Abstract
Mycoplasma hyopneumoniae is the causative pathogen of porcine enzootic pneumonia, an economically significant disease that disrupts the mucociliary escalator in the swine respiratory tract. Expression of Mhp107, a P97 paralog encoded by the gene mhp107, was confirmed using ESI-MS/MS. To investigate the function of Mhp107, three recombinant proteins, F1Mhp107, F2Mhp107, and F3Mhp107, spanning the N-terminal, central, and C-terminal regions of Mhp107 were constructed. Colonization of swine by M. hyopneumoniae requires adherence of the bacterium to ciliated cells of the respiratory tract. Recent studies have identified a number of M. hyopneumoniae adhesins that bind heparin, fibronectin, and plasminogen. F1Mhp107 was found to bind porcine heparin (KD ∼90 nm) in a dose-dependent and saturable manner, whereas F3Mhp107 bound fibronectin (KD ∼180 nm) at physiologically relevant concentrations. F1Mhp107 also bound porcine plasminogen (KD = 24 nm) in a dose-dependent and physiologically relevant manner. Microspheres coated with F3Mhp107 mediate adherence to porcine kidney epithelial-like (PK15) cells, and all three recombinant proteins (F1Mhp107-F3Mhp107) bound swine respiratory cilia. Together, these findings indicate that Mhp107 is a member of the multifunctional M. hyopneumoniae adhesin family of surface proteins and contributes to both adherence to the host and pathogenesis.
Keywords: Fibronectin, Heparin Binding protein, Multifunctional Protein, Plasminogen, Surface Plasmon Resonance (SPR), Mycoplasma Hyopneumoniae, Adhesin
Introduction
Mycoplasma hyopneumoniae is the etiological agent of porcine enzootic pneumonia. During colonization, M. hyopneumoniae adheres to ciliated epithelial cells of the swine respiratory tract (1). Colonization of respiratory tissues initially induces a loss of cilia and impeded ciliary function, followed by damage to epithelial cells (14, 17, 23). The mechanisms leading to this damage remain largely unknown (2).
Gene and protein paralogs are related through genetic duplication events and often retain similar but not necessarily identical functions. M. hyopneumoniae expresses a family of surface protein paralogs, some of which have been shown to contribute to adherence to the host. Research with M. hyopneumoniae is complicated by a lack of genetic tools available to manipulate this genome-reduced pathogen (3, 4). P97, also referred to as the cilium adhesin, was the first protein implicated in adherence (5). P97 is encoded by mhp183, the first gene in a two-gene operon with mhp182. The mhp182 gene encodes P102, a cell surface-located protein (5). Genes mhp183 and mhp182 have six paralogs within the M. hyopneumoniae genome (6) that may be differentially expressed and provide a reservoir of antigenic and functional variation (4, 7–10). Pathogens may utilize differential expression to avoid recognition by the host immune system (5). Several Mycoplasma species possess adhesins in gene families or multigene operons that display antigenic variation (11–13).
All of the currently described M. hyopneumoniae adhesins belong to the P97/P102 paralog family, with the exception of P159, which is nonetheless located immediately upstream of a P97 paralog. The mhp183 gene is expressed as a 125-kDa preprotein that undergoes a posttranslational cleavage event to create the functional 97-kDa protein P97 (14, 15). P97 is directly involved in the adherence of M. hyopneumoniae to host respiratory cilia (9) and contains two heparin-binding domains (8). Heparin is a glycosaminoglycan able to inhibit adherence of M. hyopneumoniae to swine cilia in vitro (16). Recruitment of heparin to the mycoplasmal surface provides a possible bridging molecule that may bind a range of extracellular matrix components, thereby increasing the potential of M. hyopneumoniae to colonize the host (8, 17). However, P97 is not exclusively responsible for adherence of M. hyopneumoniae, as binding to cilia is still observed when adherence via P97 is blocked (15). Recent research has uncovered several other proteins, including P159 (18), P216 (4), Mhp271 (7), and P116 (10) as M. hyopneumoniae adhesins. Of these, P159, P216, and Mhp271 are heparin-binding proteins (4, 7, 18).
The host extracellular matrix contains several possible ligands for M. hyopneumoniae adherence (16). The extracellular matrix component fibronectin is often central to the adherence of pathogenic bacteria to host cells. Adherence to fibronectin may also trigger cytoskeletal rearrangements and host cell invasion (19, 20). Fibronectin-binding domains have been identified in Mhp271 (7) and P116 (10). The domain of P116 able to bind fibronectin also contains a C-terminal lysine that mediates adherence to plasminogen (10). Plasmin(ogen) is present in both plasma and basement membranes; active plasmin is a serine protease capable of hydrolyzing extracellular matrix components including fibronectin, collagen, and laminin (21). Damage to cells by plasmin(ogen) can also expose extracellular matrix components, increasing the potential for adherence by a colonizing pathogen (22). There is evidence that, like heparin, plasminogen may also function as a bridging molecule to contribute to bacterial attachment (23). All of the proteins in the P97/P102 paralog family studied thus far have been characterized as adhesins. These exhibit a common mechanism of utilizing extracellular matrix components to contribute to the binding of host cilia by M. hyopneumoniae. An ability to recognize and adhere to components of the extracellular matrix is broadly applicable to a wide range of bacterial species, enabling bacteria to anchor to the host, thus establishing colonization (24).
