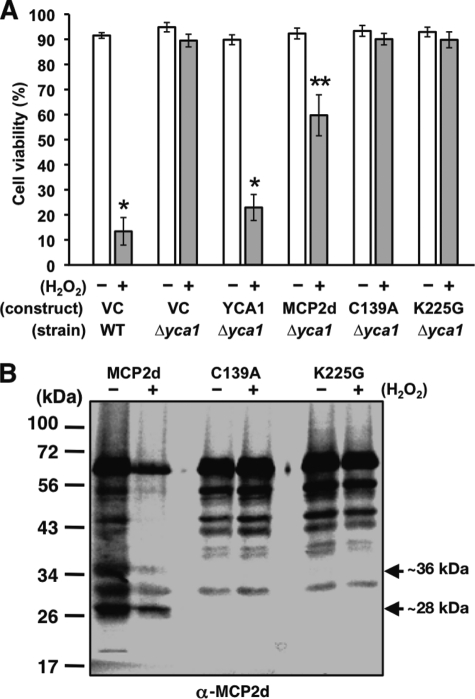
We further addressed the question of whether AtMCP2d might be able to complement the function of YCA1 in H2O2-mediated yeast cell death. When the yeast cell death program was stimulated with 1.2 mm H2O2, the wild-type strain drastically lost cell viability, showing ∼12% survival, whereas the yca1Δ strain did not show a significant loss of cell viability (Fig. 8A). However, the yca1Δ strain lost cell viability similar to the wild-type when YCA1 was overproduced (∼20% viability). Under a similar condition, the wild-type form of AtMCP2d was able to reduce cell viability (∼60%), whereas the two mutant forms (C139A and K225G) did not (Fig. 8A). Consistent with these properties, immunoblot analysis with yeast strains expressing each form of AtMCP2d revealed that the wild-type form accumulates one self-processed product (∼28 kDa) at a much higher level compared with the other two forms (Fig. 8B). These results indicate that AtMCP2d can partially complement the cell death function of YCA1 in H2O2-mediated yeast cell death, and its cell death activity depends on the catalytic activation with specific processing at Lys-225.
FIGURE 8.
Self-processing of AtMCP2d-V5-His6 at Lys-225 is essential for cell death-inducing activity in yca1Δ cells when challenged with H2O2. A, cell viability of yca1Δ cells expressing AtMCP2d, C139A, K225G, YCA1, and the corresponding vector control after 24 h of galactose shift in combination with or without 1.2 mm H2O2 treatment. Aliquots of the cells were subjected to a trypan blue dye exclusion assay to estimate the cell viability. Data are represented as the mean cell viability (%) ± S.E. (error bars) (n = 5). Asterisks above the column indicate Student's t values. *, p < 0.005; **, p < 0.01. B, immunoblot analysis of AtMCP2d, C139A, and K225G. At 24 h after galactose shift with or without 1.2 mm H2O2 treatment, cell extracts from each cell culture were subjected to immunoblot analysis using anti-MCP2d antibody. Controls harboring vector control did not show any cross-reactive bands with anti-MCP2d antibody (data not shown). Bands corresponding to specific bands in the MCP2d sample are indicated with an arrowhead. The positions of molecular mass markers are shown on the left.
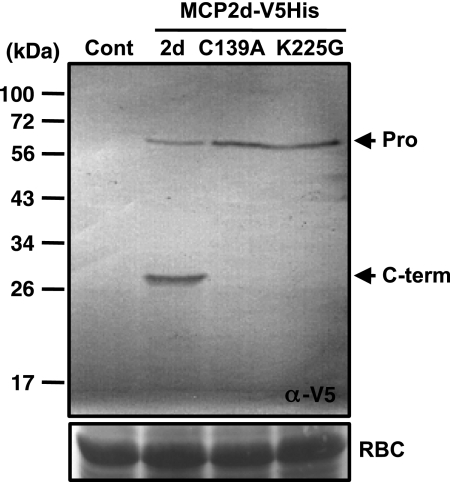
Lys-225 Is Critical for the Autocatalytic Processing of AtMCP2d in Tobacco Cells
We also transiently expressed C-terminally V5- and hexahistidine-tagged versions of AtMCP2d, whose expressions are controlled under a CaMV 35S promoter, in tobacco leaf cells via agrobacterium infiltration. Immunoblot analysis using anti-V5 antibody revealed that self-processing of the wild-type form depends absolutely on its catalytic center, Cys-139, and that mutation of Lys-225 to Gly-225 (K225G) results in a similar loss of the self-cleavage activity despite the presence of an intact catalytic site, Cys-139 (Fig. 9). This provided additional evidence to support the critical importance of the autolytic processing in the function and activity of AtMCP2d in vivo.
