Abstract
The copper-transporting P1B-type ATPases (Cu-ATPases) ATP7A and ATP7B are key regulators of physiological copper levels. They function to maintain intracellular copper homeostasis by delivering copper to secretory compartments and by trafficking toward the cell periphery to export excess copper. Mutations in the genes encoding ATP7A and ATP7B lead to copper deficiency and toxicity disorders, Menkes and Wilson diseases, respectively. This report describes the interaction between the Cu-ATPases and clusterin and demonstrates a chaperone-like role for clusterin in facilitating their degradation. Clusterin interacted with both ATP7A and ATP7B in mammalian cells. This interaction increased under conditions of oxidative stress and with mutations in ATP7B that led to its misfolding and mislocalization. A Wilson disease patient mutation (G85V) led to enhanced ATP7B turnover, which was further exacerbated when cells overexpressed clusterin. We demonstrated that clusterin-facilitated degradation of mutant ATP7B is likely to involve the lysosomal pathway. The knockdown and overexpression of clusterin increased and decreased, respectively, the Cu-ATPase-mediated copper export capacity of cells. These results highlight a new role for intracellular clusterin in mediating Cu-ATPase quality control and hence in the normal maintenance of copper homeostasis, and in promoting cell survival in the context of disease. Based on our findings, it is possible that variations in clusterin expression and function could contribute to the variable clinical expression of Menkes and Wilson diseases.
Keywords: Apolipoproteins, ATPases, Copper, Metals, Protein Degradation, ATP7A, ATP7B, Apolipoprotein J, Clusterin, Molecular Chaperones
Introduction
Menkes (MD)2 and Wilson (WD) diseases are genetically inherited disorders of copper metabolism. They illustrate the essential requirement for copper and the dire consequences of its dysregulation (1). MD is an X-linked copper deficiency disorder that arises due to mutation of ATP7A (OMIM 309400). The primary site of the defect are the intestinal enterocytes, which take up dietary copper, but absorption into the portal circulation is impaired due to defective ATP7A-mediated copper efflux (2, 3). Consequently, systemic copper deficiency accounts for the wide range of neurological and developmental defects (1). WD is an autosomal recessive disorder of copper toxicity that results from mutation of ATP7B (OMIM 277900). Defective ATP7B-mediated biliary copper excretion leads to chronic copper accumulation in the liver and often in the brain. Patients develop liver failure and/or neurological symptoms such as movement disorders, seizures, and psychosis (1).
ATP7A and ATP7B are copper-transporting P1B-type ATPases (Cu-ATPases) and are key regulators of physiological copper status. They are large transmembrane proteins with six N-terminal metal-binding domains (MBDs). Each MBD comprises ∼70 amino acids, contains the highly conserved copper-binding motif GMXCXXC, and binds one copper ion. ATP7A and ATP7B carry out ATP-dependent copper translocation across cellular membranes for metallation of secretory cuproenzymes and for efflux of excess copper as a protective mechanism (reviewed in Ref. 4). ATP7A and ATP7B reside at the trans-Golgi network, where they receive copper from the copper chaperone ATOX1. With increased intracellular copper levels, ATP7A and ATP7B traffic to exocytic vesicles near the basolateral or apical membranes, respectively (2, 3, 5, 6). When copper levels are restored, they recycle back to the trans-Golgi network (5).
For both MD and WD, there is a high degree of clinical variability (7, 8). For MD, ∼170 different mutations in ATP7A have been reported, whereas ∼400 different mutations of ATP7B have been described (Human Gene Mutation Database). Although genotype/phenotype correlations have been difficult, it is generally accepted that the clinical expression of these diseases and patient response to treatment depends on the type of mutation in ATP7A/ATP7B, which in turn can affect the levels and activity of ATP7A/ATP7B, their post-translational modifications, protein-protein interactions, cellular localization, and/or ability to traffic in response to copper (7–9). For both MD and WD, there are reports of identical mutations, even among siblings, conferring variable clinical expression (10, 11), thus implicating other factors in determining the clinical phenotype. Environmental factors and allelic variants of modifying genes may contribute to the diversity of patient symptoms and onset of disease. COMMD1 is a protein that is absent in Bedlington terrier dogs with a copper toxicosis syndrome similar to WD (12). Heterozygosity for a specific silent missense mutation in COMMD1 possibly resulted in an earlier onset of WD in patients with known ATP7B mutations (13). Hence, COMMD1 may represent a modifier gene affecting the clinical presentation of WD (13).
Clusterin (Apolipoprotein J) was among the potential ATP7B-interacting partners identified previously in a yeast two-hybrid screen (14). Clusterin is a 449-amino acid, heterodimeric glycoprotein that interacts with a variety of proteins. It is ubiquitously expressed and present in most body fluids with the highest expression in the brain (15). Clusterin has been implicated in pathological conditions in which oxidative stress plays a central role such as aging, neurodegenerative diseases, and cancer progression (16). Based on its large repertoire of unrelated binding partners, it has been also implicated in diverse physiological processes such as lipid transport, cell differentiation, regulation of apoptosis, clearance of cellular debris, and stabilization of misfolded proteins (16). Both cytoprotective and cytotoxic roles for clusterin have been reported. Functionally, clusterin is similar to the small heat shock proteins with chaperone-like activity, binding to stressed and misfolded proteins (17). Much of the work on clusterin has focused on its role as an extracellular chaperone (18, 19), binding to exposed hydrophobic regions of proteins and maintaining them in a state competent for subsequent refolding by other chaperones, e.g. HSP70 (20–22).