A putative 120-kDa P97 paralog (Mhp107) is encoded by the gene mhp107. The contribution of P97 and related paralogs to adherence and colonization by M. hyopneumoniae is the basis for our interest in establishing whether Mhp107 is a member of this multifunctional adhesin family. This study utilized proteomic analyses to determine expression and surface location of Mhp107. Characterization of the binding of recombinant Mhp107 fragments to heparin, fibronectin, and plasminogen was undertaken. The ability of Mhp107 to interact with porcine kidney epithelial-like (PK15) cells and swine cilia was established, implying a role for Mhp107 in adherence and colonization of M. hyopneumoniae.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
M. hyopneumoniae strains were grown in Friis medium and harvested as described previously (25). M. hyopneumoniae strain 232 (26) was used throughout this study unless stated otherwise. Additional strains of M. hyopneumoniae were isolated from various locations, including the laboratory-adapted strain J (25) and field strains OMZ407 (25), Hillcrest (isolated from New South Wales, Australia in 2003 and provided by Graeme Eamens at the Elizabeth Macarthur Agricultural Institute, New South Wales, Australia), 95MP1509 (27), 00MP1301 (27), C1735/2 (25), and 2–22421 (provided by J. Forbes-Faulkner, Oonoonba Veterinary Laboratory, Queensland, Australia). Escherichia coli TOP10 or BL21 star (DE3) (Invitrogen) were grown on Luria-Bertani agar plates or cultured in Luria-Bertani medium with shaking at 200 rpm. Ampicillin was added to media at 100 μg/ml when required.
Plasmid Constructs, Expression, and Purification
Polymerase chain reactions used PfuUltratm High-Fidelity II DNA polymerase (Stratagene) and a cooled gradient palm cycler GC1–96 (Corbett Research, Australia). Primer sequences are provided in supplemental Table S1. Amplification of mhp107 from M. hyopneumoniae strain 232 chromosomal DNA and subsequent cloning using the pET151/D-TOPO® cloning kit (Invitrogen) to construct pET151:mhp107 was performed following the manufacturer's instructions. As TGA encodes tryptophan in M. hyopneumoniae, site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) was performed to change in-frame TGA codons to TGG in the pET151:mhp107 construct. Regions of the pET151:mhp107 construct were then amplified and cloned using the pET151/D-TOPO® cloning kit to generate the recombinant polyhistidine fusion proteins described in this study (F1Mhp107-F3Mhp107). DNA sequencing was performed on the resulting recombinant plasmid constructs to ensure conformity to mhp107. Expression of recombinant proteins was in E. coli strain BL21 star (DE3). Recombinant plasmid constructs were transformed into this strain, and once cell growth reached an A600 of 0.8, protein expression was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside. Purification of proteins was performed under denaturing conditions using nickel-nitrilotriacetic acid-agarose in accordance with the manufacturer's instructions (Qiagen). Purified proteins were dialyzed into 0.01% SDS in PBS (10 mm sodium phosphate, 137 mm sodium chloride, 2 mm potassium phosphate, 2.7 mm potassium chloride (pH 7.4)).
Polyclonal Antisera Production and Western Blot Analysis
Antisera against recombinant Mhp107 protein fragments were raised in New Zealand White rabbits with three injections of antigen at 2-week intervals, using a method described previously (8). Serum was separated from clotted blood by centrifugation at 2,500 × g for 10 min. The resulting serum was centrifuged at 16,000 × g for 5 min to pellet remaining red blood cells. Antisera were affinity-purified using the antigen protein bound to the nitrocellulose membrane according to Seymour et al. (10). The method described by Cole et al. (28) was utilized for Western blot analysis.
Proteomic Analyses
Detection of Mhp107 expression by M. hyopneumoniae was undertaken using ESI-MS/MS analysis of M. hyopneumoniae proteins separated by SDS-PAGE of M. hyopneumoniae aqueous phase proteins. Extraction of the M. hyopneumoniae aqueous phase proteins using Triton X-114 was performed as described by Wise and Kim (29), with initial overnight cell lysis at 4 °C in 1% Triton X-114, 10 mm Tris (pH 8.0), 150 mm NaCl, and 1 mm EDTA. ESI2-MS/MS analysis was performed as described previously (10); identified peptides are detailed in supplemental Table S2. Immunoblot analyses were used in addition to ESI-MS/MS analysis to further elucidate Mhp107 expression. Immunoblots were performed with M. hyopneumoniae cell lysates, and aqueous phase proteins were probed with the purified polyclonal antisera. Immunoblot analysis was also employed to investigate variations in the expression and processing of Mhp107 among M. hyopneumoniae field strains. Immunoblots were performed using cell lysates probed with purified polyclonal antisera. The subcellular location of Mhp107 was identified using trypsin-treated intact M. hyopneumoniae as described previously (30). Trypsin treatment was performed with trypsin concentrations of 0–150 μg/ml. Cell lysates were analyzed by immunoblot with the purified polyclonal antisera; a control blot was probed with L7/L12, a ribosomal protein found in the cytosol (18), to verify the integrity of M. hyopneumoniae cells following trypsin treatment. All immunoblot analyses were performed alongside negative control blots probed with affinity-purified preimmune sera.
Heparin Binding Assays
The ability of recombinant Mhp107 protein fragments to interact with heparin was assessed in microtiter plates based on the method described by Burnett et al. (18). Minor modification of this method included allowing 1 μg of recombinant protein to adsorb overnight in each well of 96-well plates (Nunc). Wells were blocked in 2% skim milk in PBS prior to 1.5-h incubation with 0–50 μg/ml biotinylated heparin (Calbiochem) with and without 1 mg/ml unlabeled heparin to determine the level of nonspecific binding observed in total binding results. Competitive binding assays were also performed according to Burnett et al. (18).
Surface Plasmon Resonance Analyses
Interactions of recombinant Mhp107 protein fragments with fibronectin and plasminogen were investigated on a Biacore T100 instrument (Biacore AB, Sweden). Preparation of recombinant proteins, ligand immobilization, and kinetics assays were performed as described previously (10). Porcine plasminogen was purified using affinity chromatography (31, 32) followed by gel filtration and ultrafiltration. The activity and purity of plasminogen was assessed with Spectrozyme PL (American Diagnostica, Inc.) (10). Fibronectin and plasminogen were used as the ligands in the surface plasmon resonance kinetics assays, and binding of recombinant Mhp107 proteins was performed over a range of concentrations (0–500 nm). Additional cycles were performed with 200 nm F1Mhp107 in the presence of 10, 50, and 100 μm ϵ-aminocaproic acid, a lysine analog, to determine whether lysine residues were responsible for the observed plasminogen binding. Analysis of F3Mhp107 association kinetics with fibronectin were determined as the sum of two exponentials for a heterogeneous surface as described by Deutscher et al. (7). All other binding data analysis used the 1:1 Langmuir binding model with BiaEvaluation software 3.1 (Biacore AB).
