Abstract
Oxygen availability is the major determinant of the metabolic modes adopted by Escherichia coli. Although much is known about E. coli gene expression and metabolism under fully aerobic and anaerobic conditions, the intermediate oxygen tensions that are encountered in natural niches are understudied. Here, for the first time, the transcript profiles of E. coli K-12 across the physiologically significant range of oxygen availabilities are described. These suggested a progressive switch to aerobic respiratory metabolism and a remodeling of the cell envelope as oxygen availability increased. The transcriptional responses were consistent with changes in the abundance of cytochrome bd and bo′ and the outer membrane protein OmpW. The observed transcript and protein profiles result from changes in the activities of regulators that respond to oxygen itself or to metabolic and environmental signals that are sensitive to oxygen availability (aerobiosis). A probabilistic model (TFInfer) was used to predict the activity of the indirect oxygen-sensing two-component system ArcBA across the aerobiosis range. The model implied that the activity of the regulator ArcA correlated with aerobiosis but not with the redox state of the ubiquinone pool, challenging the idea that ArcA activity is inhibited by oxidized ubiquinone. The amount of phosphorylated ArcA correlated with the predicted ArcA activities and with aerobiosis, suggesting that fermentation product-mediated inhibition of ArcB phosphatase activity is the dominant mechanism for regulating ArcA activity under the conditions used here.
Keywords: Gene Regulation, Membrane Proteins, Respiratory Chain, Transcription Factors, Transcription Regulation
Introduction
The bacterium and model organism Escherichia coli K-12 has three basic metabolic modes: aerobic respiration, anaerobic respiration, and fermentation (1, 2). There is a hierarchy in which aerobic respiration is preferred to anaerobic respiration, which in turn is preferred to fermentation (1). This hierarchy reflects the relative amounts of energy that can be conserved by these metabolic modes, and oxygen availability is the major signal that governs which metabolic mode is adopted.
Many environments, both natural (host intestinal tract) and man-made (bioreactors), are characterized by the presence of oxygen gradients and/or regions of variable oxygen availability (3, 4). Thus, how patterns of gene expression adapt across the range of physiologically relevant oxygen availabilities is important for the efficiency of biotechnological processes that use E. coli as a cell factory and for competitiveness in natural environments (4). However, obtaining reproducible data from E. coli cultures at low oxygen tensions is technically demanding, and the overwhelming majority of the relevant literature reports the results of experiments with fully aerobic or anaerobic cultures. Furthermore, as indicated by Alexeeva et al. (5), in the relatively few attempts to study E. coli grown at intermediate oxygen tensions, it was apparent that neither dissolved oxygen tension nor the gas input to a chemostat accurately describes the responses of the culture to changes in oxygen availability. This is because the response of the cultures depends on the oxygen transfer rate and fermenter geometry (6), precluding the possibility of making meaningful comparisons. To overcome these technical difficulties, the aerobiosis scale was proposed as the key variable to study the response of E. coli to changes in oxygen availability (5). Under carbon-limiting conditions, the minimum oxygen input that results in undetectable excretion of the fermentation product acetate is defined as 100% aerobiosis. As the amount of oxygen supplied to cultures decreases, the specific rate of acetate production (qacetate) increases to a maximum under anaerobic conditions (0% aerobiosis). Between these limits (0–100% aerobiosis) lies the micro-aerobic range defined by the linear decrease in qacetate as the oxygen transfer rate increases, i.e. there is an inverse correlation between qacetate and aerobiosis. Thus, analyzing cultures at fixed points on the aerobiosis scale allows the responses of E. coli K-12 to environments with different oxygen availabilities to be accurately measured and compared, but this has never been exploited for transcriptomic profiling.
