Abstract
Corin is a cardiac membrane protease that activates natriuretic peptides. It is unknown how corin function is regulated. Recently, soluble corin was detected in human plasma, suggesting that corin may be shed from cardiomyocytes. Here we examined soluble corin production and activity and determined the proteolytic enzymes responsible for corin cleavage. We expressed human corin in HEK 293 cells and detected three soluble fragments of ∼180, ∼160, and ∼100 kDa, respectively, in the cultured medium by Western blot analysis. All three fragments were derived from activated corin molecules. Similar results were obtained in HL-1 cardiomyocytes. Using protease inhibitors, ionomycin and phorbol myristate acetate stimulation, small interfering RNA knockdown, and site-directed mutagenesis, we found that ADAM10 was primarily responsible for shedding corin in its juxtamembrane region to release the ∼180-kDa fragment, corresponding to the near-entire extracellular region. In contrast, the ∼160- and ∼100-kDa fragments were from corin autocleavage at Arg-164 in frizzled 1 domain and Arg-427 in LDL receptor 5 domain, respectively. In functional studies, the ∼180-kDa fragment activated atrial natriuretic peptide, whereas the ∼160- and ∼100-kDa fragments did not. Our data indicate that ADAM-mediated shedding and corin autocleavage are important mechanisms regulating corin function and preventing excessive, potentially hazardous, proteolytic activities in the heart.
Keywords: Membrane Enzymes, Membrane Proteins, Peptide Hormones, Proteolytic Enzymes, Serine Protease, Corin
Introduction
Natriuretic peptides act as a cardiac endocrine mechanism to regulate blood volume and pressure (1, 2). Corin is a serine protease that activates atrial natriuretic peptides (ANP)2 and B-type natriuretic peptides in the heart (3, 4). The physiological importance of corin function has been shown in mouse models, in which disruption of the corin gene causes hypertension and cardiac hypertrophy (5, 6). In humans, population studies have identified polymorphisms (T555I/Q568P) in the corin gene in African Americans who had high blood pressure and cardiac hypertrophy (7–9). The polymorphisms were shown to alter corin protein structure and impair its biological activity in functional studies (10). In patients with heart failure (HF), the corin variants were associated with poor natriuretic peptide processing and worse clinical outcomes (11).
Corin belongs to the type II transmembrane serine protease family, a subclass of trypsin-like enzymes defined by an N-terminal transmembrane domain and a C-terminal protease domain (12, 13). The transmembrane domain anchors the proteases on the cell surface, localizing the biological activities at specific sites. Under physiological and/or pathological conditions, type II transmembrane serine proteases can be shed from the cell surface. Soluble forms of enteropeptidase, hepsin and matriptases, for example, have been reported (14–17). Recently, soluble corin was detected in human blood, indicating that corin is shed from the heart (18–20). Interestingly, plasma corin levels were lower in patients with severe HF compared with those of normal controls or patients with mild HF (18, 21, 22), suggesting that corin shedding from the cells may be important in regulating corin function and that altered corin shedding and/or cleavage may play a role in HF.
The mechanism responsible for corin shedding was unknown. In this study, we studied corin shedding in cultured HEK 293 cells and HL-1 cardiomyocytes. We identified soluble corin fragments released either by ADAM (a disintegrin and metalloprotease)-mediated shedding or by corin autocleavage. We further examined the biological activity of these corin fragments in pro-ANP processing assays.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK 293 cells were grown in DMEM with 10% FBS. The mouse atrial HL-1 myocytes (23) were cultured in Claycomb medium (Sigma) with 10% FBS, 100 μm norepinephrine, and 4 mm l-glutamine in gelatin-coated plates. All cells were cultured at 37 °C in humidified incubators with 5% CO2 and 95% air.
Plasmid Constructs
Plasmids expressing human wild-type (WT) corin, active site mutant S985A, activation cleavage site mutant R801A, and frizzled-like domain 1 deletion mutant (ΔFz1) and rat corin were described previously (10, 24, 25). Plasmids expressing human corin mutants R134A, R164A, R180A, R213A, R239A, R244A, and R427A were made by PCR-based site-directed mutagenesis using WT corin plasmid as a template. The full-length mouse corin cDNA was amplified using mRNAs from heart samples and cloned in pcDNA vector (Invitrogen). Recombinant corin proteins expressed by these plasmids contained a C-terminal V5 tag to facilitate protein detection in Western blotting.