PK15 Cell Adherence Assay
The ability of recombinant Mhp107 fragments to promote binding to PK15 cells was determined using protein-coated microspheres. Coating of Fluoresbrite® polychromatic red polystyrene microspheres (Polysciences) was performed according to Seymour et al. (10), and adherence assays were also performed as described previously (10, 18). Briefly, PK15 cells were grown to confluence before varying periods of incubation with protein-coated microspheres. PK15 cells were washed, fixed, and stained before visualization by confocal microscopy. One-way analyses of variance with a Dunnett's comparison against results from BSA-coated beads were performed with GraphPad Prism version 4 software (GraphPad) to determine significance.
Cilia Binding Assay
The cilia-binding ability of recombinant Mhp107 fragments was investigated with an adherence assay used previously to identify M. hyopneumoniae adhesins. Microtiter plates were coated with purified cilia from tracheal mucosa of specific pathogen-free swine as described previously (16). Mhp107 proteins were incubated with swine cilia, and bound protein was detected colorimetrically as described previously (4). Assays were performed in triplicate and repeated twice. Statistical analysis with an analysis of variance with a Tukey's comparison was performed. Wells containing no protein were used as negative controls. A cilia-binding protein, P159 (F4P159), (18) was used as a positive control.
RESULTS
Proteomic Analyses
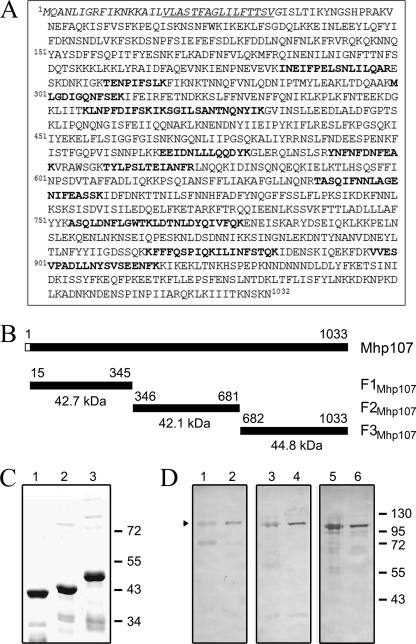
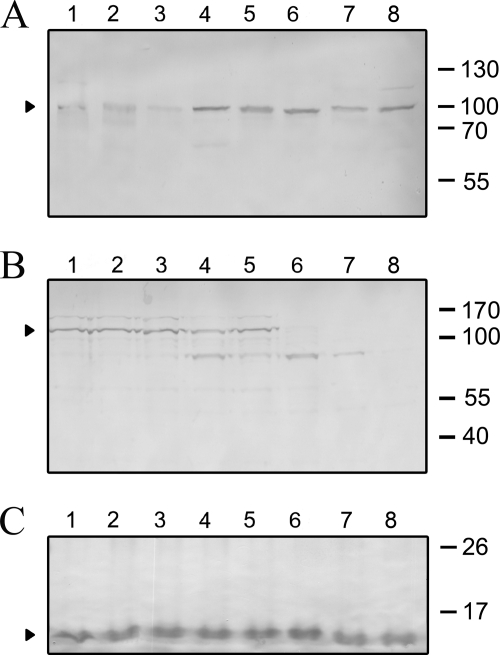
The hypothetical protein Mhp107 (GenBanktm accession number AAV27416) is a P97 paralog encoded by mhp107. Mhp107 consists of 1032 amino acids with a predicted pI of 8.81. Analysis with SignalP 3.0 server identifies a signal peptide located between amino acids 1–34 with a probability of 0.952. Prediction by TMPred detects a transmembrane domain region between amino acids 17–33 with a score of 2408. MS/MS analysis of M. hyopneumoniae proteins extracted from an SDS-PAGE gel slice containing proteins in the mass range of 79–104 kDa detected peptides that mapped to Mhp107 (Fig. 1A), demonstrating expression of Mhp107. This observation corroborates evidence of in vivo transcription of mhp107 (6). Attempts to express full-length Mhp107 in E. coli were unsuccessful (data not shown); consequently, Mhp107 was expressed as three distinct adjoining polyhistidine-tagged proteins (Fig. 1, B–C). Antisera raised against these fragments were used to characterize M. hyopneumoniae expression of Mhp107 by immunoblot analysis of the M. hyopneumoniae proteome (Fig. 1D). A 104-kDa protein was detected by antisera raised against each of the Mhp107 fragments. Immunoblot analysis did not indicate extensive proteolytic processing of Mhp107, despite extensive processing of other M. hyopneumoniae adhesins (10, 18, 33). Immunoblots with Mhp107 sera recognized a ∼100-kDa Mhp107 protein in the M. hyopneumoniae field strains tested (Fig. 2A), showing that the expression of Mhp107 in vitro is common to M. hyopneumoniae strains isolated from distinct locations. Only minor differences in processing of Mhp107 were observed among the field strains. The cellular location of Mhp107 was investigated via trypsin treatment of M. hyopneumoniae cells. As a result of trypsin treatment, the 104-kDa protein recognized by Mhp107 antisera was observed to be degraded, suggesting that the protein is surface-located (Fig. 2B). As a control to confirm the integrity of M. hyopneumoniae cells throughout trypsin treatment, sera against the intracellular protein L7/L12 was used to confirm that L7/L12 remained intact (18) (Fig. 2C). Hence, Mhp107 is expressed in a number of M. hyopneumoniae strains and is present at the mycoplasmal surface with an observed size of 104 kDa.
FIGURE 1.