The remodeling of metabolism associated with oxygen availability and switching between alternative metabolic modes requires widespread changes in gene expression (7–11). Two major transcription factors are implicated in this process: the indirect oxygen sensor ArcBA and the direct oxygen sensor FNR (reviewed in Ref. 12). Under anaerobic conditions FNR is a dimeric iron-sulfur protein that binds at specific DNA sequences to activate the expression of “anaerobic genes” and to repress the expression of “aerobic genes” (7, 8, 13). FNR senses the presence of oxygen by disassembly of its oxygen-sensitive iron-sulfur cluster, converting the protein to a non-DNA-binding monomer (13). Thus, FNR activity is controlled directly by the reaction of oxygen with the iron-sulfur cluster (14–16). In contrast to FNR, ArcA is the response regulator of a two-component system. When oxygen availability is low, the membrane-bound sensor, ArcB, is autophosphorylated and activates ArcA by transfer of the phospho-group (reviewed in Ref. 12). ArcA phosphorylation initiates a change in oligomeric state, resulting in DNA binding (17). ArcB does not sense oxygen directly but is thought to sense the redox state of the ubiquinone/ubiquinol pool in the aerobic respiratory chain because ubiquinone inhibits ArcB autokinase activity (18, 19). It has also been shown that metabolites associated with fermentation (e.g. the reduced fermentation products acetate, d-lactate, pyruvate, and NADH) inhibit the ability of ArcB to dephosphorylate ArcA (20). Thus, when the ubiquinone pool is more oxidized and the concentrations of fermentation products are low (i.e. aerobic respiratory conditions), ArcBA is switched off. However, the significance of the redox state of the ubiquinone pool for the activity of ArcA has recently been questioned (21). Nevertheless, it is clear that FNR and ArcA work together to coordinate gene expression in response to the availability of oxygen (2).
To deepen our understanding of how E. coli adapts to environments with limited oxygen availabilities, transcript profiles and biochemical measurements across the physiologically relevant range of oxygen availabilities were obtained. Thus, for the first time, carefully controlled chemostat cultures have been used to systematically study the effects of oxygen availability on the transcriptome, redox state of the ubiquinone/ubiquinol pool, outer membrane protein profiles, and terminal oxidase proteins of E. coli K-12. The activity of ArcA inferred from the transcript profiles using TFInfer (22) did not correlate with the redox state of the ubiquinone/ubiquinol pool. This prompted experiments to measure the amounts of phosphorylated ArcA, which did correlate well with ArcA activity predicted by the model. This implies that the redox state of the ubiquinone pool is not a major factor in switching ArcA between active and inactive states under the conditions used here, but rather that ArcA activity decreases as aerobiosis increases.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
E. coli K-12 substrain MG1655 (23) and MG1655 ΔarcA, constructed using the standard linear transformation protocol (24), were used in this study. Steady-state continuous cultures were established in a 2-liter Labfors chemostat (Infors-HT, Bottmingen, Switzerland) at 37 °C, a 0.2 h−1 dilution rate, a 950-ml working volume, and a gas flow rate of 1 liter/min. The pH was maintained at 6.9 by titration with 1 m sodium hydroxide. Continuous cultures were grown in a defined minimal glucose-limited medium based on that of Evans et al. (25) with the exception that 2 mm nitrilotriacetic acid was used as the metal ion chelator instead of citrate. The medium was supplemented with 30 μg/liter sodium selenite, and 20 mm glucose was used as the carbon source. Mass flow controllers allowed the correct ratio of air and pure N2 gas to be mixed to produce the desired oxygen input in the chemostat. To determine aerobiosis levels within the chemostat, acetate calibration was carried out as described previously (5).
Transcriptional Profiling
Chemostat culture samples (two biological and two technical replicates at each aerobiosis value) for transcriptional profiling were directly transferred into RNAprotect (Qiagen) to rapidly stabilize the mRNA. Total RNA was prepared using the RNeasy RNA purification kit (Qiagen) according to the manufacturer's instructions (including the DNase treatment step). RNA was directly converted to Cy3-labeled cDNA, and genomic DNA was converted to Cy5-labeled cDNA, which were then combined and hybridized to each microarray using an indirect reference style of hybridization (26). Labeling protocols are described on the Institute of Food Research website. Hybridizations were carried out overnight with E. coli K-12 V2 OciChipTM microarrays (Ocimum Biosolutions, Hyderabad, India) following the manufacturer's instructions. Slides were scanned in a DNA microarray scanner (Agilent, Santa Clara, CA) with subsequent feature extraction using ImaGene v5.1 (Biodiscovery, El Segundo, CA) and data analysis using the rank products technique (27). Transcriptomic data have been deposited with ArrayExpress (ID E-MTAB-285) and in SysMO-DB.
Measurement of Biomass and Organic Acids
Dry cell weight was estimated after centrifuging 10 ml of culture broth (3000 × g, 20 min, 4 °C), resuspending the pellet in 5 ml of deionized water, centrifuging (3000 × g, 15 min, 4 °C), and drying the pellet for 24 h at 105 °C. For measuring extracellular metabolite concentrations by 1H NMR, cell-free supernatant fractions were analyzed in a volume of 500 μl containing 450 μl of sample, 50 μl of D2O, and 1 mm trimethylsilyl propionate as a standard. The samples were analyzed in 5-mm diameter tubes at 298 K. All spectra were acquired on a Bruker DRX-500 spectrometer operating at 500 MHz. The H2O signal was reduced by presaturation for 2 s applied during the recycle time. Carbon decoupling was applied during acquisition to suppress 13C-labeled satellites. Spectra were processed and peaks were quantified by integration using Topspin. Chemical shifts and concentrations were established by reference to trimethylsilyl propionate.