In this study, we investigated the role of clusterin in regulating the activity of ATP7A and ATP7B and its participation in copper homeostasis. We demonstrated an interaction between clusterin and the Cu-ATPases that was modulated by oxidative stress and mutations in ATP7B that led to its misfolding. We further demonstrated that clusterin has a role in facilitating the degradation of misfolded or unstable ATP7B, and its overexpression and knockdown affected intracellular copper levels. Therefore, clusterin plays a key role in Cu-ATPase quality control, which is necessary for maintaining normal copper homeostasis and cell survival in the context of disease. Our findings support the possibility that clusterin gene (CLU) variations that affect its expression, function or efficiency could be one mechanism that underlies the variable clinical phenotypes of MD and WD. Given the emphasis on extracellular clusterin, the Cu-ATPases also serve as a valuable model for deciphering the role of intracellular clusterin in protein turnover.
EXPERIMENTAL PROCEDURES
Constructs and Reagents
ATP7B cDNA constructs encoding WT ATP7B, ATP7B(MBD 1–6del), and ATP7B(MBD 3–5del), and N-terminal FLAG-tagged WT-ATP7B-FLAG and ATP7B-G85V-FLAG were generated previously (23, 24). Clusterin cDNA in pIREShyg1 (Clontech) was kindly provided by Saverio Bettuzzi (University of Parma). pTriEx/ATP7B-N was kindly provided by Svetlana Lutsenko (Johns Hopkins University, Baltimore, MD). Clusterin siRNA was purchased from Dharmacon (catalogue no. L-019513-00-0005). The metal response element (MRE)-luciferase and Renilla reporter constructs were generated previously (25).
Cell Culture and Transfection
HepG2, HEK293T, and CHO-K1 cells were cultured as described (5). Human neuroblastoma M17 cells were cultured at 37 °C in Opti-Mem® reduced serum medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum. Where indicated, the growth medium was supplemented with 50–200 μm CuCl2, 50–500 μm bathocuproinedisulfonic acid (BCS), 50–500 μm D-penicillamine, 0.5–2 mm hydrogen peroxide, 2–10 mm diamide, 53 μm cycloheximide, 5 μm lactacystin, or 5 μm NH4Cl, prior to harvesting cells. HEK293T cells were transfected by calcium phosphate precipitation as described previously (25). The generation of stably transfected CHO-K1 cell lines was described previously (23).
Western Blotting and Immunoprecipitation
Mammalian cells were harvested and lysed as described (26). For co-immunoprecipitations, protein G-coupled magnetic beads (Dynabeads Protein G, Invitrogen) were used according to recommended protocols. Briefly, ∼3 mg of total cell protein was applied to Dynabeads prepared with immobilized antibody, either sheep anti-ATP7B (NC36, ammonium sulfate precipitated) (23) (∼50 μg), sheep anti-ATP7A (R17-BX, affinity purified) (27) (∼3 μg), rabbit anti-ZnT1 or preimmune serum. Protein complexes were eluted with 1 m glycine, pH 2.0, neutralized in 1 m Tris-HCl, pH 8.0, and analyzed by Western blotting together with ∼50 μg total protein extracts.
Protein extracts were fractionated by SDS-PAGE and immunoblotted with the following antibodies: sheep anti-ATP7B (diluted 1/1000) (23), rabbit anti-(mouse) Atp7b (diluted 1/1000) (28), sheep anti-ATP7A (diluted 1/1000) (27), or goat anti-clusterin (diluted 1/1000) (Sigma or Santa Cruz Biotechnology). To control for protein loading either mouse monoclonal anti-β-actin (diluted 1/5000) (Sigma) or rabbit anti-β-tubulin antibodies (diluted 1/5000) (Abcam) were used. FLAG-tagged proteins were detected using HRP-conjugated mouse anti-FLAG M2 antibody (diluted 1/5000) (Sigma). Secondary antibodies used included HRP-conjugated sheep anti-rabbit IgG (Millipore), rabbit anti-goat IgG (Sigma), and sheep anti-mouse IgG (Dako) (diluted 1/10,000). Proteins were detected using Immobilon Western HRP Substrate Peroxide Solution (Millipore), and the images were captured using the Luminescent Image Analyzer LAS-3000 (Fujifilm).
Preparation of Mouse Liver Tissues
Adult mice were sacrificed by CO2 asphyxiation, and the livers were harvested and frozen on dry ice. Liver tissue protein extracts were prepared in lysis buffer (26), fractionated by SDS-PAGE, and immunoblotted with the appropriate antibodies.
Luciferase Reporter Assay
The luciferase reporter assay was carried out essentially as described previously (25). Briefly, HEK293T cells were seeded into a 96-well plate and transfected with the Renilla-luciferase and MRE-luciferase reporter constructs, then co-transfected with the pEBB-FLAG empty vector, pIREShyg1/clusterin, clusterin siRNA, pEBB/WT-ATP7B-FLAG, pEBB/WT-ATP7B-FLAG plus pIREShyg1/clusterin, or pEBB/WT-ATP7B-FLAG plus clusterin siRNA. After 24 h, the cells were either left untreated or treated with 150 μm CuCl2 (16 h). The cells were rinsed with PBS, harvested in passive lysis buffer (Promega) according to the manufacturer's protocol, and assayed by luminometry (Centro LB960, Berthold Technologies) using the Dual-Luciferase® reporter assay system (Promega) for firefly and Renilla luciferase activity. The relative light units were calculated by dividing firefly luciferase measurements by Renilla luciferase measurements. All values are expressed as fold induction relative to the pEBB empty vector control or pEBB/WT-ATP7B-FLAG incubations (set as 1).