Proteomic analyses of Mhp107. A, expression of Mhp107 occurs in vitro. ESI-MS/MS analysis of M. hyopneumoniae proteins extracted from an SDS-PAGE gel identified the amino acids shown in bold. Amino acids in italics and underlined italics indicate the predicted signal peptide sequence and transmembrane sequence, respectively. B, schematic representation of Mhp107, the putative 120-kDa protein encoded by mhp107. A predicted transmembrane and signal peptide sequence is depicted by the unfilled box. The position of the recombinant Mhp107 protein fragments (F1Mhp107-F3Mhp107) produced for this study are shown with amino acid positions indicated. The recombinant fragments include a 3.8-kDa polyhistidine region encoded by the expression vector. C, Coomassie-stained 12% SDS-PAGE gel of purified polyhistidine-tagged Mhp107 fragments. F1Mhp107 (lane 1), F2Mhp107 (lane 2), and F3Mhp107 (lane 3). D, immunoblot analyses using affinity-purified Mhp107 antisera. M. hyopneumoniae strain 232 whole cell lysates (lanes 1, 3, and 5) or aqueous phase (lanes 2, 4, and 6) were analyzed by immunoblot. Blots were probed with affinity-purified F1Mhp107 (lanes 1 and 2), F2Mhp107 (lanes 3 and 4), and F3Mhp107 (lanes 5 and 6) antisera.
FIGURE 2.
Immunoblot analyses of Mhp107. A, immunoblotting detected Mhp107 in M. hyopneumoniae field strains. Blots of M. hyopneumoniae whole cell lysates were probed with pooled affinity-purified F1Mhp107, F2Mhp107, and F3Mhp107 antisera. M. hyopneumoniae lysates were prepared from the following strains: 232 (lane 1), J (lane 2), OMZ407 (lane 3), Hillcrest (lane 4), 95MP1509 (lane 5), 00MP1301 (lane 6), 2–22421 (lane 7), and C1735/2 (lane 8). B and C, Mhp107 is a surface-located protein. Immunoblot analysis of M. hyopneumoniae strain 232 incubated with 0 (lane 1), 0.5 (lane 2), 1 (lane 3), 3 (lane 4), 5 (lane 5), 10 (lane 6), 50 (lane 7), or 150 (lane 8) μg/ml trypsin prior to preparation as whole cell lysates. Blots were probed with pooled affinity-purified F1Mhp107, F2Mhp107, and F3Mhp107 antisera (B) or anti-L7/L12, a cytosolic ribosomal protein (C).
F1Mhp107 Binds Heparin
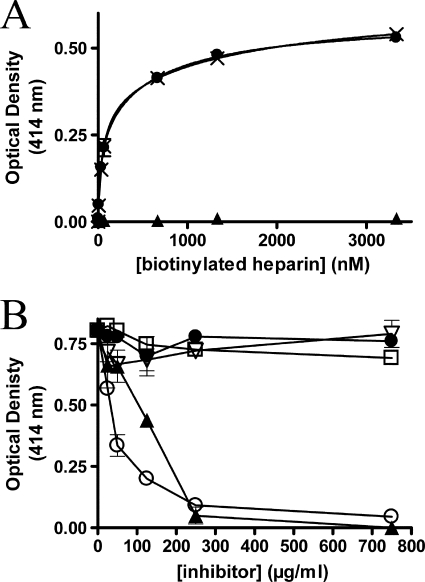
Heparin is able to inhibit the adherence of M. hyopneumoniae to cilia and is a potential ligand for M. hyopneumoniae (16, 18). The ability to bind heparin is common to several members of the P97/P102 adhesin family (4, 7, 8, 18). F1Mhp107 contains a motif (SKKKLKKL) that matches a putative heparin-binding consensus sequence, XBBBXXBX, where X is any amino acid and B is a basic residue (8). The interaction of heparin with immobilized F1Mhp107 was observed (Fig. 3A). Binding was dose-dependent with a KD of 87 ± 9 nm. F2Mhp107 and F3Mhp107 were unable to bind heparin. Fucoidan is a highly sulfated polysaccharide and was able to inhibit the ability of F1Mhp107 to bind heparin in competition assays (Fig. 3B). F1Mhp107 bound heparin in the presence of excess mucin and chondroitin sulfates A and B, suggesting that the high degree of sulfation of fucoidan may disrupt binding between F1Mhp107 and heparin.
FIGURE 3.
F1Mhp107 binds heparin. Microtiter plate assays were used to analyze binding of immobilized Mhp107 proteins to heparin. A, nonspecific (▴), total (×), and specific (●) binding of biotinylated heparin to F1Mhp107. Error bars represent the mean ± S.D. of triplicate readings. B, competitive binding assays showing inhibition of heparin binding by F1Mhp107 using a range of unlabeled polysaccharides; heparin (○), fucoidan (▴), mucin (▿), chondroitin sulfate A (□), and chondroitin sulfate B (●). Error bars represent the mean ± S.D. of triplicate readings.