FIGURE 9.
Transient expression of AtMCP2d-V5-His6 and its variants in tobacco leaf cells via agrobacterim-mediated transformation. A. tumefaciens GV3101 carrying the binary vector expressing AtMCP2d-V5-His6 or its variants (C139A or K225G) was used to infiltrate tobacco leaves. Crude extracts were extracted from each of the infiltrated area 2 days postinfiltration and then subjected to immunoblot analysis using anti-V5 epitope antibody. Proenzyme (Pro) and C-terminal fragment (C-term) are shown on the right. Ponceau S staining intensities of RBCL protein (RBC) in the same samples confirm equal loading in the individual lanes. The positions of molecular mass markers are shown on the left.
DISCUSSION
In the present study, we provide further evidence that the endopeptidase (GRRase) and autocatalytic processing activities of rAtMCP2d are strictly Ca2+-dependent, requiring a millimolar range of Ca2+ to stimulate its latent activities (Fig. 1). Although the latent endopeptidase activity of rAtMCP2d can be stimulated much more strongly by Ca2+ at higher concentrations of 10–50 mm, Ca2+ also dramatically attenuated the activity of rAtMCP2d in a time- and enzyme concentration-dependent manner. We found that processed forms, including low molecular mass polypeptides (26–32 kDa) are observed at the later stages of the reaction, and they are quite stable even after prolonged incubation (Fig. 2B). However, these processed forms of rAtMCP2d at the later stage of reaction were catalytically inactive. This time-dependent inactivation of the enzyme can be explained by further cleavage of rAtMCP2d subsequent to its initial activation involving the Lys-225 site and is consistent with the sequential appearance of several intermediates and final processed forms that are probably mediated by intra- and intermolecular proteolytic events (Fig. 4). However, whether and how these subsequent cleavage sites are involved in the inactivation process remains to be established. Although higher Ca2+ concentrations facilitate zymogen activation via acceleration of the catalytic activity without significant changes in substrate affinity (supplemental Table S2), it also leads to enzyme inactivation concomitantly. These results support the idea that AtMCP2f and AtMCP2d are enzymatically distinct members in the type II subfamily of Arabidopsis MCPs because AtMCP2f does not require Ca2+ for its activation, and no inactivation via self-cleavage was reported (6).
Intermolecular mechanisms of autocatalytic processing for zymogen activation have been demonstrated for a number of proteases, including caspases and arginine-specific gingipain (gingipain-R) (24, 25). Overexpressed rAtMCP2d in bacteria or yeast cells appears to spontaneously undergo self-processing, depending on its active site, Cys139, which is similar to the case of caspases. Although the mechanism is not well understood, it is assumed that direct association of recombinant caspases at high concentrations can facilitate intermolecular cleavage (26–28). However, Ca2+ was found to have no effect on the activity of any of the caspases at concentrations up to 100 mm (27). On the other hand, purified rAtMCP2d is largely inactive without added Ca2+ stimuli (Fig. 1). Although initiation of the activation process in vivo has remained an unsolved question, rAtMCP2d proenzymes produced in bacteria may exhibit minor catalytic activity, which could potentially initiate a cascade reaction for proteolytic processing (Fig. 2A). When two proenzyme molecules come into close contact, one active proenzyme molecule can cleave the second molecule. The behavior of Ca2+-dependent rAtMCP2d activity in vitro appears to be a reflection of the complexity of the mechanism for processing of proenzyme and for its further autolysis (Figs. 1 and 2). In fact, intermolecular processing of rAtMCP2d can occur only at a minor level under our assay conditions (Fig. 4A), and we found that initial activation of rAtMCP2d by Ca2+ is mediated predominantly by an intramolecular mechanism (Fig. 4, B and C). During this process, Ca2+-stimulated proenzyme is rapidly cleaved at Lys-225 by itself. At the later step of the reaction, however, AtMCP2d activation is not stable, and the protease undergoes further autolysis. In contrast, gingipain-R from Porphyromous gingivalis, which has a similar overall protein fold to caspases and has been suggested to be an ancestor of caspase-related cysteine protease, was shown to be synthesized as a zymogen containing N-terminal and C-terminal extensions and is converted to its stable active form by sequential autolytic processing of these two extensions (29, 30). Interestingly, gingipain-R requires DTT for its activation and a submillimolar range of Ca2+ for stabilization (29, 31, 32). Thus, MCPs and gingipain-R belong to a caspase-related protease family and may have evolved distinct mechanisms to control their activation from the latent zymogen.