Transfection, Immunoprecipitation, and Western Blotting
Plasmids were transfected into HEK 293 and HL-1 cells using FuGENE (Roche Diagnostics) or Lipofectamine 2000 (Invitrogen) reagents, as described previously (25, 26). Conditioned medium was collected after 48–72 h, and recombinant proteins were immunoprecipitated with an anti-V5-antibody. Cells were lysed in a buffer containing 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% Nonidet P-40 (v/v), and a protease inhibitor mixture (Sigma, 1:100). Protein samples were analyzed by Western blotting with (reducing) or without (non-reducing) 2.5% β-mercaptoethanol in a sample buffer, as described previously (25, 26). ADAM10, ADAM17, and GAPDH proteins in cell lysate were analyzed by Western blotting using commercial antibodies (Millipore).
Effects of Protease Inhibitors on Corin Shedding
To identify proteases involved in corin cleavage, a panel of inhibitors including benzamidine (trypsin-like serine protease inhibitor, 5 mm), GM6001 (metalloproteinase inhibitor, 50 μm), TAPI-1 (ADAM inhibitor, 50 μm), N-acetyl-leucyl-leucyl-methionine (cysteine protease inhibitor, 50 μm), and a mixture of protease inhibitors (1:400) were added to corin-expressing cells in separate wells. The conditioned medium was collected after 19 h. Soluble corin fragments were analyzed by immunoprecipitation and Western blotting.
Effects of Ionomycin and PMA on Corin Shedding
Corin-expressing cells were treated with vehicle (dimethyl sulfoxide), ionomycin (2.5 μm) or phorbol 12-myristate 13-acetate (PMA) (200 ng/ml) for 30–45 min. Soluble corin in the conditioned medium and cellular corin in the lysate were analyzed by immunoprecipitation and Western blotting. The intensity of protein bands on Western blots was quantified using a densitometer (Bio-Rad). The ratio of soluble/cellular corin, an indicator of corin shedding, was calculated.
Analysis of ADAM10 and ADAM17 mRNA Expression
Total RNAs were isolated from HEK 293 and HL-1 cells with TRIzol reagents (Invitrogen). First-strand cDNAs were synthesized using the SuperScriptTM III kit (Invitrogen). Specific oligonucleotide primers were used in RT-PCR to amplify human or mouse ADAM10 and ADAM17 mRNAs in HEK 293 or HL-1 cells, respectively. In these experiments, negative controls included reactions done without cDNA templates. PCR products were separated on 1% agarose gels and visualized by ethidium bromide staining under UV light.
Knockdown ADAM10 and ADAM17 Expression
Six single siRNAs targeting different sequences of human and mouse ADAM10 genes (three siRNAs each) and pooled siRNAs for human and mouse ADAM10 and ADAM17 genes (Dharmacon) were used to transfect corin-expressing HEK 293 and HL-1 cells, respectively, at concentrations of 25–50 nm, according to the manufacturer's instructions. A non-targeting siRNA pool (Dharmacon) was used as a negative control, and siRNAs pools against the human and mouse GAPDH genes (Dharmacon) were used as positive controls. After transfection, cell culture medium was changed, and the cells were incubated for 72 h. The conditioned medium was collected, and the cell lysate was prepared. Soluble and cellular corin proteins were analyzed by immunoprecipitation and Western blotting.
Pro-ANP Processing Assay
Conditioned medium containing recombinant human pro-ANP was made from transfected cells. Conditioned medium containing soluble corin from transfected HEK 293 or HL-1 cells was collected, concentrated with an Amicon device (Millipore), and incubated with the pro-ANP medium at 37 °C. Control medium without soluble corin was used as a negative control, and cell lysate with full-length corin was used as a positive control. Pro-ANP and ANP in the medium were analyzed by immunoprecipitation and Western blotting, as described previously (24, 25).
RESULTS
Soluble Corin Fragments from HEK 293 and HL-1 Cells
To study corin cleavage from the cell surface, we expressed human corin in HEK 293 cells and mouse HL-1 cardiomyocytes and analyzed soluble corin fragments in the conditioned medium. By immunoprecipitation and Western blotting, we detected three distinct corin fragments of ∼180, ∼160, and ∼100 kDa, respectively, in both cell types (Fig. 1A, left lanes). These fragments were smaller than the full-length corin in cell lysates (Fig. 1A, right lanes), indicating that they were cleaved forms of corin.
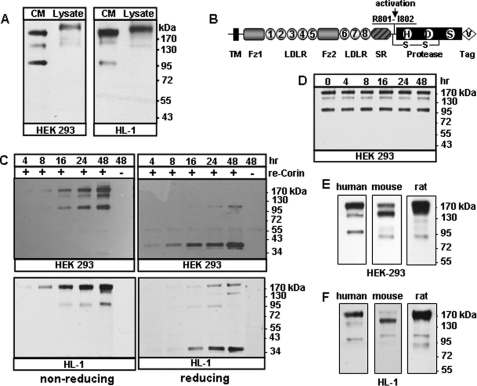
FIGURE 1.