Binding of F3Mhp107 to Fibronectin
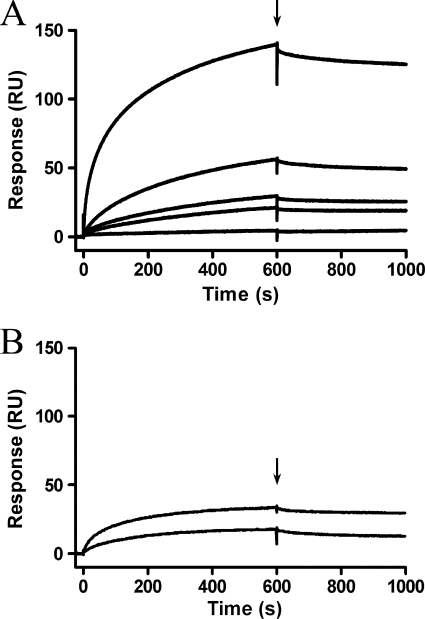
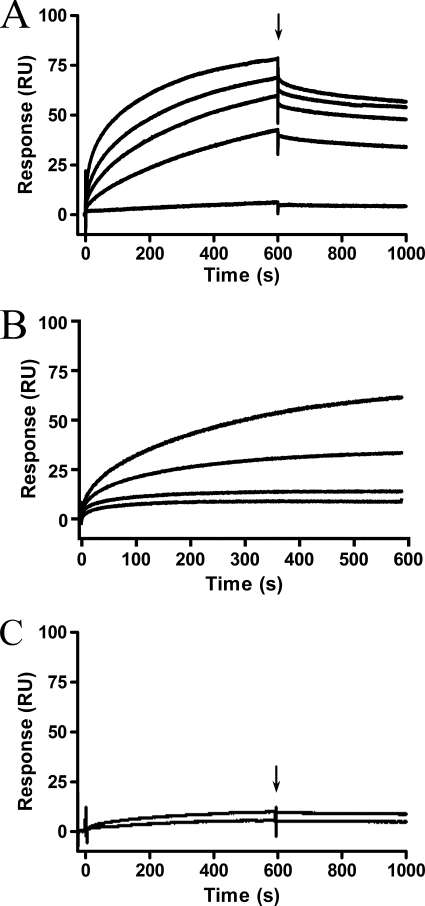
M. hyopneumoniae is a fibronectin-binding pathogen that expresses surface-located fibronectin-binding proteins P116 and Mhp271 (7, 10). The ability to bind fibronectin has been reported in other Mycoplasma species as a key virulence factor (34–36). Investigation into the fibronectin binding ability of Mhp107 fragments revealed only F3Mhp107 as a fibronectin-binding domain (Fig. 4). Using surface plasmon resonance, it was determined that F3Mhp107 is able to bind to fibronectin in a dose-dependent manner with a physiologically relevant KD of 174 ± 80 nm; ka = (4.8 ± 0.8) × 105 m−1s−1. Binding of F3Mhp107 to fibronectin was very stable, resulting in a slow dissociation rate. No previously described fibronectin binding motif was identified; however, analysis of Mhp107 with PONDR VSL1 (37) revealed regions of disorder within F3Mhp107. Disordered regions in proteins often contain binding sites that become ordered upon binding to target molecules (38).
FIGURE 4.
F3Mhp107 binds fibronectin. Surface plasmon resonance was used to analyze binding of Mhp107 proteins to immobilized fibronectin. Mhp107 proteins were injected over the immobilized fibronectin with a flow rate of 10 μl/min for 600 s. The end of the injection period is indicated by an arrow, at which stage any dissociation of the Mhp107 proteins from the ligand can be observed. A, the sensorgrams of F3Mhp107 at concentrations of 10 (bottom), 50, 100, 200, and 500 (top) nm binding fibronectin. B, sensorgrams of 200 nm F1Mhp107 (top) and F2Mhp107 (bottom) injected over immobilized fibronectin.
F1Mhp107 Binds Plasminogen
M. hyopneumoniae cells are able to bind plasminogen (10). Plasminogen contains five kringle domains with lysine-binding sites (39); consequently, a majority of plasminogen-binding proteins have a binding site containing lysine residues, and often the binding site is a C-terminal lysine (39). Although Mhp107 does not possess a C-terminal lysine residue, the Mhp107 amino acid sequence contains 124 lysines, which prompted investigation of the ability of P120 fragments to bind plasminogen. Using surface plasmon resonance, F1Mhp107 was found to bind plasminogen (Fig. 5A). F1Mhp107 binds plasminogen in a dose-dependent manner with a KD of 24 ± 2 nm; ka = (7.0 ± 0.7) × 104 m−1s−1. The ability of F1Mhp107 to bind plasminogen was inhibited by the lysine analog ϵ-aminocaproic acid (Fig. 5B), supporting the role of lysine residues in plasminogen binding by F1Mhp107. P116 is the only other M. hyopneumoniae protein known to interact with plasminogen. However, the plasminogen binding site in P116 is a C-terminal lysine (10). F1Mhp107 does not contain a terminal lysine or a binding site motif described within other plasminogen-binding proteins, therefore it is anticipated that a novel binding motif exists within the F1Mhp107 region of Mhp107.
FIGURE 5.
F1Mhp107 binds plasminogen. Surface plasmon resonance was used to analyze binding of Mhp107 proteins to immobilized porcine plasminogen. Mhp107 proteins were injected at a flow rate of 10 μl/min for 600 s. A, F1Mhp107 was injected over the immobilized plasminogen at concentrations of 10 (bottom), 50, 100, 200, and 500 (top) nm. The end of the injection period is indicated by an arrow, at which stage dissociation of F1Mhp107 from the ligand can be observed. B, sensorgrams of 200 nm F1Mhp107 binding to immobilized plasminogen in the presence of 0, 10, 50, and 100 μm ϵ-aminocaproic acid. The analyte was injected with a flow rate of 10 μl/min for 600 s. C, sensorgrams of 200 nm F2Mhp107 (top) and F3Mhp107 (bottom) injected over immobilized plasminogen. The end of the injection period is indicated by an arrow.
Adherence of PK15 Cells and Swine Cilia by Mhp107 Fragments
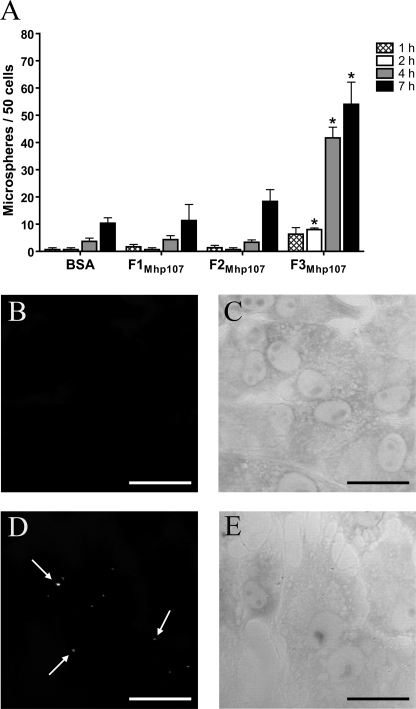
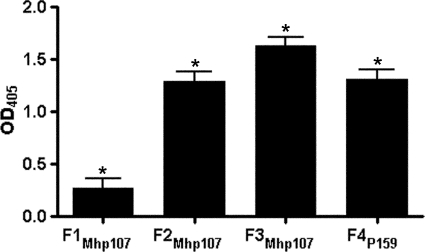
Binding of M. hyopneumoniae to swine cilia is a prerequisite for colonization in the host. The PK15 cell line is useful as a model for adherence to swine epithelia (4, 10, 18, 40). Both PK15 cells and swine respiratory cilia were used in this study to investigate the role of Mhp107 in adherence. Protein-coated fluorescent microspheres were used to determine the ability of Mhp107 fragments to promote binding to PK15 cells. It was found that F3Mhp107 was able to promote adherence of microspheres to PK15 cells (Fig. 6A–E). An assay based on that developed by Zhang et al. (16) was employed to investigate the ability of Mhp107 fragments to bind swine respiratory cilia (Fig. 7). A recombinant fragment of a M. hyopneumoniae adhesin (F4P159) (18) was used as a positive control protein. Each of the Mhp107 fragments described in this study is able to bind swine respiratory cilia.