Using a biotinylated peptidyl inhibitor as an active site affinity-labeling reagent, we found that autocatalytic processing of rAtMCP2d can be delayed in the presence of this inhibitor, and sequential processing of the enzyme into several fragments can be captured (Figs. 5 and 6). This labeling is specific because it can be efficiently competed by adding non-biotinylated inhibitors, such as FPR-cmk and Z-FK-tbmk, but not the inactive inhibitor analog Z-FA-cmk (supplemental Fig. S6). Biotinylated bands ranging from 26 to 32 kDa probably corresponded to fragments containing the N-terminal domain of rAtMCP2d and the catalytic dyad, based on our N-terminal peptide sequencing results (Fig. 2B). The biotinylated ∼36 kDa fragment may be an active form of rAtMCP2d(1–225) (Fig. 6B). However, it was difficult to trace the sequential event for the activation and autolytic processes in detail because the disappearance of the proenzyme is very rapid (Fig. 2B), and intermolecular as well as intramolecular proteolytic events probably occur concomitantly to generate various possible fragments (Figs. 5 and 6). Based on the fragmentation pattern of activated rAtMCP2d, it is likely that multiple alternative internal cleavage sites are present in and around the conserved p20 and p10 domains. The determination of the sites of cleavage in biotinylated AtMCP2d during these processes will be an important step in the future to resolving the mechanism of this enzyme's processing and inactivation by Ca2+.
Previous studies on the mechanism of autocatalytic processing of AtMCP2f by Vercammen et al. (6) indicated that processing at Arg-183 is needed for its endopeptidase activity. No alternative autoprocessing site that can substitute for this residue was identified; thus, the autoprocessing is specific at the single site of Arg-183 in AtMCP2f. Similar to the case with AtMCP2f, we found that specific processing at Lys-225 of AtMCP2d is a prerequisite for Ca2+-induced catalytic activation and self-processing in vitro (Fig. 2E) and in vivo (Figs. 7–9). Interestingly, activation and inactivation of rAtMCP2d in the presence of Ca2+ cannot be inhibited by the presence of excess amounts of peptidyl substrate (Boc-GRR-MCA) or BSA in the reaction mixture, which should present excess cleavage substrate target sites in trans. This observation suggests that autocatalytic processing of rAtMC2d is more efficient than intermolecular peptidyl substrate cleavage. It is noteworthy that Lys-225 of AtMCP2d is located at a conserved region in all type II MCPs, including AtMCP2f and mcII-Pa (see Fig. 2D and supplemental Fig. S1A). This specific cleavage results in separation of two predicted domains (catalytic N-terminal domain and C-terminal region), which are termed as p20- and p10-like regions via a linking loop, and is based on a predicted structural model of AtMCP2f that was inferred from the three-dimensional structure of human caspase-8 (5). It seems very likely that plant type II MCPs can be expressed as inactive zymogens composed of two catalytic domains: p20-like and p10-like regions that are proteolytically separated by itself (autolytic processing) or by other MCPs (processing in trans) that are required for their conversion into mature enzymes. Our analysis using C-terminal truncated forms of rAtMCP2d (AtMCP2d(1–190), AtMCP2d(1–225), and AtMCP2d(1–271)) revealed that they do not display any obvious GRRase activity and autolysis in the presence or absence of Ca2+ (Fig. 3 and supplemental Fig. S4), thus supporting the idea that the p10-like domain is also required for catalytic activity of rAtMCP2d after its specific processing at Lys-225. However, how the C-terminal domain of AtMCP2d contributes to catalytic regulation remains unclear.