Measurement of Total Copper
Copper in cell pellets of transfected HEK293T cells was measured as described (5). Cells were seeded into 25-cm2 flasks and transfected with 5 μg of plasmid DNA for 48 h. The medium was supplemented with 150 μm CuCl2 for the final 16 h, and then cell pellets were prepared for copper measurements. Copper in cell pellets was measured by inductively coupled plasma MS (UltraMass 700, Varian). Within each experiment, the average of triplicate determinations was used for comparisons.
Statistical Analysis
Unless otherwise indicated, data are reported as means ± S.E., where n = at least 3. Statistical analysis was carried out using Student's t test.
RESULTS
Fluctuations in Intracellular Copper Promotes Clusterin Interaction with ATP7B
Coimmunoprecipitation experiments using HepG2 cells verified the yeast two-hybrid data and showed that endogenous ATP7B and clusterin interact in mammalian cells (Fig. 1A). The anti-ATP7B antibody immunoprecipitated both ATP7B and clusterin, whereas an unrelated antibody anti-ZnT1, and preimmune serum did not precipitate either protein (Fig. 1A). The two bands sometimes observed in the clusterin immunoblots may represent different glycosylated forms of clusterin as it is known to be heavily glycosylated (29). In the reverse experiment, an anti-clusterin antibody also was able to immunoprecipitate ATP7B and clusterin (data not shown). ATP7B-clusterin interaction in mouse liver was confirmed by co-immunoprecipitation experiments using a mouse anti-Atp7b antibody (Fig. 1B). Although clusterin was identified in a yeast two-hybrid screen with the ATP7B N terminus as a bait (14), it also interacted with ATP7B lacking the N terminus (MBD 1–6del), and hence the interaction was not dependent on the MBDs (supplemental Fig. 1). When HepG2 intracellular copper levels were manipulated either by copper supplementation (CuCl2, 24 h) or chelation (BCS and D-penicillamine, 72 h) (Fig. 1, C and D, respectively), the degree of ATP7B-clusterin interaction increased in a dose-dependent manner. The additional lower molecular weight band detected in the ATP7B immunoblots likely represents a degradation product of ATP7B that was previously characterized (30). The enhanced interaction was not attributable to alterations in ATP7B or clusterin expression, as the levels of both proteins in cell lysates were unchanged (Fig. 1, C and D). Therefore, intracellular copper levels influence ATP7B and clusterin interaction, with any variance from the basal level stimulating an increased interaction.
FIGURE 1.
The interaction between ATP7B and clusterin. Coimmunoprecipitation (co-IP) of ATP7B and clusterin from HepG2 cells (A), mouse liver tissues (B), HepG2 cells supplemented with CuCl2 (0–200 μm for 24 h) (C), and HepG2 cells incubated with BCS/D-penicillamine (D-Pen; 0–500 μm for 72 h) (D). Proteins were fractionated and immunoblotted with anti-ATP7B, anti-(mouse) Atp7b, anti-clusterin, and anti-β-actin antibodies. Densitometric analysis of immunoprecipitated clusterin is shown (C(ii) and D(ii)), expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different from control. *, p < 0.05.
Oxidative Stress Promotes Clusterin Interaction with ATP7B
Clusterin activity is initiated in response to cellular stresses (17, 20), and both copper excess and deficiency are known to cause oxidative stress in cells (31, 32). To determine whether the interaction is modulated directly by oxidative stress, co-immunoprecipitation experiments were repeated using HepG2 cells treated with either H2O2 (20 min) or diamide (15 min) (Fig. 2, A and B, respectively). Both oxidizing agents increased the ATP7B-clusterin interaction in a dose-dependent manner. The levels of hemoxygenase-1, a commonly used marker of oxidative stress, increased after treating the cells with H2O2, diamide, CuCl2, and BCS/D-penicillamine (Fig. 2C). These results suggested that both copper excess and deficiency enhance the interaction between ATP7B and clusterin through the generation of oxidative stress, rather than representing a direct copper-specific effect.
FIGURE 2.
The interaction between ATP7B and clusterin increases under oxidative stress. Coimmunoprecipitation (Co-IP) of ATP7B and clusterin from HepG2 cells following treatment with H2O2 (0–2 mm for 20 min) (A) (i) and diamide (0–10 mm for 15 min) (B) (i). Cell lysates and co-immunoprecipitated proteins were fractionated and immunoblotted with anti-ATP7B, anti-clusterin, and anti-β-actin antibodies. C (i), hemoxygenase-1 (HO-1) expression levels in HepG2 cells treated with CuCl2 (0–200 μm for 24 h), BCS/D-penicillamine (D-Pen; 0–200 μm for 72 h), H2O2 (0–2 mm for 20 min) or diamide (0–10 mm for 15 min). Cell lysates were fractionated and immunoblotted with anti-hemoxygenase-1 and anti-β-actin antibodies. Densitometric analyses of immunoprecipitated clusterin (A(ii) and B(ii)) or hemoxygenase-1 in cell lysates (C(ii)) are shown, expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different from control. *, p < 0.05.