Measurement of ArcA Phosphorylation
ArcA phosphorylation levels were measured using Phos-tagTM-acrylamide gel electrophoresis and subsequent Western immunoblotting. Culture samples were taken from the chemostat directly into formic acid (1 m final concentration) to stabilize phospho-Asp residues. Aliquots were pelleted by centrifugation (10,000 × g, 30 s), resuspended in 50 μl of 1 m formic acid, and stored at −80 °C. Protein sample processing was then carried out as described previously (28) with the proteins resolved on a 37.5 μm Phos-tagTM-10% acrylamide gel using initial A600 values to calculate loading volumes. Gel preparation, electrophoresis, and electrophoretic transfer of the protein onto nitrocellulose membrane (including the additional EDTA step) were carried out following the manufacturer's instructions for Phos-tagTM-acrylamide (NARD Institute, Ltd., Amagasaki City, Japan). ArcA immunodetection was carried out using rabbit ArcA polyclonal antiserum and the ECL system (GE Healthcare). ArcA protein was purified as described previously (21).
Determination of Cytochrome bd and bo′ Content
Culture samples were collected directly into chloramphenicol (final concentration of 50 mg/ml). The bacteria were isolated by centrifugation and then resuspended in 0.1 m sodium phosphate buffer (pH 7.0) for spectroscopy. Electronic absorption spectra were measured using a custom-built SDB4 dual-wavelength scanning spectrophotometer (University of Pennsylvania School of Medicine Biomedical Instrumentation Group and Current Designs, Philadelphia, PA) (29). Spectra were collected for several different sample dilutions. Cytochrome bd and bo′ concentrations were determined from the dithionite-reduced plus CO − dithionite-reduced difference absorption spectra using Δϵ642–622 = 12.6 mm−1 cm−1 and Δϵ416–430 = 145 mm−1 cm−1, respectively (30). The numbers of cytochrome bd and bo′ molecules/cell were calculated using the dry cell weight of the original culture and a conversion factor of 2.8 × 10−13 g of dry weight/cell (31). Note that cytochrome bd quinol oxidases I and II are effectively identical spectroscopically, and thus, the values obtained are of the total heme d. Also, the CO-bound form of numerous heme-containing proteins contributes to absorbance in the Soret region, making accurate quantification of cytochrome bo′ difficult. Therefore, the numbers of cytochrome bo′ molecules are, at best, an approximation of the true value. For each aerobiosis value, cytochrome numbers were calculated from two independent chemostat cultures: biological replicates A and B. Biological replicate A contained nA technical replicates (dilutions) and similarly for B. Ai and Bi represent the individual values in A and B for different dilutions. First, the means of the biological replicates were calculated (mean A = sum Ai/nA, and mean B = sum Bi/nB), followed by the mean of the overall experiment (mean = (nA*mean A + nB*mean B)/(nA + nB)). The biological variance was then computed (biological variance = (1/(2 − 1))*((mean − mean A)2 + (mean − mean B)2)). The biological standard deviation is the square root of this value.
Quinone Extraction and Analysis
The ubiquinone/ubiquinol extraction and measurement methodology was adapted from Bekker et al. (32). Samples (20 ml) from chemostat cultures grown at various aerobiosis levels were taken using a rapid sampling device (33) and quenched in 60 ml of ice-cold methanol. The ubiquinone/ubiquinol content from 8 ml of methanol/culture mixture was extracted by adding 6 ml of petroleum ether (40–60 °C). Samples were vortexed for 1 min and centrifuged at 900 × g for 2 min. The upper phase (petroleum ether) was removed into a clean tube and evaporated under constant nitrogen flow. The lower phase was again treated with 6 ml of petroleum ether, vortexed, and centrifuged, and the process was repeated to back-extract any remaining ubiquinones. The extracted and dried ubiquinone/ubiquinol mixture was resuspended in 50 μl of 99.9% ethanol using a glass rod. A 30-μl sample of this solution was loaded onto a reverse-phase LiChrosorb (Chrompack) 10-RP18 column (4.6-mm inner diameter, 250-mm length). The column was equilibrated with methanol as the mobile phase. Ubiquinone/ubiquinol content was analyzed using the GE Healthcare gradient pump 2249 HPLC system. Peaks were identified spectroscopically (32), and the amount of ubiquinones was calculated from their corresponding area using a standard curve for ubiquinone-10. Methanol, ethanol, and petroleum ether were of analytical grade. The Ubiquinone/ubiquinol content of each chemostat was analyzed in duplicate.