Recently, recombinant MCP of Trypanosoma brucei, MCA2, produced in bacteria was shown to have an arginine/lysine-specific, Ca2+-dependent proteolytic activity with autocatalytic processing at two internal cleavage sites (Lys-55 and Lys-268), although this processing is not necessary or sufficient for the enzyme to be active (11). It remains unclear whether or not self-processing of recombinant MCA2 at these two sites will lead to enzyme inactivation. Our present work strongly suggests that self-processing of some MCPs from plants, yeast, and protozoa may also lead to inactivation of the enzyme upon exposure to Ca2+. We note that rapid self-inactivation of MCPs has not been reported for MCPs previously. Although the binding of protease inhibitors, such as serpin-1 has been reported for AtMCP2f (19), the relevance of this association to enzyme activity control in vivo remains uncertain (33). The mechanisms for tight control of the different MCPs are probably critical for maintaining proper activation of key processes in vivo, such as PCD. Having an efficient and rapid self-inactivation pathway inherently linked to its conversion from the proenzyme form into catalytically active forms should ensure that activated AtMCP2d will have a short functional half-life. We speculate that this can provide an exquisitely sensitive switch that can rapidly and specifically respond to changes in the levels of different inductive signals. This is analogous to the desensitization control of well characterized signaling pathways, such as those related to rhodopsin-activated visual signaling and acetylcholine receptor mediated pathways (34, 35). To further examine this hypothesis, the identification of critical proteolytic target site(s) for the autoinactivation of AtMCP2d would be important to define the mode of catalytic regulation of this protease in vivo.
Calcium ion plays a pivotal role as second messenger in plant cells, and changes in cytosolic free calcium concentrations ([Ca2+]cyt) are apparent during the transduction of a wide array of abiotic and biotic signals (21–23). Although the [Ca2+] required for the activation of AtMCP2d is remarkably higher than the estimated physiological concentration of Ca2+ in the cytoplasm of plant cells, which is usually in the submicromolar levels (36), all plant calcium-dependent proteases, including MCPs, that have been characterized so far require greater than millimolar levels of Ca2+ for optimal enzyme activation in vitro (6–12, 37–40). One plausible scenario for the high [Ca2+] requirement is that Ca2+ may be able to mimic a physiological factor(s) or signaling molecules that normally regulate catalytic activation of plant proteases. Alternatively, these proteases may also interact with another physiological factor(s) in vivo that can reduce the necessary dose of [Ca2+] for enzyme activation. The mechanism as well as the biological role of Ca2+ in activation of these plant proteases remain to be determined.
In conclusion, our present work revealed that simultaneous activation and spontaneous inactivation mechanisms exist in the regulation of Ca2+-dependent AtMCP2d activity under our in vitro assay conditions. However, how AtMCP2d is converted into the mature form with an active status remains unclear at the molecular level. Direct identification of multiple self-processing sites and the use of site-directed mutagenesis to dissect the functional relevance of these cleavage sites will allow us to investigate how AtMCP2d proenzymes convert into the active and inactive forms in detail. The demonstration here of the critical importance of Lys-225 in the Ca2+-dependent cleavage activation process is the first step toward this goal. In vivo activation and processing of AtMCP2d is probably more complex because its activity can be affected by changes in environmental factors, such as pH, ion strength, redox status, and free Ca2+ level in the cytosol. In addition, a potential protein partner(s) that may be critical for facilitating the conversion of the AtMCP2d zymogen would also be a likely candidate(s) for its control. Further studies will be needed to provide insights into the regulation of MCP activity in plants.
Acknowledgments
We thank Dr. Frank Madeo (University of Graz, Austria) for kindly providing the yeast and E. coli strains and Joel Nott (Iowa State University) for protein sequencing service.
This work was supported by National Science Foundation Grant IOS-0744709.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S6.
- PCD
- programmed cell death
- AMC
- 7-amino-4-methylcoumarin
- AtMCP
- A. thaliana MCP
- rAtMCP2d
- recombinant AtMCP2d
- cmk
- chloromethylketone
- biotin-FPR-cmk
- biotin-X-d-Phe-Pro-Arg-chloromethylketone
- Boc
- t-butyloxycarbonyl
- MCA
- 4-methylcoumaryl-7-amide
- MCP
- metacaspase
- Z
- benzyloxycarbonyl
- tbmk
- 2,4,6-trimethylbenzoyloxymethyl ketone.