Detection of soluble corin fragments. A, recombinant human corin proteins in the conditioned medium (CM) and cell lysate (Lysate) from transfected HEK 293 and HL-1 cells were analyzed by Western blotting. B, corin protein domains. The transmembrane (TM), frizzled-like (Fz), LDL receptor (LDLR), scavenger receptor (SR), and protease domains are shown. The activation cleavage site, active site residues His (H), Asp (D), and Ser (S), C-terminal V5 tag (V), and the disulfide bond connecting the propeptide and the protease domain are indicated. C, soluble corin fragments in the medium from HEK 293 (top) or HL-1 (bottom) cells expressing recombinant (re) corin collected over time were analyzed by Western blotting under non-reducing and reducing conditions. The ∼40-kDa bands in the right panels represent the activated corin protease domain. D, the conditioned medium was collected from transfected HEK 293 cells and incubated at 37 °C over time. Corin fragments were analyzed by Western blotting. E and F, human, mouse and rat soluble corin fragments in the conditioned medium from transfected HEK 293 and HL-1 cells, respectively. Data are from representative experiments repeated at least three times.
Corin is made as a zymogen. Cleavage at Arg-801 activates corin (3, 27). The activated protease domain, ∼40 kDa in length, is linked to the propeptide region by a disulfide bond (Fig. 1B). To examine the activation status of the soluble corin fragments, we analyzed the conditioned medium from the transfected HEK 293 and HL-1 cells by Western blotting under non-reducing and reducing conditions. As shown in Fig. 1C, soluble corin fragments appeared in the medium after ∼4–8 h and accumulated over time. Under reducing conditions (right panels), most of the corin fragments were reduced to ∼40 kDa, indicating that the ∼180-, ∼160-, and ∼100-kDa fragments were activated two-chain molecules linked by the disulfide bond.
To examine if the ∼160- and ∼100-kDa bands were derived from additional cleavage of the ∼180-kDa fragment after it was released from the cells, the conditioned medium was collected and incubated at 37 °C over time. Western-blot analysis showed that all three fragments remained at similar levels without the cells (Fig. 1D), suggesting that these fragments came directly from the cells and were stable in the conditioned medium.
We also examined the shedding of mouse and rat corin. In Western-blot analysis, soluble corin fragments were detected in the conditioned medium from the transfected HEK 293 (Fig. 1E) and HL-1 cells (Fig. 1F). Mouse corin had three major soluble fragments, similar to that of human corin, whereas rat corin had one predominant fragment of ∼180 kDa and two additional fragments of smaller size.
Effects of Protease Inhibitors
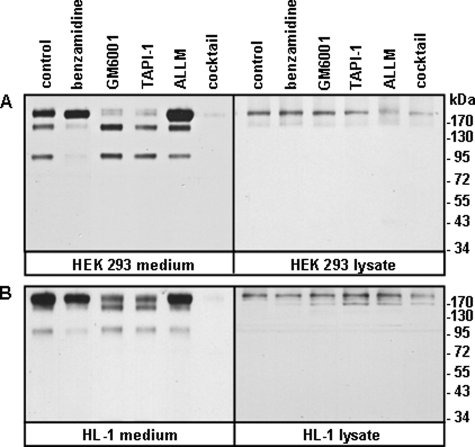
To identify the enzyme(s) responsible for corin shedding, we tested the effects of protease inhibitors on soluble corin generation in HEK 293 and HL-1 cells. As shown in Fig. 2, A and B (left panels), the level of the ∼180-kDa fragment was markedly reduced in the presence of a metalloproteinase inhibitor (GM6001) or an ADAM inhibitor (TAPI-1), but not trypsin-like serine protease (benzamindine) or cysteine protease (N-acetyl-leucyl-leucyl-methionine) inhibitors. In contrast, the levels of the ∼160- and ∼100-kDa fragments were inhibited by benzamidine but not the other inhibitors. As a positive control, all three fragments were inhibited by a protease inhibitor mixture. As another control, recombinant corin expression in HEK 293 and HL-1 cells was not inhibited by the inhibitors (Fig. 2, A and B, right panels). The results indicated that the ∼180-kDa fragment was cleaved by ADAM protease(s), whereas the ∼160- and ∼100-kDa fragments were cleaved by trypsin-like serine protease(s).
FIGURE 2.
Effects of protease inhibitors. HEK 293 (A) and HL-1 (B) cells expressing recombinant human corin were incubated with vehicle (control), benzamidine, GM6001, TAPI-1, N-acetyl-leucyl-leucyl-methionine, or protease inhibitor mixture. Soluble corin fragments in the conditioned medium were analyzed by Western blotting (left panels). Corin proteins in cell lysate were verified (right panels). Data are from representative experiments repeated at least three times.