Clusterin Interacts with ATP7A
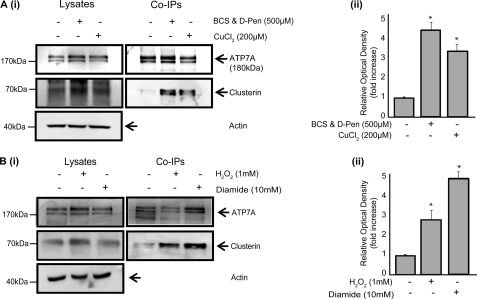
Co-immunoprecipitation experiments carried out using the ATP7A-expressing M17 neuroblastoma cells revealed that clusterin also interacts with ATP7A (Fig. 3). Similar to ATP7B, this interaction was substantially increased when cells were exposed to fluctuations in copper levels (Fig. 3A) or to oxidative stress mediated by H2O2 or diamide (Fig. 3B).
FIGURE 3.
The interaction betwen ATP7A and clusterin. Coimmunoprecipitation (Co-IP) of ATP7A and clusterin from M17 cells supplemented with CuCl2 (200 μm for 24 h) (A) (i) or BCS/D-penicillamine (D-Pen; 500 μm for 72 h) and H2O2 (0–2 mm for 20 min) or diamide (0–10 mm for 15 min) (B) (i). Cell lysates and coimmunoprecipitated proteins were fractionated by SDS-PAGE and immunoblotted with anti-ATP7A, anti-clusterin, and anti-β-actin antibodies. Densitometric analysis of immunoprecipitated clusterin is shown (A(ii) and B(ii)), expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different from control. *, p < 0.05.
Clusterin Binds to Misfolded ATP7B Protein
We investigated the possibility that clusterin interacts to a greater extent with misfolded, unstable ATP7B variants. We generated N-terminally truncated variants of ATP7B, which localized throughout the cell in a reticulated pattern, indicative of misfolded protein that is retained in the endoplasmic reticulum (23) (Fig. 4). Retention in the endoplasmic reticulum is common for misfolded ATP7B variants (7, 24), which often regain their subcellular localization and function when synthesized at a permissive temperature of 30 °C (5, 7, 33). Western blot and immunofluorescence analysis of CHO-K1 cells that expressed these truncated variants in which MBDs 1–6 (MBD 1–6del) or 3–5 (MBD 3–5del) were deleted (Fig. 4A), verified that these deletions caused temperature-sensitive instability (supplemental Fig. 2).
FIGURE 4.
Increased clusterin binding to unstable ATP7B mutants MBD1–6del and MBD3–5del. A, schematic representation of the N terminus of WT-ATP7B, MBD1–6del, and MBD3–5del. B (i), coimmunoprecipitation (Co-IP) of clusterin and WT-ATP7B, MBD1–6del, or MBD3–5del from CHO-K1 cells cultured at either 37 or 30 °C. Cell lysates and coimmunoprecipitated proteins were fractionated and immunoblotted with anti-ATP7B, anti-clusterin, and anti-β-actin antibodies. B (ii) densitometric analysis of immunoprecipitated clusterin relative to immunoprecipitated ATP7B is shown, expressed as relative optical density, and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different. *, p < 0.05.
To determine whether ATP7B instability influenced its interaction with clusterin, we investigated clusterin binding to the truncated ATP7B variants at both 30 °C and 37 °C (Fig. 4B). For the truncated variants, while their expression levels were increased at 30 °C compared with 37 °C, the amount of clusterin that co-precipitated with them was markedly decreased at 30 °C compared with 37 °C (Fig. 4B). We concluded that more clusterin binds to unstable than to stable ATP7B proteins.
Clusterin Facilitates Degradation of ATP7B
Previous studies suggested that clusterin both increases protein stability and targets proteins for degradation. To determine whether clusterin interaction with ATP7B provides stability or facilitates its degradation, we investigated the impact of clusterin overexpression on WT-ATP7B and a mutant ATP7B protein (ATP7B-G85V) derived by introducing a WD patient mutation (Human Gene Mutation Database) (Fig. 5). This mutant protein is unstable and localizes to the endoplasmic reticulum, characteristic of a misfolded ATP7B protein (24). HEK293T cells were co-transfected with plasmids encoding clusterin and either WT-ATP7B (Fig. 5, A and B) or ATP7B-G85V (Fig. 5, C and D). Although there is slight variability in transient expression levels of clusterin between independent experiments, clusterin levels are clearly increased above endogenous levels, and the overall trends relating to its effects on ATP7B levels are clearly apparent. Levels of WT-ATP7B were decreased when clusterin was overexpressed compared with cells only expressing WT-ATP7B (Fig. 5, A and B, compare lanes 1 and 4). This decrease was more pronounced for ATP7B-G85V co-expressed with clusterin compared with ATP7B-G85V alone (Fig. 5, C and D, compare lanes 1 and 4). Clusterin co-expression in the presence of the protein synthesis inhibitor cycloheximide led to a further decrease in the level of ATP7B proteins (Fig. 5, A–D, lanes 4 and 5). We concluded that clusterin facilitates ATP7B degradation.
FIGURE 5.
Clusterin facilitates the degradation of ATP7B. HEK293T cells were transfected with cDNA constructs encoding WT-ATP7B-FLAG (A and B) and ATP7B-G85V-FLAG (C and D) without and with clusterin cDNA constructs. Cells were incubated with cycloheximide (CHX; 8 h) in the absence and presence of lactacystin (C; 8 h) or NH4Cl (D; 20 min). Cell lysates were immunoblotted with anti-FLAG, anti-clusterin, and anti-β-actin antibodies. Densitometric analysis of ATP7B levels are expressed as relative optical density and represent the mean ± S.E. (n = 3).