Analysis of Outer Membrane Protein Profiles
This was achieved as described (34). Samples were taken from cultures grown at the indicated aerobiosis values. After the cell pellets were washed with 10 mm HEPES (pH 7.4), the cells were lysed in a French pressure cell. Unbroken bacteria were removed by centrifugation (15,000 × g, 10 min). The inner and outer membrane fractions were obtained by ultracentrifugation of the supernatants. After resuspension of the membrane pellet in 10 mm HEPES (pH 7.4), the protein content was measured, and then 20 μl of a 30% solution of Sarkosyl was added per mg of protein. After the mixtures were shaken at 200 rpm for 30 min, the samples were transferred to centrifuge tubes and centrifuged at 125,000 × g for 60 min at 4 °C. After the pellets were washed in 10 mm HEPES (pH 7.4), they were resuspended in 200 μl of deionized water. Outer membrane proteins were separated by electrophoresis on 10% SDS-polyacrylamide gels containing 8 m urea. Polypeptides were transferred to PVDF membrane, and after briefly staining with Coomassie Blue, the N-terminal amino acid sequences were determined by Edman degradation using an Applied Biosystems protein sequencer.
RESULTS AND DISCUSSION
Steady-state chemostat cultures of E. coli MG1655 were established at 0, 31, 56, 85, and 115% aerobiosis for transcript profiling and measurement of ubiquinone/ubiquinol pools as described under “Experimental Procedures.” Comparison of the transcript profiles at predetermined points on the aerobiosis scale with that obtained under anaerobic conditions (0% aerobiosis) revealed that 76 operons exhibited significantly altered expression. These included 21 transcripts encoding central metabolic functions (Table 1), only four transcripts associated with the oxidative stress response (Table 2), seven transcripts encoding transcription factors (Table 3), four transcripts encoding outer membrane proteins and one encoding an adhesin (Table 4), and 10 other genes of unknown function (Table 5).
TABLE 1.
Transcripts encoding proteins of central metabolism that were present in altered abundance at different aerobiosis values
The data shown are for the first gene in the operon except where indicated by boldface type. The -fold change (by at least 2-fold at one or more aerobiosis values) is the ratio of transcript levels under the indicated conditions compared with transcript levels at 0% aerobiosis (p ≤ 0.05). Relevant regulatory proteins are indicated: (−) denotes negative regulation, (+) denotes positive regulation, and (−/+) denotes dual regulation. The table was compiled from references in the text and the EcoCyc Database (35).
| Operon | Product | -Fold change in abundance relative to anaerobic (0% aerobiosis) steady state |
Relevant regulatory proteins | |||
|---|---|---|---|---|---|---|
| 31% | 56% | 85% | 115% | |||
| aceBA | Malate synthase/isocitrate lyase | 1.5 | 2.0 | 2.5 | 2.8 | ArcA (−), CRP (−), FruR (+), IclR (−), IHF (+) |
| acnB | Aconitase B | 1.2 | 1.9 | 2.5 | 3.9 | ArcA (−), CRP (+), Fis (−), FruR (−) |
| acs-yjcH-actP | Acetyl-CoA synthetase/acetate transporter | 1.0 | 1.1 | 1.2 | 3.7 | CRP (+), Fis (−), IHF (−), NarL (−) |
| cydAB | Cytochrome bd-I oxidase | 2.2 | 6.4 | 2.5 | 1.4 | ArcA (+), FNR (−), FruR (+), H-NS (−) |