ADAM-mediated Corin Shedding
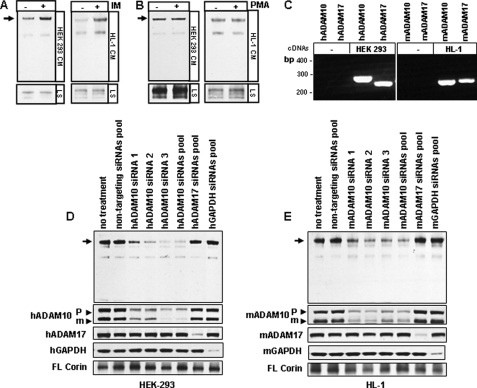
ADAM proteases are important in membrane protein shedding (28, 29). Among them, ADAM10 and ADAM17 are major sheddases whose activities are enhanced differentially by different stimuli. ADAM10 activity is stimulated by ionomycin but not PMA, whereas ADAM17 activity is stimulated by both ionomyocin and PMA (30, 31). We found that the ∼180-kDa fragment production in HEK 293 and HL-1 cells was enhanced by ionomyocin but not PMA (Fig. 3, A and B), suggesting that ADAM10 is a likely corin sheddase. By RT-PCR, we confirmed ADAM10 and ADAM17 mRNA expression in HEK 293 and HL-1 cells (Fig. 3C). In these cells, transfection of single or pooled siRNAs targeting different sequences of human or mouse ADAM10 gene inhibited the ∼180-kDa fragment production (Fig. 3, D and E). In contrast, transfection of siRNA pools against human or mouse ADAM17 or GAPDH genes had little effect. The reduction of ADAM10, ADAM17, and GAPDH protein expression was confirmed in the transfected cells (Fig. 3, D and E, lower panels). The results supported ADAM10 as a likely sheddase for the ∼180-kDa corin fragment.
FIGURE 3.
ADAM-mediated corin shedding. HEK 293 and HL-1 cells expressing recombinant corin were treated with vehicle, ionomycin (IM) (A) or PMA (B). Corin fragments in the conditioned medium (CM) and cell lysate (LS) were analyzed by Western blotting. C, expression of human (h) and mouse (m) ADAM10 and ADAM17 mRNAs in parental HEK 293 and HL-1 cells was confirmed by RT-PCR. HEK 293 (D) and HL-1 (E) cells expressing corin were transfected with single or pooled siRNAs against the human or mouse ADAM10 or ADAM17 genes. The production of the ∼180-kDa fragment (arrows) in the medium was reduced in ADAM10 siRNA-treated cells, as shown by Western blotting. Controls included non-targeting siRNAs pool or siRNAs pools against the human and mouse GAPDH genes. The reduction of ADAM10 (proform (P) and mature form (m) (arrow heads)), ADAM17, and GAPDH proteins in transfected cells was shown by Western blotting. Full-length (FL) corin in cell lysate also was shown. Data are from representative experiments that were repeated at least three times.
ADAM-mediated Shedding Site
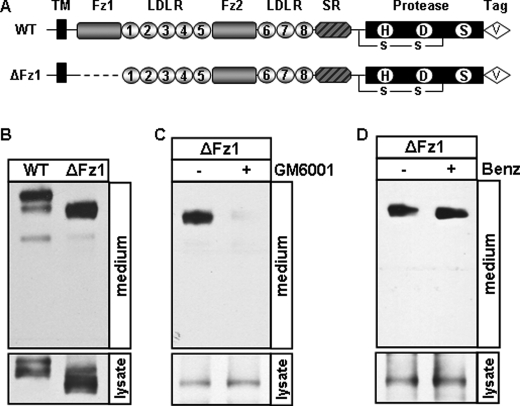
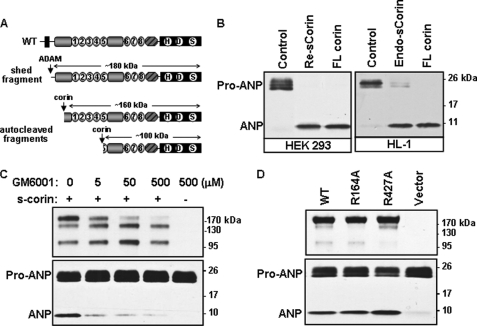
ADAM proteases typically cleave their substrates between the membrane domain and the first globular extracellular domain (32). Based on the apparent molecular mass, consistently, the ∼180-kDa fragment was predicted to be cleaved within the juxtamembrane region between the transmembrane and Fz1 domains. We verified this hypothesis with corin Fz1 domain deletion mutant ΔFz1 (Fig. 4A), which was expected to yield a shorter (<180 kDa) shed fragment. As shown in Fig. 4B, the ∼180-kDa fragment was shortened to ∼160 kDa. This shortened fragment was inhibited when the cells expressing the ΔFz1 mutant were cultured with GM6001 but not benzamidine (Fig. 4, C and D), indicating that the ∼160-kDa fragment was from ADAM-mediated shedding and supporting that the cleavage occurred in its juxtamembrane region before Fz1 domain.