FIGURE 6.
F3Mhp107 promotes adherence to PK15 cells. Fluorescent microspheres coated with recombinant Mhp107 proteins were incubated with PK15 cells. BSA-coated microspheres were employed as a negative control. A, graphical summary of the ability of proteins to promote microsphere adherence to PK15 cells. *, results differing significantly from the BSA-negative control when a one-way analysis of variance is performed (p < 0.01). B–E, representative confocal microscopy images of microspheres adhered to PK15 cells. BSA- (B) and F3Mhp107-coated (D) microspheres bound to PK15 cells following a 4-h incubation. C and E are corresponding transmission images of B and D, respectively. Arrows indicate microspheres. Scale bars = 25 μm.
FIGURE 7.
Mhp107 fragments bind porcine respiratory cilia. Recombinant Mhp107 proteins were incubated with swine cilia in a microtiter plate-based binding assay. The M. hyopneumoniae adhesin F4P159 acts as a positive binding protein. Data represents the mean ± S.E. of the response above background of triplicate measurements replicated twice. *, results differing significantly from the background (p < 0.05).
DISCUSSION
The gene mhp107 encodes Mhp107, a putative 120-kDa protein and a P97 cilium adhesin paralog. ESI-MS/MS analysis of Mhp107 demonstrated the presence of Mhp107 in the M. hyopneumoniae proteome, consistent with evidence of mhp107 transcription (6). Adams et al. (6) reported mhp107 transcription in vivo during M. hyopneumoniae infection. Immunoblots of M. hyopneumoniae field strains probed with Mhp107 antisera recognized a ∼100-kDa protein comparable in size to that observed in whole cell immunoblots and trypsin digest blots. The surface location and presence of Mhp107 in multiple field strains corroborates evidence for the expression of Mhp107 and a potential role of Mhp107 as an adhesin.
Mhp107 contains a heparin-binding consensus sequence (8) and is able to bind specifically to heparin. Heparin inhibits the adherence of M. hyopneumoniae to porcine cilia and PK15 cells (16, 18), and a number of adhesins from the P97/P102 paralog family are heparin-binding proteins (4, 7, 8, 18). A structurally similar proteoglycan, heparan sulfate, is found in the extracellular matrix and on cell surfaces, including the epithelial cells of the swine respiratory tract (41, 42). Heparin is also able to act as a bridging molecule, providing a mechanism to bind extracellular matrix components, including fibronectin, vitronectin, and laminin (43). Mhp107 thus joins the P97/P102 adhesin family, which contains other adhesin proteins with the capacity to bind heparin. As with F1Mhp107, the interaction between each of these other adhesins and heparin is blocked by fucoidan (4, 7, 8, 18). Such inhibition of binding by fucoidan suggests that these proteins bind heparin through electrostatic interactions. The negatively charged sulfate groups of fucoidan compete with heparin to interact with basic amino acids in these M. hyopneumoniae adhesin proteins. The basic residues in the identified putative heparin binding motif 204SKKKLKKL211 within F1Mhp107 may contribute to this interaction. The ability of heparin to inhibit binding of M. hyopneumoniae to cilial cells coupled with several M. hyopneumoniae adhesins able to bind heparin provides compelling evidence of the important role for heparin or heparan sulfate binding in the colonization process.
Recent reports indicate that M. hyopneumoniae binds both fibronectin and plasminogen (7, 10). The ability to interact with fibronectin promotes virulence in a large number of bacterial species (44), including the evolutionally related streptococci (45, 46). Here we have used surface plasmon resonance to demonstrate that F3Mhp107 binds fibronectin in a dose-dependent manner with a physiologically relevant KD value. Mhp271 and P116 are also M. hyopneumoniae proteins with a fibronectin binding capability (7, 10); F3Mhp107 binds fibronectin with a similar affinity to F2271. Other mycoplasmal species that express fibronectin-binding proteins include Mycoplasma penetrans (20), Mycoplasma pneumoniae (47), and Mycoplasma gallisepticum (34). Fibronectin-binding proteins can provide a mechanism for adherence to host cells and tissues during colonization and enable some bacteria to trigger cytoskeletal rearrangements, thereby facilitating invasion (20, 44, 48, 49). The ability of Mhp271, P116, and Mhp107 to bind fibronectin suggests that the interaction of M. hyopneumoniae with fibronectin is a key virulence determinant and contributes significantly to the pathogenesis of infection.