REFERENCES
- 1. Kumar S. (2007) Cell Death Differ. 14, 32–43 [DOI] [PubMed] [Google Scholar]
- 2. Uren A. G., O'Rourke K., Aravind L. A., Pisabarro M. T., Seshagiri S., Koonin E. V., Dixit V. M. (2000) Mol. Cell 6, 961–967 [DOI] [PubMed] [Google Scholar]
- 3. Watanabe N., Lam E. (2004) Mol. Plant Pathol. 5, 65–70 [DOI] [PubMed] [Google Scholar]
- 4. Sanmartín M., Jaroszewski L., Raikhel N. V., Rojo E. (2005) Plant Physiol. 137, 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vercammen D., Declercq W., Vandenabeele P., Van Breusegem F. (2007) J. Cell Biol. 179, 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vercammen D., van de Cotte B., De Jaeger G., Eeckhout D., Casteels P., Vandepoele K., Vandenberghe I., Van Beeumen J., Inzé D., Van Breusegem F. (2004) J. Biol. Chem. 279, 45329–45336 [DOI] [PubMed] [Google Scholar]
- 7. Watanabe N., Lam E. (2005) J. Biol. Chem. 280, 14691–14699 [DOI] [PubMed] [Google Scholar]
- 8. Bozhkov P. V., Suarez M. F., Filonova L. H., Daniel G., Zamyatnin A. A., Jr., Rodriguez-Nieto S., Zhivotovsky B., Smertenko A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14463–14468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. González I. J., Desponds C., Schaff C., Mottram J. C., Fasel N. (2007) Int. J. Parasitol. 37, 161–172 [DOI] [PubMed] [Google Scholar]
- 10. Lee N., Gannavaram S., Selvapandiyan A., Debrabant A. (2007) Eukaryot. Cell 6, 1745–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moss C. X., Westrop G. D., Juliano L., Coombs G. H., Mottram J. C. (2007) FEBS Lett. 581, 5635–5639 [DOI] [PubMed] [Google Scholar]
- 12. He R., Drury G. E., Rotari V. I., Gordon A., Willer M., Farzaneh T., Woltering E. J., Gallois P. (2008) J. Biol. Chem. 283, 774–783 [DOI] [PubMed] [Google Scholar]
- 13. Coornaert B., Baens M., Heyninck K., Bekaert T., Haegman M., Staal J., Sun L., Chen Z. J., Marynen P., Beyaert R. (2008) Nat. Immunol. 9, 263–271 [DOI] [PubMed] [Google Scholar]
- 14. Rebeaud F., Hailfinger S., Posevitz-Fejfar A., Tapernoux M., Moser R., Rueda D., Gaide O., Guzzardi M., Iancu E. M., Rufer N., Fasel N., Thome M. (2008) Nat. Immunol. 9, 272–281 [DOI] [PubMed] [Google Scholar]
- 15. Madeo F., Herker E., Maldener C., Wissing S., Lächelt S., Herlan M., Fehr M., Lauber K., Sigrist S. J., Wesselborg S., Fröhlich K. U. (2002) Mol. Cell 9, 911–917 [DOI] [PubMed] [Google Scholar]
- 16. Silva R. D., Sotoc A. R., Johansson B., Ludovico P., Sansonetty F., Silva M. T., Peinado J. M., Côrte-Real M. (2005) Mol. Microbiol. 58, 824–834 [DOI] [PubMed] [Google Scholar]
- 17. Hauptmann P., Riel C., Kunz-Schughart L. A., Fröhlich K. U., Madeo F., Lehle L. (2006) Mol. Microbiol. 59, 765–778 [DOI] [PubMed] [Google Scholar]
- 18. Guaragnella N., Pereira C., Sousa M. J., Antonacci L., Passarella S., Côrte-Real M., Marra E., Giannattasio S. (2006) FEBS Lett. 580, 6880–6884 [DOI] [PubMed] [Google Scholar]