| cyoA–E | Cytochrome bo oxidase | 1.2 | 1.8 | 4.0 | 7.2 | ArcA (−), CRP (+), FNR (−), Fur (−), GadE (+), PdhR (−) |
| dctA | Dicarboxylate transport | 1.3 | 1.4 | 2.1 | 2.8 | ArcA (−), CRP (+), DcuR (+) |
| fdoGHI | Formate dehydrogenase | 1.9 | 3.3 | 3.6 | 4.5 | |
| fumA | Fumarase | 1.0 | 1.5 | 1.9 | 3.6 | ArcA (−), CRP (+), FNR (−) |
| gapA | Glyceraldehyde-3-phosphate dehydrogenase | 0.6 | 0.8 | 0.4 | 0.3 | CRP (+), FruR (−) |
| glcDEFG | Glycolate oxidase | 1.0 | 1.1 | 1.2 | 2.2 | ArcA (−), GlcC (+), IHF (+) |
| gldA | Glycerol dehydrogenase | 0.6 | 0.7 | 0.5 | 0.4 | |
| gltIJKL | Glutamate transporter | 1.4 | 2.1 | 2.1 | 2.8 | FhlDC (+) |
| hyaA-F | Hydrogenase-1 | 0.7 | 0.5 | 0.4 | 0.3 | AppY (+), ArcA (+), Fis (−), IscR (−), NarL (−), NarP (−) |
| hycABCDEFGHI | Hydrogenase-3 | 0.6 | 0.4 | 0.4 | 0.4 | FhlA (+), IHF (+,) ModE (+), NssR (−) |
| icd | Isocitrate dehydrogenase | 1.8 | 1.6 | 2.0 | 3.3 | ArcA (−), FruR (+) |
| mglBAC | Galactose transporter | 1.5 | 3.6 | 4.1 | 6.1 | CRP (+), FlhDC (−), GalS (−), GalR (−) |
| nadK | NAD kinase | 1.3 | 2.4 | 2.1 | 2.7 | |
| narGHIJ | Nitrate reductase | 0.6 | 0.3 | 0.3 | 0.3 | Fis (−), FNR (+), IHF (+), NarL (+), RstA (+) |
| oppABCDF | Oligopeptide transporter | 0.6 | 0.6 | 0.5 | 0.4 | ArcA (−), Lrp (−), ModE (−) |
| pdhR-aceE-aceF-lpdA | Pyruvate dehydrogenase | 1.1 | 2.0 | 2.2 | 2.0 | CRP (+), FNR (−/+), PdhR (−) |
| sdhC-sdhD-sdhA-sdhB | Succinate dehydrogenase | 2.4 | 5.9 | 8.7 | 11.1 | ArcA (−/+), CRP (+), FNR (−), Fur (+) |
| sucABCD | Succinyl-CoA synthetase | 1.4 | 2.2 | 2.2 | 3.6 | ArcA (−), FNR (−), IHF (−) |
| yqaB | Fructose-1-phosphatase | 1.2 | 2.3 | 2.6 | 2.5 | |
TABLE 2.
Transcripts encoding proteins of the oxidative stress response that were present in altered abundance at different aerobiosis values
The -fold change (by at least 2-fold at one or more aerobiosis values) is the ratio of transcript levels under the indicated conditions compared with transcript levels at 0% aerobiosis (p ≤ 0.05). Relevant regulatory proteins are indicated: (−) denotes negative regulation, and (+) denotes positive regulation. The table was compiled from references in the text and the EcoCyc Database (35).
| Gene | Product | -Fold change in abundance relative to the anaerobic (0% aerobiosis) steady state |
Relevant regulatory proteins | |||
|---|---|---|---|---|---|---|
| 31% | 56% | 85% | 115% | |||
| grxB | Glutaredoxin-2 | 0.5 | 0.7 | 0.8 | 0.4 | |
| tpx | Lipid hydroperoxide peroxidase | 0.8 | 3.3 | 1.7 | 2.3 | ArcA (−), FNR (−) |
| msrB | Methionine sulfoxide reductase | 1.4 | 2.6 | 2.7 | 3.2 | Predicted FNR box |
| ychH | Role in hydrogen peroxide resistance | 1.5 | 2.7 | 2.1 | 2.4 | CRP (+) |
TABLE 3.
Transcripts encoding transcription factors that were present in altered abundance at different aerobiosis values
The -fold change (by at least 2-fold at one or more aerobiosis values) is the ratio of transcript levels under the indicated conditions compared with transcript levels at 0% aerobiosis (p ≤ 0.05). Relevant regulatory proteins are indicated: (−) denotes negative regulation, and (+) denotes positive regulation. The table was compiled from references in the text and EcoCyc Database (35).