F1Mhp107 is also a plasminogen-binding protein. Inhibition of plasminogen binding by a lysine analog suggests that the internal lysine residues of this protein contribute to the observed plasminogen binding ability of F1Mhp107. Bound plasminogen is more susceptible to activation to plasmin, a broad-spectrum serine protease (50). Bacterial interactions with plasmin(ogen) have been implicated in host tissue damage (39, 51, 52) and may contribute to tissue damage observed in the respiratory tract of swine during M. hyopneumoniae infection. The plasminogen binding affinity of F1Mhp107 is physiologically relevant, characterized by a relatively slow association and dissociation rate. The plasminogen binding characteristics observed in this study are similar to those reported for the M. hyopneumoniae plasminogen-binding protein P116 (10). Damage to host epithelial cells by plasmin(ogen) can expose extracellular matrix components such as fibronectin to enhance adherence by pathogens. Deposition of fibronectin also occurs during wound repair at the interface of epithelial cells and the extracellular matrix (22, 53). Typically, plasminogen-binding proteins are multifunctional surface proteins (39) and have been reported to contribute to virulence in pathogens of the respiratory tract (54–58).
The use of in vitro cellular models to investigate adherence has underpinned the characterization of P97, P216, P159, and P116 as M. hyopneumoniae adhesins (4, 10, 18, 59). During disease, M. hyopneumoniae adheres to and colonizes ciliated cells of the swine respiratory tract (1). In vitro, M. hyopneumoniae is able to bind the porcine kidney epithelial-like cell line PK15, prompting the use of PK15 cells as an eukaryotic cell binding model (40). We have demonstrated the ability of Mhp107 fragments to adhere to swine cilia and to promote adherence to PK15 cells. These findings support a role for Mhp107 in the adherence of the bacterium to the host. In recent years since the initial discovery of P97 as the cilium adhesin, P216, P159, Mhp271, and P116 from the P97/P102 paralog family have been described as multifunctional adhesins (4, 7, 10, 18, 59). The genetic relationships among these protein paralogs indicate that they are likely to have similar or related functions. Ongoing research has provided mounting evidence of the importance of these proteins in mediating M. hyopneumoniae adherence to and colonization of the host. Here, we have described Mhp107 as a multifunctional cilial adhesin found on the surface of M. hyopneumoniae. Furthermore, the ability of this adhesin to interact with heparin, fibronectin and plasminogen indicates that Mhp107 may be an important factor in the virulence and disease potential of M. hyopneumoniae.
Acknowledgments
We thank Paul Young and Jake Matic for technical assistance and Bradley W. Van De Woestyne for assistance with statistical analysis of cilia binding data.
This work was supported by the Australian Postgraduate Award (to L. M. S.) and an Australian Research Council Linkage Project grant (LP776771).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- ESI
- electrospray ionization.
REFERENCES
- 1. Razin S. (1999) Biosci. Rep. 19, 367–372 [DOI] [PubMed] [Google Scholar]
- 2. Zhang Q., Young T. F., Ross R. F. (1994) Infect. Immun. 62, 4367–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Minion F. C. (2002) Front. Biosci. 7, d1410–1422 [DOI] [PubMed] [Google Scholar]
- 4. Wilton J., Jenkins C., Cordwell S. J., Falconer L., Minion F. C., Oneal D. C., Djordjevic M. A., Connolly A., Barchia I., Walker M. J., Djordjevic S. P. (2009) Mol. Microbiol. 71, 566–582 [DOI] [PubMed] [Google Scholar]
- 5. Hsu T., Minion F. C. (1998) Gene 214, 13–23 [DOI] [PubMed] [Google Scholar]
- 6. Adams C., Pitzer J., Minion F. C. (2005) Infect. Immun. 73, 7784–7787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deutscher A. T., Jenkins C., Minion F. C., Seymour L. M., Padula M. P., Dixon N. E., Walker M. J., Djordjevic S. P. (2010) Mol. Microbiol. 78, 444–458 [DOI] [PubMed] [Google Scholar]
- 8. Jenkins C., Wilton J. L., Minion F. C., Falconer L., Walker M. J., Djordjevic S. P. (2006) Infect. Immun. 74, 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsu T., Artiushin S., Minion F. C. (1997) J. Bacteriol. 179, 1317–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seymour L. M., Deutscher A. T., Jenkins C., Kuit T. A., Falconer L., Minion F. C., Crossett B., Padula M., Dixon N. E., Djordjevic S. P., Walker M. J. (2010) J. Biol. Chem. 285, 33971–33978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baseggio N., Glew M. D., Markham P. F., Whithear K. G., Browning G. F. (1996) Microbiology 142, 1429–1435 [DOI] [PubMed] [Google Scholar]
- 12. Dallo S. F., Baseman J. B. (1991) Microb. Pathog. 10, 475–480 [DOI] [PubMed] [Google Scholar]
- 13. Reddy S. P., Rasmussen W. G., Baseman J. B. (1995) J. Bacteriol. 177, 5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu T., Minion F. C. (1998) Infect. Immun. 66, 4762–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Q., Young T. F., Ross R. F. (1995) Infect. Immun. 63, 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Q., Young T. F., Ross R. F. (1994) Infect. Immun. 62, 1616–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duensing T. D., Wing J. S., van Putten J. P. (1999) Infect. Immun. 67, 4463–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burnett T. A., Dinkla K., Rohde M., Chhatwal G. S., Uphoff C., Srivastava M., Cordwell S. J., Geary S., Liao X., Minion F. C., Walker M. J., Djordjevic S. P. (2006) Mol. Microbiol. 60, 669–686 [DOI] [PubMed] [Google Scholar]