To determine the involvement of the proteasomal and lysosomal degradation pathways, lactacystin and NH4Cl were used to inhibit these pathways, respectively. With lactacystin treatment, there was partial rescue of ATP7B-G85V levels but not WT-ATP7B(Fig. 5, A and C, lanes 2 and 3). With clusterin overexpression in combination with lactacystin treatment, there was no evidence of rescue but a further reduction in WT-ATP7B and ATP7B-G85V levels (Fig. 5, A and C, lanes 3 and 6 and lanes 5 and 6). We concluded that the proteasomal degradation pathway is unlikely to play a significant role in clusterin-facilitated ATP7B proteolysis. NH4Cl treatment also resulted in partial rescue of ATP7B-G85V levels both without and with clusterin overexpression, implicating the lysosomal pathway in clusterin-facilitated ATP7B degradation (Fig. 5D). A similar rescue effect was evident with WT-ATP7B but only when clusterin was overexpressed (Fig. 5B, lanes 4–6). The incomplete rescue (back to basal levels) was likely due to the fact that clusterin overexpression preceded NH4Cl treatment, so that ATP7B degradation was already underway before NH4Cl was added. These data confirmed that clusterin facilitates the degradation of misfolded ATP7B and indicated that the lysosomal pathway is likely to be involved in this clusterin-facilitated degradation of ATP7B.
Clusterin Influences Cellular Copper Concentrations
To determine whether clusterin levels affected intracellular copper levels, an MRE-luciferase reporter assay was employed (25). The MRE-luciferase reporter acts as a copper sensor that responds to bioavailable cytosolic copper (25). HEK293T cells were transiently transfected with the MRE-luciferase reporter and the Renilla-luciferase control constructs. In addition, these cells were co-transfected with constructs encoding WT-ATP7B-FLAG, clusterin, clusterin siRNA, or an empty vector as a control. Transfected cells were incubated without added copper or with 150 μm CuCl2 (16 h) before luminescence was measured. Under basal conditions, there was an increase in reporter gene induction when clusterin was overexpressed compared with vector-transfected cells and a decrease in induction when clusterin gene expression was knocked down (Fig. 6A). Following copper supplementation, there was a 10-fold increase in reporter activity, although the pattern of response remained similar. Therefore, increased clusterin expression led to increased cytosolic copper, possibly due to a reduction in Cu-ATPase molecules. Reduced clusterin expression led to increased copper export and less cytosolic copper available to activate reporter gene expression. When ATP7B was overexpressed, the effects of variations in clusterin expression on cytosolic copper levels were similar but only evident following copper supplementation when enhanced Cu-ATPase-mediated copper export capacity was required (Fig. 6B). ATP7B and clusterin levels following clusterin overexpression and knockdown are shown (Fig. 6C).
FIGURE 6.
Clusterin levels affect the copper export capacity of ATP7B. A, HEK293T cells were transfected with the MRE-luciferase reporter construct and co-transfected with the pEBB-FLAG empty vector, pIREShyg1/clusterin, or clusterin siRNA. Cells were either left untreated or incubated with 150 μm CuCl2 (16 h). Luminescence was measured, and data are expressed as fold induction relative to the pEBB-FLAG empty vector. B, HEK293T cells were transfected with the MRE-luciferase reporter construct and co-transfected with pEBB/WT-ATP7B-FLAG, pEBB/WT-ATP7B-FLAG plus pIREShyg1/clusterin or pEBB/WT-ATP7B-FLAG plus clusterin siRNA. Luminescence was measured and data are expressed as fold induction relative to WT-ATP7B-FLAG. C, cell lysates following the above transfections were fractionated and immunoblotted with anti-FLAG, anti-clusterin, and anti-β-tubulin antibodies. D, densitometric analysis of ATP7B and clusterin levels expressed as relative optical density and representing the mean ± S.E. (n = 3). Asterisks indicate values that are significantly different. *, p < 0.05. E and F, cells transfected as above were left untreated or were supplemented with 150 μm CuCl2 (16 h) and then harvested for copper analysis by inductively coupled plasma MS. Data are expressed as the mean ± S.E. (n = 4). Asterisks indicate values that are significantly different. *, p < 0.05 versus empty vector and ATP7B alone controls. KD, knockdown.
This experiment was repeated, but total copper levels were measured following transfection with clusterin constructs and copper supplementation. Copper levels increased substantially when clusterin was overexpressed. However, in contrast to cytosolic copper levels, there was a modest increase in total copper when clusterin was knocked down, relative to the vector control, both in the presence and absence of exogenously expressed ATP7B (Figs. 6E and 5F). Together, these results suggested that clusterin levels can affect the copper export capacity of the cell through regulating the levels of functionally active Cu-ATPase molecules.