| Gene | Product | -Fold change in abundance relative to the anaerobic (0% aerobiosis) steady state |
Relevant regulatory proteins | |||
|---|---|---|---|---|---|---|
| 31% | 56% | 85% | 115% | |||
| gadE | Regulator of pH homeostasis | 0.8 | 0.2 | 0.1 | 0.2 | CRP (−), EvgA (+), GadE (+), GadW (+), GadX (+), YdeO (+) |
| gadX | Regulator of pH homeostasis | 0.7 | 0.5 | 0.4 | 0.4 | ArcA (+), CRP (−), H-NS (−), Fis (−), FNR (−), GadE (+), GadW (−), RcsB (−), RutR (−), TorR (−) |
| hcaR | Regulator of 3-phenylpropionic acid catabolism | 0.8 | 1.5 | 1.2 | 2.0 | HcaR (−) |
| mhpR | Regulator of 3-hydroxyphenyl propionate degradation | 1.0 | 1.0 | 1.2 | 2.3 | |
| nac | Regulator of nitrogen assimilation | 0.8 | 2.9 | 1.6 | 1.0 | Nac (−), NtrC (+) |
| ygeK | Predicted transcriptional regulator | 0.9 | 2.1 | 1.0 | 1.1 | |
| yiaG | Predicted transcription factor | 1.0 | 0.7 | 0.4 | 0.3 | |
TABLE 4.
Transcripts encoding outer membrane proteins that were present in altered abundance at different aerobiosis values
The -fold change (by at least 2-fold at one or more aerobiosis values) is the ratio of transcript levels under the indicated conditions compared with transcript levels at 0% aerobiosis (p ≤ 0.05). Relevant regulatory proteins are indicated: (−) denotes negative regulation, (+) denotes positive regulation, and (−/+) denotes dual regulation. The table was compiled from references in the text and the EcoCyc Database (35).
| Gene | Product | -Fold change in abundance relative to the anaerobic (0% aerobiosis) steady state |
Relevant regulatory proteins | |||
|---|---|---|---|---|---|---|
| 31% | 56% | 85% | 115% | |||
| ompF | Outer membrane porin-1a | 1.8 | 3.7 | 2.9 | 3.3 | CpxR (−), CRP (+), EnvY (+), Fur (+), IHF (−/+), Lrp (+), OmpR (−/+), RstA (−) |
| ompW | Outer membrane porin W | 4.9 | 8.2 | 7.1 | 4.5 | FNR (+) |
| phoE | Outer membrane phosphoporin E | 1.6 | 2.9 | 2.6 | 3.0 | |
| slp | Outer membrane lipoprotein | 1.0 | 0.5 | 0.3 | 0.4 | GadW (+), GadX (+), MarA (−) |
| sfmH | Predicted adhesin | 1.0 | 1.9 | 2.3 | 2.0 | |
TABLE 5.
Transcripts encoding proteins of unknown function that were present in altered abundance at different aerobiosis values
The -fold change (by at least 2-fold at one or more aerobiosis values) is the ratio of transcript levels under the indicated conditions compared with transcript levels at 0% aerobiosis (p ≤ 0.05). Relevant regulatory proteins are indicated. The table was compiled from references in the text and the EcoCyc Database (35).
| Gene | Product | -Fold change in abundance relative to the anaerobic (0% aerobiosis) steady state |
Relevant regulatory proteins | |||
|---|---|---|---|---|---|---|
| 31% | 56% | 85% | 115% | |||
| Maximum abundance under fully aerobic conditions | ||||||
| yjhO | Predicted endoglucanase | 1.3 | 1.4 | 1.7 | 2.2 | |
| Decreased abundance as aerobiosis increases | ||||||
| yadO | Predicted ATP-dependent helicase | 0.1 | 0.1 | 0.1 | 0.1 | |
| ydfZ | Conserved protein | 0.5 | 0.6 | 0.3 | 0.3 | Predicted FNR box |
| yfaV | Predicted transporter | 0.8 | 0.9 | 0.9 | 0.4 | |
| yfbD | Conserved protein | 0.7 | 0.4 | 0.4 | 0.6 | |
| ytfL | Predicted inner membrane protein | 0.7 | 0.3 | 0.3 | 0.4 | |
| yjfM | Conserved protein | 0.4 | 0.6 | 0.7 | 0.4 | |
| Maximum abundance at intermediate aerobiosis values | ||||||
| yhfV | Predicted hydrolase | 1.2 | 2.8 | 1.0 | 1.0 | |
| yoaI | Predicted protein | 1.3 | 1.1 | 3.2 | 1.6 | |
| yoeE | CP4-44 prophage | 0.9 | 1.2 | 2.5 | 1.3 | |
The changes in the abundance of transcripts encoding central metabolic functions across the aerobiosis scale were predictable. Expression of citric acid cycle and glyoxylate shunt genes was enhanced as the degree of aerobiosis increased. This is consistent with the relief of ArcA-mediated repression (Table 1). Induction of the glyoxylate shunt with increased aerobiosis will tend to lower energy production by bypassing steps of the citric acid cycle that yield 2 NADH molecules, FADH2, and ATP. Lower NADH production and enhanced abundance of the nadK transcript (encoding NAD kinase) (Table 1) are likely to promote the production of the antioxidant NADPH, at the expense of the pro-oxidant NADH, by NADP+-dependent dehydrogenases, as suggested for Pseudomonas fluorescens (36). Thus, the profiles of “metabolic” transcripts and the lack of induction of OxyR- and SoxRS-regulated transcripts (37) as aerobiosis increased (Table 2) suggest that remodeling metabolism is sufficient to manage endogenous oxidative stress under the conditions investigated here.