- 19. Chaussee M. S., Cole R. L., van Putten J. P. M. (2000) Infect. Immun. 68, 3226–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Girón J. A., Lange M., Baseman J. B. (1996) Infect. Immun. 64, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dubreuil J. D., Giudice G. D., Rappuoli R. (2002) Microbiol. Mol. Biol. Rev. 66, 617–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scarselli M., Serruto D., Montanari P., Capecchi B., Adu-Bobie J., Veggi D., Rappuoli R., Pizza M., Aricò B. (2006) Mol. Microbiol. 61, 631–644 [DOI] [PubMed] [Google Scholar]
- 23. Pancholi V., Fontan P., Jin H. (2003) Microb. Pathog. 35, 293–303 [DOI] [PubMed] [Google Scholar]
- 24. Patti J. M., Allen B. L., McGavin M. J., Höök M. (1994) Annu. Rev. Microbiol. 48, 585–617 [DOI] [PubMed] [Google Scholar]
- 25. Scarman A. L., Chin J. C., Eamens G. J., Delaney S. F., Djordjevic S. P. (1997) Microbiology 143, 663–673 [DOI] [PubMed] [Google Scholar]
- 26. Bereiter M., Young T. F., Joo H. S., Ross R. F. (1990) Vet. Microbiol. 25, 177–192 [DOI] [PubMed] [Google Scholar]
- 27. Madsen M. L., Oneal M. J., Gardner S. W., Strait E. L., Nettleton D., Thacker E. L., Minion F. C. (2007) J. Bacteriol. 189, 7977–7982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cole J. N., Djordjevic S. P., Walker M. J. (2008) Methods Mol. Biol. 425, 295–311 [DOI] [PubMed] [Google Scholar]
- 29. Wise K. S., Kim M. F. (1987) J. Bacteriol. 169, 5546–5555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilton J. L., Scarman A. L., Walker M. J., Djordjevic S. P. (1998) Microbiology 144, 1931–1943 [DOI] [PubMed] [Google Scholar]
- 31. Sanderson-Smith M., Batzloff M., Sriprakash K. S., Dowton M., Ranson M., Walker M. J. (2006) J. Biol. Chem. 281, 3217–3226 [DOI] [PubMed] [Google Scholar]
- 32. Andronicos N. M., Ranson M., Bognacki J., Baker M. S. (1997) Biochim. Biophys. Acta 1337, 27–39 [DOI] [PubMed] [Google Scholar]
- 33. Djordjevic S. P., Cordwell S. J., Djordjevic M. A., Wilton J., Minion F. C. (2004) Infect. Immun. 72, 2791–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. May M., Papazisi L., Gorton T. S., Geary S. J. (2006) Infect. Immun. 74, 1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yavlovich A., Rottem S. (2007) FEMS Microbiol. Lett. 266, 158–162 [DOI] [PubMed] [Google Scholar]
- 36. Balasubramanian S., Kannan T. R., Baseman J. B. (2008) Infect. Immun. 76, 3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Obradovic Z., Peng K., Vucetic S., Radivojac P., Dunker A. K. (2005) Proteins 61, 176–182 [DOI] [PubMed] [Google Scholar]
- 38. Uversky V. N., Dunker A. K. (2010) Biochim. Biophys. Acta 1804, 1231–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lähteenmäki K., Kuusela P., Korhonen T. K. (2001) FEMS Microbiol. Rev. 25, 531–552 [DOI] [PubMed] [Google Scholar]
- 40. Zielinski G. C., Young T., Ross R. F., Rosenbusch R. F. (1990) Am. J. Vet. Res. 51, 339–343 [PubMed] [Google Scholar]
- 41. Erlinger R. (1995) Cell Tissue Res. 281, 473–483 [DOI] [PubMed] [Google Scholar]
- 42. Rabenstein D. L. (2002) Nat. Prod. Rep. 19, 312–331 [DOI] [PubMed] [Google Scholar]
- 43. Jackson R. L., Busch S. J., Cardin A. D. (1991) Physiol. Rev. 71, 481–539 [DOI] [PubMed] [Google Scholar]
- 44. Joh D., Wann E. R., Kreikemeyer B., Speziale P., Höök M. (1999) Matrix Biol. 18, 211–223 [DOI] [PubMed] [Google Scholar]
- 45. Ramachandran V., McArthur J. D., Behm C. E., Gutzeit C., Dowton M., Fagan P. K., Towers R., Currie B., Sriprakash K. S., Walker M. J. (2004) J. Bacteriol. 186, 7601–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Delvecchio A., Currie B. J., McArthur J. D., Walker M. J., Sriprakash K. S. (2002) Epidemiol. Infect. 128, 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dallo S. F., Kannan T. R., Blaylock M. W., Baseman J. B. (2002) Mol. Microbiol. 46, 1041–1051 [DOI] [PubMed] [Google Scholar]
- 48. Dziewanowska K., Patti J. M., Deobald C. F., Bayles K. W., Trumble W. R., Bohach G. A. (1999) Infect. Immun. 67, 4673–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Secott T. E., Lin T. L., Wu C. C. (2004) Infect. Immun. 72, 3724–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Plow E. F., Herren T., Redlitz A., Miles L. A., Hoover-Plow J. L. (1995) FASEB J. 9, 939–945 [DOI] [PubMed] [Google Scholar]
- 51. Sanderson-Smith M. L., Dinkla K., Cole J. N., Cork A. J., Maamary P. G., McArthur J. D., Chhatwal G. S., Walker M. J. (2008) FASEB J. 22, 2715–2722 [DOI] [PubMed] [Google Scholar]
- 52. McKay F. C., McArthur J. D., Sanderson-Smith M. L., Gardam S., Currie B. J., Sriprakash K. S., Fagan P. K., Towers R. J., Batzloff M. R., Chhatwal G. S., Ranson M., Walker M. J. (2004) Infect. Immun. 72, 364–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Coraux C., Roux J., Jolly T., Birembaut P. (2008) Proc. Am. Thorac. Soc. 5, 689–694 [DOI] [PubMed] [Google Scholar]
- 54. Pancholi V., Fischetti V. A. (1998) J. Biol. Chem. 273, 14503–14515 [DOI] [PubMed] [Google Scholar]
- 55. Sanderson-Smith M. L., Dowton M., Ranson M., Walker M. J. (2007) J. Bacteriol. 189, 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bergmann S., Rohde M., Hammerschmidt S. (2004) Infect. Immun. 72, 2416–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yavlovich A., Higazi A. A., Rottem S. (2001) Infect. Immun. 69, 1977–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cork A. J., Jergic S., Hammerschmidt S., Kobe B., Pancholi V., Benesch J. L., Robinson C. V., Dixon N. E., Aquilina J. A., Walker M. J. (2009) J. Biol. Chem. 284, 17129–17137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Minion F. C., Adams C., Hsu T. (2000) Infect. Immun. 68, 3056–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]