DISCUSSION
This study demonstrated that clusterin facilitates the degradation of stressed, unstable, and misfolded forms of the Cu-ATPases. Clusterin interacted with endogenous ATP7A and ATP7B, and the amount of interacting clusterin increased when cells were stressed by altered copper levels or by treatment with oxidizing agents H2O2 or diamide. This result together with increased clusterin binding to unstable and misfolded ATP7B variants, and clusterin interaction with a truncated ATP7B protein lacking the copper-binding N terminus, indicated that this interaction is not copper-dependent but rather is favored by protein misfolding. Clusterin interaction with a wide range of proteins has been extensively documented in the literature, and it is now apparent that the primary function of clusterin underlying its many interactions is its chaperone action that involves binding to misfolded proteins (19). There is compelling evidence that the chaperone activity of clusterin involves hydrophobic interactions with its targets and is likely to be important in the quality control of protein folding (16, 17, 21, 22) A recent report demonstrated that clusterin formed high molecular weight soluble complexes with citrate synthetase, fibrinogen, and glutathione S-transferase, binding to exposed hydrophobic regions on these proteins following exposure to stress (20). Clusterin also interacts with COMMD1 and promotes its ubiquitination and subsequent degradation via the proteasomal pathway (34), highlighting the fact that clusterin may target different proteins for degradation by either or both of the proteasomal and lysosomal pathways. Our observations are consistent with a chaperone-like role for clusterin binding to misfolded Cu-ATPase molecules and mediating their quality control.
We confirmed previous observations (24) that the WD patient mutation, G85V led to the production of a less stable ATP7B protein. Clusterin targeted this variant ATP7B for degradation that was partially mediated by the lysosomal pathway. This conclusion was based on three observations: (i) more clusterin bound to mutated and unstable ATP7B than to stable protein; (ii) clusterin overexpression led to a more substantial decrease in the amount of mutant ATP7B-G85V compared with the WT-ATP7B protein; and (iii) incubation of cells with an inhibitor of the lysosomal degradation pathway, NH4Cl, limited ATP7B degradation in the presence of clusterin, compared with little or no effect observed with lactacystin, a proteasomal pathway inhibitor. COMMD1, which is absent in Bedlington terrier dogs with a copper toxicosis syndrome similar to WD (12), also interacts with ATP7A (7) and ATP7B (24) and showed increased binding to misfolded ATP7B variants (including G85V and G591D) arising from WD patient mutations (24). However, COMMD1 targeted the mutant ATP7B proteins for degradation partially via the proteasomal pathway (24). Therefore, ATP7B may be a substrate for both the proteasomal and lysosomal pathways via COMMD1 and clusterin.
Both ATP7A and ATP7B are expressed in HEK293T (35) cells, making them a useful model to study the effects of clusterin on intracellular copper. Clearly, variations in clusterin expression affected the levels of both cytosolic and total copper. Increased clusterin levels led to increased cytosolic and total cellular copper levels. This observation was consistent with reduced copper export (via ATP7A) and/or sequestration (via ATP7B) (35), respectively, resulting from a reduction in Cu-ATPase levels when clusterin was overexpressed. Clusterin knockdown led to decreased cytosolic copper levels, but a modest increase in total cellular copper. Decreased Cu-ATPase degradation with reduced clusterin levels likely led to reduced cytosolic copper levels consistent with enhanced sequestration and export. However, in HEK293T cells, the total cellular copper represents the balance between copper sequestration and copper export. As ATP7B is thought to be primarily involved in copper sequestration (5, 35), enhanced ATP7B-mediated copper sequestration may account for the modest increase in total copper and is also consistent with the overall higher total copper levels seen with increased ATP7B levels (Fig. 6, E and F). These data suggest that WT Cu-ATPase molecules that are recognized by clusterin are likely to be misfolded to some extent but still retain some level of activity. Note that the observed changes in intracellular copper levels with alterations in clusterin levels cannot be attributed with clusterin-mediated effects on the major copper importer Ctr1. That is, if clusterin overexpression led to reduced Ctr1 levels, then intracellular copper levels would be expected to decrease, and if clusterin knockdown led to increased Ctr1 levels, copper levels would increase.
The role of clusterin as an extracellular chaperone is well established (16, 19, 36). For example, its interaction with β-amyloid promotes β-amyloid clearance and uptake (via cell surface megalin receptor) and subsequent degradation (36). Recent association of single nucleotide polymorphisms at the CLU gene locus with Alzheimer disease (37, 38) supports the suggestion that both CLU and APOE may act as modifying genes to cooperatively regulate the deposition and clearance of β-amyloid (39), which may affect the onset and/or clinical expression of the disease. Similarly clusterin and COMMD1, by affecting the efficiency of clearance of mutant Cu-ATPase molecules, may play a role in modifying the clinical expression of Menkes and Wilson diseases. Whether they function cooperatively or redundantly remains to be established. Recently, the pharmacological folding chaperones such as 4-phenylbutyrate and curcumin showed potential to rescue the folding defects of ATP7B harboring patient mutations (40) and may overcome some of the effects of variations in these potential modifying genes.
Our data support a role for clusterin in copper homeostasis and adds the Cu-ATPases to the growing list of clusterin substrates. Intracellular (nuclear and cytosolic) forms of clusterin exist, but the mechanisms responsible for the derivation of these forms remain poorly defined, and less is known about its intracellular role (16, 19). Clusterin potentially represents the only chaperone that regulates protein stability both extra- and intracellularly (16). Therefore, the Cu-ATPases provide a new model toward understanding the role of intracellular clusterin. This work also provides an insight into the mechanisms of Cu-ATPase quality control, which is necessary for the maintenance of normal copper homeostasis and cell survival in disease states in the context of continuous synthesis of mutant Cu-ATPase molecules. Overall, the results from this study support the possibility that variations in clusterin alleles could contribute to the variability in the clinical expression of Menkes and Wilson diseases.