- 19. Vercammen D., Belenghi B., van de Cotte B., Beunens T., Gavigan J. A., De Rycke R., Brackenier A., Inzé D., Harris J. L., Van Breusegem F. (2006) J. Mol. Biol. 364, 625–636 [DOI] [PubMed] [Google Scholar]
- 20. Belenghi B., Romero-Puertas M. C., Vercammen D., Brackenier A., Inzé D., Delledonne M., Van Breusegem F. (2007) J. Biol. Chem. 282, 1352–1358 [DOI] [PubMed] [Google Scholar]
- 21. Lam E. (2004) Nat. Rev. Mol. Cell Biol. 5, 305–315 [DOI] [PubMed] [Google Scholar]
- 22. Lecourieux D., Ranjeva R., Pugin A. (2006) New Phytol. 171, 249–269 [DOI] [PubMed] [Google Scholar]
- 23. Hofius D., Tsitsigiannis D. I., Jones J. D., Mundy J. (2007) Semin. Cancer Biol. 17, 166–187 [DOI] [PubMed] [Google Scholar]
- 24. Shi Y. (2002) Mol. Cell 9, 459–470 [DOI] [PubMed] [Google Scholar]
- 25. Potempa J., Sroka A., Imamura T., Travis J. (2003) Curr. Protein Pept. Sci. 4, 397–407 [DOI] [PubMed] [Google Scholar]
- 26. Zhou Q., Snipas S., Orth K., Muzio M., Dixit V. M., Salvesen G. S. (1997) J. Biol. Chem. 272, 7797–7800 [DOI] [PubMed] [Google Scholar]
- 27. Stennicke H. R., Salvesen G. S. (1997) J. Biol. Chem. 272, 25719–25723 [DOI] [PubMed] [Google Scholar]
- 28. Koeplinger K. A., Mildner A. M., Leone J. W., Wheeler J. S., Heinrikson R. L., Tomasselli A. G. (2000) Protein Expr. Purif. 18, 378–387 [DOI] [PubMed] [Google Scholar]
- 29. Mikolajczyk J., Boatright K. M., Stennicke H. R., Nazif T., Potempa J., Bogyo M., Salvesen G. S. (2003) J. Biol. Chem. 278, 10458–10464 [DOI] [PubMed] [Google Scholar]
- 30. Eichinger A., Beisel H. G., Jacob U., Huber R., Medrano F. J., Banbula A., Potempa J., Travis J., Bode W. (1999) EMBO J. 18, 5453–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Margetts M. B., Barr I. G., Webb E. A. (2000) Protein Expr. Purif. 18, 262–268 [DOI] [PubMed] [Google Scholar]
- 32. Chen Z., Potempa J., Polanowski A., Wikstrom M., Travis J. (1992) J. Biol. Chem. 267, 18896–18901 [PubMed] [Google Scholar]
- 33. Lampl N., Budai-Hadrian O., Davydov O., Joss T. V., Harrop S. J., Curmi P. M., Roberts T. H., Fluhr R. (2010) J. Biol. Chem. 285, 13550–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okada T., Ernst O. P., Palczewski K., Hofmann K. P. (2001) Trends Biochem. Sci. 26, 318–324 [DOI] [PubMed] [Google Scholar]
- 35. Giniatullin R., Nistri A., Yakel J. L. (2005) Trends Neurosci. 28, 371–378 [DOI] [PubMed] [Google Scholar]
- 36. Bush D. S. (1995) Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 95–122 [Google Scholar]
- 37. Reddy A. S., Safadi F., Beyette J. R., Mykles D. L. (1994) Biochem. Biophys. Res. Commun. 199, 1089–1095 [DOI] [PubMed] [Google Scholar]
- 38. Safadi F., Mykles D. L., Reddy A. S. (1997) Arch. Biochem. Biophys. 348, 143–151 [DOI] [PubMed] [Google Scholar]
- 39. Wang C., Barry J. K., Min Z., Tordsen G., Rao A. G., Olsen O. A. (2003) J. Biol. Chem. 278, 34467–34474 [DOI] [PubMed] [Google Scholar]
- 40. Kahn S., Verma G., Sharma S. (2010) J. Plant Physiol. 167, 855–861 [DOI] [PubMed] [Google Scholar]