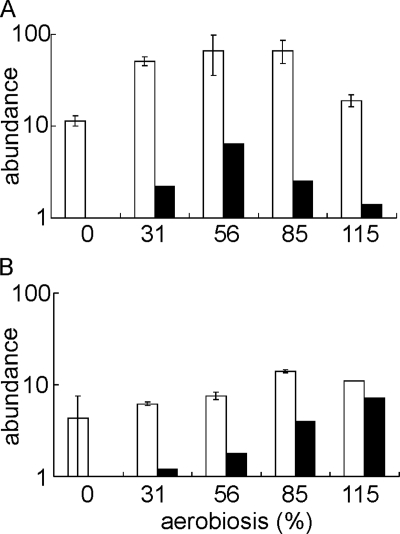
Recall that oxygen availability has a profound effect on the energetics of E. coli, which has two well characterized terminal oxidases: cytochrome bd-I (encoded by cydAB) and cytochrome bo′ (encoded by cyoABCDE) (38). The cydAB operon was maximally expressed at 56% aerobiosis, whereas the cyoABCDE operon was maximally expressed under fully aerobic conditions (Table 1). Measurements of the cytochrome bd and cytochrome bo′ content of cultures grown at different oxygen availabilities indicated that the changes in abundance of the cydAB and cyoA–E transcripts were reflected in changes in cellular content of cytochrome bd and cytochrome bo′ (Fig. 1). This is consistent with the previously measured oxygen affinities of the two major oxidases; cytochrome bd-I has an extraordinarily high oxygen affinity (Km = 3–8 nm) (39), whereas cytochrome bo′ has Km values in the range 0.05–0.2 μm (40). The high oxygen affinity of cytochrome bd is presumably the basis of the recently discovered ability of E. coli to grow on non-fermentable substrates at nanomolar oxygen concentrations (41). The shifts in oxidase expression seen in this study are thus consistent with a progressive switch from fermentative metabolism to micro-aerobic respiration (using cytochrome bd-I as the terminal oxidase) and then to fully aerobic respiration (using cytochrome bo′ as the terminal oxidase) as oxygen availability was increased.
FIGURE 1.
The cytochrome bd and bo′ content of cultures of E. coli MG1655 across the aerobiosis scale. The graphs show the amounts of cytochrome bd (A) and cytochrome bo′ (B) (molecules/cell × 10−3 on a log10 scale; open bars) calculated as described under “Experimental Procedures” and the -fold changes in the abundance of the cydA (A) and cyoA (B) transcripts relative to that at 0% aerobiosis (data from Table 1) shown on a log10 scale (closed bars) in cultures grown at the indicated aerobiosis points.
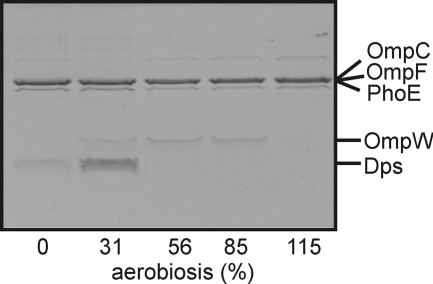
The profiles of transcripts encoding the outer membrane proteins OmpF, OmpW, PhoE, Slp, and SfmH changed across the aerobiosis scale (Table 4). The ompF, phoE, and sfmH transcripts exhibited maximum abundance at aerobiosis values ≥56%, whereas the abundance of the slp transcript decreased at aerobiosis values ≥56%, and the ompW transcript was maximal at 56% aerobiosis, with lower expression in the presence of more or less oxygen (Table 4). Analysis of outer membrane protein profiles confirmed that changing aerobiosis resulted in remodeling of the cell envelope (Fig. 2). Particularly striking was the correlation between the transcript and protein profiles for OmpW, both being maximal in the mid-aerobiosis range (Fig. 2 and Table 4). Expression of ompW is inhibited by the small RNA RybB, which is under the control of the alternative sigma factor σE (42). It has also been suggested that the oxygen-responsive transcription factor FNR activates expression of ompW (7), although direct evidence for FNR interaction with the ompW promoter is lacking. There are two putative FNR sites (both 9/10 matches to the consensus) located at −81.5 and −126.5 relative to the predicted ompW transcript start. Tandem FNR sites have been previously associated with micro-aerobic expression patterns (43), and hence, the tandem FNR sites in the ompW promoter region provide a plausible explanation for the observed ompW transcript profile.