Acknowledgments
We thank Peter van den Berghe (University Medical Center, Utrecht, the Netherlands) for critically reading the manuscript, Svetlana Lutsenko (Johns Hopkins University, Baltimore, MD) for kindly providing the pTriEx/ATP7B-N plasmid, Saverio Bettuzzi (University of Parma, Italy) for kindly providing the full-length clusterin cDNA in pIREShyg1, Leigh Ackland (Deakin University) for providing the anti-ZnT1 antibody, and Roxana Llanos and Alison Blake (Deakin University) and Willianne Vonk (University Medical Center, Utrecht, the Netherlands) for technical support.
This work was supported by grants from The National Health and Medical Research Council of Australia (NHMRC; to S. L. and J. F. B. M.), an NHMRC biomedical research fellowship (to M. A. C.), and Dutch Digestive Disease Foundation Grant WS02-34, The Netherlands Organisation for Scientific Research (NWO) Program Grant 912.04.106, and NWO Earth and Life Sciences Grant 817.02.022 (to L. W. J. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Figs. 1 and 2.
- MD
- Menkes disease
- WD
- Wilson disease
- BCS
- bathocuproinedisulfonic acid
- Cu-ATPase
- copper-transporting P1B-type ATPase
- MBD
- metal binding domain
- MRE
- metal-responsive element.
REFERENCES
- 1. Danks D. M. (1995) in The Metabolic and Molecular Basis of Inherited Disease (Scriver C. R., Beaudet A. L., Sly W. M., Valle D. eds) pp. 2211–2235, McGraw-Hill, New York [Google Scholar]
- 2. Monty J. F., Llanos R. M., Mercer J. F., Kramer D. R. (2005) J. Nutr. 135, 2762–2766 [DOI] [PubMed] [Google Scholar]
- 3. Nyasae L., Bustos R., Braiterman L., Eipper B., Hubbard A. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1181–1194 [DOI] [PubMed] [Google Scholar]
- 4. Lutsenko S., Barnes N. L., Bartee M. Y., Dmitriev O. Y. (2007) Physiol. Rev. 87, 1011–1046 [DOI] [PubMed] [Google Scholar]
- 5. Cater M. A., La Fontaine S., Shield K., Deal Y., Mercer J. F. (2006) Gastroenterology 130, 493–506 [DOI] [PubMed] [Google Scholar]
- 6. Guo Y., Nyasae L., Braiterman L. T., Hubbard A. L. (2005) Am. J. Physiol. Gastrointest. Liver. Physiol. 289, G904–916 [DOI] [PubMed] [Google Scholar]
- 7. de Bie P., Muller P., Wijmenga C., Klomp L. W. (2007) J. Med. Genet. 44, 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tümer Z., Møller L. B. (2010) Eur. J. Hum. Genet. 18, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. La Fontaine S., Mercer J. F. (2007) Arch. Biochem. Biophys. 463, 149–167 [DOI] [PubMed] [Google Scholar]
- 10. Borm B., Møller L. B., Hausser I., Emeis M., Baerlocher K., Horn N., Rossi R. (2004) J. Pediatr. 145, 119–121 [DOI] [PubMed] [Google Scholar]
- 11. Duc H. H., Hefter H., Stremmel W., Castañeda-Guillot C., Hernández A., Cox D. W., Auburger G. (1998) Eur. J. Hum. Genet. 6, 616–623 [DOI] [PubMed] [Google Scholar]
- 12. van de Sluis B. J., Breen M., Nanji M., van Wolferen M., de Jong P., Binns M. M., Pearson P. L., Kuipers J., Rothuizen J., Cox D. W., Wijmenga C., van Oost B. A. (1999) Hum. Mol. Genet. 8, 501–507 [DOI] [PubMed] [Google Scholar]
- 13. Stuehler B., Reichert J., Stremmel W., Schaefer M. (2004) J. Mol. Med. 82, 629–634 [DOI] [PubMed] [Google Scholar]
- 14. Lim C. M., Cater M. A., Mercer J. F., La Fontaine S. (2006) J. Biol. Chem. 281, 14006–14014 [DOI] [PubMed] [Google Scholar]
- 15. Aronow B. J., Lund S. D., Brown T. L., Harmony J. A., Witte D. P. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 725–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trougakos I. P., Gonos E. S. (2006) Free. Radic. Res. 40, 1324–1334 [DOI] [PubMed] [Google Scholar]
- 17. Humphreys D. T., Carver J. A., Easterbrook-Smith S. B., Wilson M. R. (1999) J. Biol. Chem. 274, 6875–6881 [DOI] [PubMed] [Google Scholar]
- 18. Wilson M. R., Yerbury J. J., Poon S. (2008) Mol. Biosyst. 4, 42–52 [DOI] [PubMed] [Google Scholar]
- 19. Wyatt A., Yerbury J., Poon S., Dabbs R., Wilson M. R. (2009) Adv. Cancer. Res. 104, 89–114 [DOI] [PubMed] [Google Scholar]
- 20. Wyatt A. R., Yerbury J. J., Wilson M. R. (2009) J. Biol. Chem. 284, 21920–21927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poon S., Rybchyn M. S., Easterbrook-Smith S. B., Carver J. A., Pankhurst G. J., Wilson M. R. (2002) J. Biol. Chem. 277, 39532–39540 [DOI] [PubMed] [Google Scholar]
- 22. Poon S., Easterbrook-Smith S. B., Rybchyn M. S., Carver J. A., Wilson M. R. (2000) Biochemistry 39, 15953–15960 [DOI] [PubMed] [Google Scholar]