FIGURE 4.
ADAM-mediated cleavage in the juxtamembrane domain. A, illustration of WT corin and the Fz1 deletion mutant (ΔFz1). HEK 293 cells expressing WT corin and the ΔFz1 mutant incubated without (B) or with GM6001 (C) or benzamidine (Benz) (D). Soluble corin fragments in the medium were analyzed by Western blotting. Lysate was included as a control. Data are from representative experiments that were repeated at least three times.
Soluble Corin Fragments Produced by Corin Autocleavage
Studies with protease inhibitors suggested that the ∼160- and ∼100-kDa fragments were generated by serine protease(s) (Fig. 2). Because corin is a serine protease, it is possible that these fragments were produced by corin autocleavage. We tested this hypothesis using corin mutant S985A, in which the catalytic Ser residue was replaced by Ala, and mutant R801A, in which the activation cleavage site residue Arg was replaced by Ala (Fig. 5A). Both mutations abolish corin catalytic activity (24, 27). As shown in Fig. 5, B and C, the conditioned medium from HEK 293 and HL-1 cells expressing these mutants contained only the ∼180-kDa fragment but no ∼160- and ∼100-kDa fragments, indicating that these two smaller fragments were generated by corin autocleavage.
FIGURE 5.
Soluble corin fragments from the S985A and R801A mutants. A, illustration of WT corin, active site mutant S985A, and activation cleavage site mutant R801A. Soluble corin fragments from transfected HEK 293 (B) and HL-1 (C) cells expressing WT corin and the S985A and R801A mutants were analyzed by Western blotting (left panels). Cell lysate was included as a control for corin expression levels (right panels). Data are from representative experiments that were repeated at least three times.
Identification of Corin Autocleavage Sites
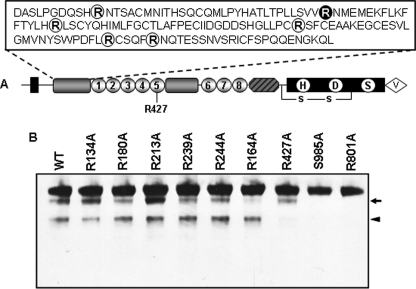
Based on their apparent molecular masses, the two corin autocleavage fragments were predicted to be cleaved in the Fz1 and LDLR5 domains, respectively (Fig. 6A). As a trypsin-like protease, corin was shown to have a preference to cleave after Arg residues (27). By site-directed mutagenesis, we made corin mutants replacing all single Arg with Ala residues within these two domains, including R134A, R164A, R180A, R213A, R239A, R244A, and R427A (Fig. 6A). Western-blot analysis of the conditioned medium from cells expressing these mutants showed that R164A mutation prevented the ∼160-kDa fragment production, whereas R427A mutation prevented the ∼100-kDa fragment production (Fig. 6B). In contrast, mutations at the other residues did not prevent the production of these two fragments. These data indicated that corin cleaved at R164 in the Fz1 domain and R427 in the LDLR5 repeat, generating the ∼160- and ∼100-kDa fragments, respectively.
FIGURE 6.
Corin autocleavage sites. A, illustration of Arg residues in the Fz1 and LDLR5 domains that were substituted by Ala in single mutants. B, soluble corin fragments in the medium from HEK 293 cells expressing WT corin and mutants were analyzed by Western blotting. The S985A and R801A mutants, in which autocleavage was abolished, were used as controls. The ∼160- and ∼100-kDa bands are indicated by an arrow or arrow head, respectively. Data are from representative experiments that were repeated at least three times.
Biological Activity of Soluble Corin Fragments
To test if soluble corin fragments are biologically active, we incubated the soluble corin fragments with human pro-ANP and analyzed pro-ANP processing. As shown in Fig. 7B (left panel), pro-ANP was converted to ANP with the conditioned medium containing soluble corin. In this experiment, the conditioned medium from parental HEK 293 cells was used as a negative control and full-length corin from cell lysate as a positive control. Similar results were found in the conditioned medium from HL-1 cells that express endogenous corin (Fig. 7B, right panel).
FIGURE 7.