- 23. Cater M. A., Forbes J., La Fontaine S., Cox D., Mercer J. F. (2004) Biochem. J. 380, 805–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Bie P., van de Sluis B., Burstein E., van de Berghe P. V., Muller P., Berger R., Gitlin J. D., Wijmenga C., Klomp L. W. (2007) Gastroenterology 133, 1316–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van den Berghe P. V., Folmer D. E., Malingré H. E., van Beurden E., Klomp A. E., van de Sluis B., Merkx M., Berger R., Klomp L. W. (2007) Biochem. J. 407, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tao T. Y., Liu F., Klomp L., Wijmenga C., Gitlin J. D. (2003) J. Biol. Chem. 278, 41593–41596 [DOI] [PubMed] [Google Scholar]
- 27. Ke B. X., Llanos R. M., Wright M., Deal Y., Mercer J. F. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R1460–1467 [DOI] [PubMed] [Google Scholar]
- 28. La Fontaine S., Theophilos M. B., Firth S. D., Gould R., Parton R. G., Mercer J. F. (2001) Hum. Mol. Genet. 10, 361–370 [DOI] [PubMed] [Google Scholar]
- 29. Kapron J. T., Hilliard G. M., Lakins J. N., Tenniswood M. P., West K. A., Carr S. A., Crabb J. W. (1997) Protein. Sci. 6, 2120–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bartee M. Y., Ralle M., Lutsenko S. (2009) Biochemistry 48, 5573–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagasaka H., Takayanagi M., Tsukahara H. (2009) J. Toxicol. Sci. 34, 229–236 [DOI] [PubMed] [Google Scholar]
- 32. Rice R. H., Vidrio E. A., Kumfer B. M., Qin Q., Willits N. H., Kennedy I. M., Anastasio C. (2009) Chem. Biol. Interact. 181, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Payne A. S., Kelly E. J., Gitlin J. D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zoubeidi A., Ettinger S., Beraldi E., Hadaschik B., Zardan A., Klomp L. W., Nelson C. C., Rennie P. S., Gleave M. E. (2010) Mol. Cancer. Res. 8, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barnes N., Bartee M. Y., Braiterman L., Gupta A., Ustiyan V., Zuzel V., Kaplan J. H., Hubbard A. L., Lutsenko S. (2009) Traffic 10, 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nuutinen T., Suuronen T., Kauppinen A., Salminen A. (2009) Brain. Res. Rev. 61, 89–104 [DOI] [PubMed] [Google Scholar]
- 37. Lambert J. C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M. J., Tavernier B., Letenneur L., Bettens K., Berr C., Pasquier F., Fiévet N., Barberger-Gateau P., Engelborghs S., De Deyn P., Mateo I., Franck A., Helisalmi S., Porcellini E., Hanon O., de Pancorbo M. M., Lendon C., Dufouil C., Jaillard C., Leveillard T., Alvarez V., Bosco P., Mancuso M., Panza F., Nacmias B., Bossù P., Piccardi P., Annoni G., Seripa D., Galimberti D., Hannequin D., Licastro F., Soininen H., Ritchie K., Blanché H., Dartigues J. F., Tzourio C., Gut I., Van Broeckhoven C., Alpérovitch A., Lathrop M., Amouyel P. (2009) Nat. Genet. 41, 1094–1099 [DOI] [PubMed] [Google Scholar]
- 38. Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M. L., Pahwa J. S., Moskvina V., Dowzell K., Williams A., Jones N., Thomas C., Stretton A., Morgan A. R., Lovestone S., Powell J., Proitsi P., Lupton M. K., Brayne C., Rubinsztein D. C., Gill M., Lawlor B., Lynch A., Morgan K., Brown K. S., Passmore P. A., Craig D., McGuinness B., Todd S., Holmes C., Mann D., Smith A. D., Love S., Kehoe P. G., Hardy J., Mead S., Fox N., Rossor M., Collinge J., Maier W., Jessen F., Schürmann B., van den Bussche H., Heuser I., Kornhuber J., Wiltfang J., Dichgans M., Frölich L., Hampel H., Hüll M., Rujescu D., Goate A. M., Kauwe J. S., Cruchaga C., Nowotny P., Morris J. C., Mayo K., Sleegers K., Bettens K., Engelborghs S., De Deyn P. P., Van Broeckhoven C., Livingston G., Bass N. J., Gurling H., McQuillin A., Gwilliam R., Deloukas P., Al-Chalabi A., Shaw C. E., Tsolaki M., Singleton A. B., Guerreiro R., Mühleisen T. W., Nöthen M. M., Moebus S., Jöckel K. H., Klopp N., Wichmann H. E., Carrasquillo M. M., Pankratz V. S., Younkin S. G., Holmans P. A., O'Donovan M., Owen M. J., Williams J. (2009) Nat. Genet. 41, 1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DeMattos R. B., Cirrito J. R., Parsadanian M., May P. C., O'Dell M. A., Taylor J. W., Harmony J. A., Aronow B. J., Bales K. R., Paul S. M., Holtzman D. M. (2004) Neuron 41, 193–202 [DOI] [PubMed] [Google Scholar]
- 40. van den Berghe P. V., Stapelbroek J. M., Krieger E., de Bie P., van de Graaf S. F., de Groot R. E., van Beurden E., Spijker E., Houwen R. H., Berger R., Klomp L. W. (2009) Hepatology 50, 1783–1795 [DOI] [PubMed] [Google Scholar]