The activity of soluble corin fragments. A, illustration of soluble corin fragments. B, pro-ANP processing activity. Conditioned medium containing recombinant (Re) or endogenous (Endo) soluble (s) corin from HEK 293 (left) or HL-1 (right) cells were incubated with pro-ANP. Control medium and cell lysate with full-length (FL) corin were included as negative and positive controls, respectively. Pro-ANP to ANP conversion was analyzed by Western blotting. C, HEK 293 cells expressing corin or parental HEK 293 cells were incubated with increasing doses of GM6001. Soluble corin fragments (top) and pro-ANP processing activity (bottom) in the medium were analyzed by Western blotting. D, soluble corin fragments (top) and pro-ANP processing activity (bottom) in the conditioned medium from HEK 293 cells expressing WT corin and mutants R164A and R427A were analyzed by Western blotting. Data are from representative experiments that were repeated at least three times.
To determine which soluble corin fragment is biologically active, we first inhibited the ADAM activity with GM6001, which prevented the ∼180-kDa fragment production (Fig. 7C, top panel). The pro-ANP processing activity was significantly inhibited with increasing concentrations of GM6001 (Fig. 7C, lower panel), indicating that the ∼180-kDa fragment was biologically active whereas the other two fragments had little activity. We also tested pro-ANP processing activity in the conditioned medium from HEK 293 cells expressing either corin mutant R164A or R427A, lacking the ∼160- or ∼100-kDa fragment, respectively (Fig. 7A). No significant reduction in pro-ANP processing activity was observed in these two mutants compared with that of WT (Fig. 7D). These data were consistent, indicating that ∼160- and ∼100-kDa fragments had little biological activity and that the majority of the observed pro-ANP processing activity was from the ∼180-kDa fragment.
DISCUSSION
Ectodomain shedding is an important posttranslational process in a variety of cell membrane proteins, including adhesion molecules, enzymes, cytokines, growth factors, and receptors (28, 29, 33, 34). Corin is a membrane protease that processes natriuretic peptides in cardiomyocytes (35, 36). To date, the mechanisms that regulate corin activity remain unclear. Recently, soluble corin was detected in human blood, suggesting that corin may be shed from the cells (18–20). In this study, we examined soluble corin production and activity and determined the proteolytic enzymes responsible for corin shedding.
In the cultured medium from transfected HEK 293 cells, we detected three distinct human corin fragments of ∼180, ∼160 and ∼100 kDa, respectively. Similar results were found when human corin was expressed in mouse atrial HL-1 cells, which retained all structural and functional characteristics of cardiomyocytes (23). Because of lacking suitable antibodies, we were unable to analyze endogenous mouse soluble corin in HL-1 cells. We expressed recombinant mouse and rat corin in HEK 293 and HL-1 cells and detected shed fragments of ∼180 kDa. Mouse and rat corin also had several soluble fragments that differed in molecular mass from those of autocleaved human fragments, which may reflect different corin sequences in these species. The data supported that corin shedding was physiologically relevant.
In experiments with protease inhibitors, ionomycin and PMA stimulation, and gene knockdown by siRNAs, we showed that the ADAM10, a major membrane protein sheddase, was most likely responsible for shedding the ∼180-kDa corin fragment. Studies have shown that ADAMs usually cleave their substrates between the membrane domain and the first globular extracellular domain (28, 29, 32, 37). The recognition mechanisms by these proteases largely depend on a substrate structure close to the cell membrane but not specific amino acid sequences. Consistent with these findings, we showed that ADAM-mediated cleavage occurred in the corin juxtamembrane domain. We also found that the ADAM-mediated shedding was not highly sequence-specific, as additional point mutations and small deletions within this region did not completely block the cleavage (data not shown). In mice, ADAM10 deficiency is lethal, resulting in early embryonic loss (38). It is unknown if corin expression in the heart is altered in these mice and if other ADAMs may compensate ADAM10 to shed corin in vivo.
Unlike the ∼180-kDa fragment, the ∼160- and ∼100-kDa fragments were generated by corin autocleavage, which was inhibited by benzamidine. Mutations at the corin active site, Ser-985, or the zymogen activation cleavage site, Arg-801, which abolish corin protease activity, prevented the production of these two fragments. By mutagenesis, we localized the autocleavage sites at Arg-164 in the Fz1 domain and Arg-427 in the LDLR5 repeat, respectively. Mutation at either Arg-164 or Arg-427 apparently did not alter the rate of cleavage at the other residue (Fig. 6). It appeared, therefore, that these two autocleavage fragments were generated independently but not sequentially.
In functional assays, the ∼180-kDa fragment, which represented the near-entire extracellular region of corin, exhibited the biological activity in processing pro-ANP. In contrast, the ∼160- and ∼100-kDa fragments had little activity. The results were consistent with our previous findings that the transmembrane domain is not required for corin to process pro-ANP, whereas the Fz1 and LDLR domains are important for the activity (24, 27). Thus, our results showed that ADAM-mediated shedding generated a long soluble corin that was biologically active. In contrast, corin cleaved itself, producing two shorter fragments that were inactive.
Excessive proteolytic activities are potentially hazardous. Regulatory mechanisms have evolved to control the activities to avoid undesired consequences. The activities of most trypsin-like proteases are controlled by endogenous inhibitors that are usually blood-bound. Surprisingly, corin-mediated pro-ANP processing was not inhibited by human plasma (27). To date, no physiological corin inhibitors have been identified. The corin shedding and autocleavage described here may represent an important mechanism in regulating corin activity. It is likely that once corin is activated and cleaves natriuretic peptides, active corin molecules are removed from the cell surface to prevent excessive proteolytic activities in the heart. This action appears to be carried out by an ADAM, most likely ADAM10. Corin inactivates the remaining molecules on the cell surface by autocleavage. Consistent with this working model, we found that most soluble fragments were derived from active corin, as indicated by a broken peptide bond at Arg-801 to Ile-802 (Fig. 1C), and that the ADAM-generated ∼180-kDa fragment appeared earlier in the medium than the other two autocleaved fragments (Fig. 1C).
Our finding that the ∼180-kDa fragment is biologically active suggests that corin may function not only in the heart but also in blood and other distant organs. Supporting this notion, two most recent studies showed natriuretic peptide processing activities in human blood (21, 39). Because plasma corin levels were low, ranging from pico- to nanogram per ml (18–20), it remains to be determined if the activity was indeed from the ∼180-kDa fragment. Interestingly, we and others found that plasma corin levels were lower in patients with severe HF compared with that in normal individuals or patients with less severe HF (18, 21, 22). We also found that cardiac corin protein expression, but not its activity, was increased in mouse and human failing hearts, indicating that corin activation is a rate-limiting step in HF (40). As shown in this study, corin shedding and autocleavage were closely related to its activation. It is possible that low levels of plasma corin may reflect inadequate corin activation in failing hearts. The reduction of the overall corin activity in the heart and blood is expected to impair the ANP pathway, which may exacerbate body fluid retention and poor cardiac function in patients with severe HF.
Acknowledgments
We thank William Claycomb for providing HL-1 cells and Carl Blobel for helpful suggestions on ADAMs.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL089298, R01HL089298-S1, and R01HD064634. This work was also supported by grants from the Ralph Wilson Medical Research Foundation, the Bakken Heart-Brain Institute, and by National Natural Science Foundation of China Grant 31070716.
- ANP
- atrial natriuretic peptide
- HF
- heart failure
- ADAM
- a disintegrin and metalloprotease
- PMO
- phorbol myristate acetate.
REFERENCES
- 1. McGrath M. F., de Bold M. L., de Bold A. J. (2005) Trends Endocrinol. Metab. 16, 469–477 [DOI] [PubMed] [Google Scholar]
- 2. Potter L. R., Abbey-Hosch S., Dickey D. M. (2006) Endocr. Rev. 27, 47–72 [DOI] [PubMed] [Google Scholar]
- 3. Yan W., Sheng N., Seto M., Morser J., Wu Q. (1999) J. Biol. Chem. 274, 14926–14935 [DOI] [PubMed] [Google Scholar]
- 4. Yan W., Wu F., Morser J., Wu Q. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8525–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan J. C., Knudson O., Wu F., Morser J., Dole W. P., Wu Q. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nigrovic P. A., Gray D. H., Jones T., Hallgren J., Kuo F. C., Chaletzky B., Gurish M., Mathis D., Benoist C., Lee D. M. (2008) Am. J. Pathol. 173, 1693–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dries D. L., Victor R. G., Rame J. E., Cooper R. S., Wu X., Zhu X., Leonard D., Ho S. I., Wu Q., Post W., Drazner M. H. (2005) Circulation 112, 2403–2410 [DOI] [PubMed] [Google Scholar]
- 8. Pan J., Hinzmann B., Yan W., Wu F., Morser J., Wu Q. (2002) J. Biol. Chem. 277, 38390–38398 [DOI] [PubMed] [Google Scholar]
- 9. Rame J. E., Drazner M. H., Post W., Peshock R., Lima J., Cooper R. S., Dries D. L. (2007) Hypertension 49, 857–864 [DOI] [PubMed] [Google Scholar]
- 10. Wang W., Liao X., Fukuda K., Knappe S., Wu F., Dries D. L., Qin J., Wu Q. (2008) Circ. Res. 103, 502–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rame J. E., Tam S. W., McNamara D., Worcel M., Sabolinski M. L., Wu A. H., Dries D. L. (2009) Circ. Heart Fail. 2, 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bugge T. H., Antalis T. M., Wu Q. (2009) J. Biol. Chem. 284, 23177–23181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hooper J. D., Clements J. A., Quigley J. P., Antalis T. M. (2001) J. Biol. Chem. 276, 857–860 [DOI] [PubMed] [Google Scholar]
- 14. Hadorn B., Steiner N., Sumida C., Peters T. J. (1971) Lancet 1, 165–166 [DOI] [PubMed] [Google Scholar]
- 15. Lin C. Y., Tseng I. C., Chou F. P., Su S. F., Chen Y. W., Johnson M. D., Dickson R. B. (2008) Front Biosci. 13, 621–635 [DOI] [PubMed] [Google Scholar]
- 16. Stirnberg M., Maurer E., Horstmeyer A., Kolp S., Frank S., Bald T., Arenz K., Janzer A., Prager K., Wunderlich P., Walter J., Gütschow M. (2010) Biochem. J. 430, 87–95 [DOI] [PubMed] [Google Scholar]
- 17. Wu Q. (2001) Front Biosci. 6, D192–200 [DOI] [PubMed] [Google Scholar]
- 18. Dong N., Chen S., Yang J., He L., Liu P., Zheng D., Li L., Zhou Y., Ruan C., Plow E., Wu Q. (2010) Circ. Heart Fail. 3, 207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dong N., Dong J., Liu P., Xu L., Shi S., Wu Q. (2010) Clin. Chim. Acta 411, 1998–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peleg A., Jaffe A. S., Hasin Y. (2009) Clin. Chim. Acta 409, 85–89 [DOI] [PubMed] [Google Scholar]
- 21. Ibebuogu U. N., Gladysheva I. P., Houng A. K., Reed G. L. (2011) Circ. Heart Fail., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shrestha K., Troughton R. W., Borowski A. G., Yandle T. G., Richards A. M., Klein A. L., Tang W. H. (2010) J. Card. Fail. 16, 621–627 [DOI] [PubMed] [Google Scholar]
- 23. Claycomb W. C., Lanson N. A., Jr., Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., Izzo N. J., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knappe S., Wu F., Madlansacay M. R., Wu Q. (2004) J. Biol. Chem. 279, 34464–34471 [DOI] [PubMed] [Google Scholar]
- 25. Liao X., Wang W., Chen S., Wu Q. (2007) J. Biol. Chem. 282, 27728–27735 [DOI] [PubMed] [Google Scholar]
- 26. Jiang J., Pristera N., Wang W., Zhang X., Wu Q. (2010) Clin. Chem. 56, 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Knappe S., Wu F., Masikat M. R., Morser J., Wu Q. (2003) J. Biol. Chem. 278, 52363–52370 [DOI] [PubMed] [Google Scholar]
- 28. Blobel C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 32–43 [DOI] [PubMed] [Google Scholar]
- 29. Reiss K., Saftig P. (2009) Semin. Cell Dev. Biol. 20, 126–137 [DOI] [PubMed] [Google Scholar]
- 30. Le Gall S. M., Bobé P., Reiss K., Horiuchi K., Niu X. D., Lundell D., Gibb D. R., Conrad D., Saftig P., Blobel C. P. (2009) Mol. Biol. Cell 20, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sahin U., Weskamp G., Kelly K., Zhou H. M., Higashiyama S., Peschon J., Hartmann D., Saftig P., Blobel C. P. (2004) J. Cell Biol. 164, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Becherer J. D., Blobel C. P. (2003) Curr. Top. Dev. Biol. 54, 101–123 [DOI] [PubMed] [Google Scholar]
- 33. Garton K. J., Gough P. J., Raines E. W. (2006) J. Leukocyte Biol. 79, 1105–1116 [DOI] [PubMed] [Google Scholar]
- 34. Murphy G. (2009) Semin. Cell Dev. Biol. 20, 138–145 [DOI] [PubMed] [Google Scholar]
- 35. Wu Q. (2007) Front Biosci. 12, 4179–4190 [DOI] [PubMed] [Google Scholar]
- 36. Wu Q., Xu-Cai Y. O., Chen S., Wang W. (2009) Kidney Int. 75, 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X., He K., Gerhart M., Huang Y., Jiang J., Paxton R. J., Yang S., Lu C., Menon R. K., Black R. A., Baumann G., Frank S. J. (2002) J. Biol. Chem. 277, 50510–50519 [DOI] [PubMed] [Google Scholar]
- 38. Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., von Figura K., Saftig P. (2002) Hum. Mol. Genet. 11, 2615–2624 [DOI] [PubMed] [Google Scholar]
- 39. Ichiki T., Huntley B. K., Heublein D. M., Sandberg S. M., McKie P. M., Martin F. L., Jougasaki M., Burnett J. C., Jr. (2011) Clin. Chem. 57, 40–47 [DOI] [PubMed] [Google Scholar]
- 40. Chen S., Sen S., Young D., Wang W., Moravec C. S., Wu Q. (2010) Am. J. Physiol. Heart Circ. Physiol. 299, H1687–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]