FIGURE 2.
Changes in outer membrane protein profiles across the aerobiosis scale. The Coomassie Blue-stained urea/SDS-polyacrylamide gel shows polypeptides present in outer membrane preparations isolated from equivalent numbers of bacteria sampled from steady-state cultures grown at the indicated points on the aerobiosis scale. The proteins were identified by N-terminal amino acid sequencing.
A surprising feature of the outer membrane profiles was the presence of Dps at the lower end of the aerobiosis scale (Fig. 2). It is noted that the abundance of the dps transcript was not significantly altered in the transcriptomic analysis. The major role of Dps is to protect the genetic information of the cell from oxidative damage by forming condensed co-crystals with chromosomal DNA in stationary phase cultures (reviewed in Ref. 44). However, Dps has also been identified as an outer membrane protein in a bacteriophage-tolerant E. coli MG1655 strain (45), and in Helicobacter pylori, where it plays a role in modulating adhesion and chemotaxis of mononuclear phagocytes (46). The presence of Dps in the outer membrane of E. coli grown at low oxygen availabilities suggests that this protein might play a secondary role in modifying the properties of the cell envelope in anaerobic/micro-aerobic environments. Alternatively, any co-crystals formed by interaction between Dps and chromosomal DNA (44) might have been pelleted along with the outer membrane fraction. However, because the growth rate was the same for all cultures, it is not obvious why this would occur only in those grown at 31% aerobiosis.
Although the roles of FNR and ArcA in mediating the changes in transcript profiles necessary for adaptation to growth at different oxygen availabilities are well established (see above), the consequent changes in physiology are expected to modulate the activities of other transcription factors that respond to specific metabolic signals that are affected by oxygen availability. For example, as aerobiosis increased, the production of weak organic acids decreased (Table 6). Accordingly, the abundance of transcripts (gadA, gadBC, and hdeAB) associated with acid resistance decreased as aerobiosis increased (Table 7). This suggests that although the external culture pH was maintained at 6.9, the generation of acidic fermentation products at the low end of the aerobiosis scale could depress intracellular pH. Induction of the gadA and gadBC genes facilitates proton consumption by glutamate decarboxylase (GadA and GadB) and exchange of the product of this reaction, 4-aminobutyrate, for excreted glutamate by GadC (47) to counteract acidification of the cytoplasm. This is accompanied by induction of the periplasmic chaperones HdeA and HdeB (48). Consistent with this interpretation, the abundance of transcripts encoding the acid-responsive regulators of gadA, gadBC, and hdeAB expression, GadE and GadX, decreased as aerobiosis increased (Tables 3 and 7).
TABLE 6.
Measurements of acidic fermentation products and biomass production for steady-state cultures of E. coli MG1655 grown at different aerobiosis values
Metabolites were quantified by NMR as described under “Experimental Procedures.” The specific rates (q) are expressed as millimoles/g of cell dry weight/h (mean ± S.D., n = 3).
| Aerobiosis | Biomass | qacetate | qformate | qsuccinate | qlactate |
|---|---|---|---|---|---|
| g/liter | |||||
| 0% | 0.34 ± 0.05 | 10.1 ± 0.3 | 8.5 ± 2.3 | 1.8 ± 0.4 | 0.10 ± 0.05 |
| 31% | 0.62 ± 0.05 | 6.2 ± 0.4 | 2.5 ± 0.5 | 1.1 ± 0.1 | 0.01 ± 0.05 |
| 56% | 0.91 ± 0.05 | 4.7 ± 0.2 | 0 | 0 | 0 |
| 85% | 1.1 ± 0.08 | 1.3 ± 0.2 | 0 | 0 | 0 |
| 115% | 1.3 ± 0.02 | 0.1 ± 0.1 | 0 | 0 | 0 |
TABLE 7.
Transcripts encoding proteins associated with acid resistance that were present in altered abundance at different aerobiosis values
The data shown are for the first gene in the operon except where indicated by boldface type. The -fold change (by at least 2-fold at one or more aerobiosis values) is the ratio of transcript levels under the indicated conditions compared with transcript levels at 0% aerobiosis (p ≤ 0.05). Relevant regulatory proteins are indicated: (−) denotes negative regulation, (+) denotes positive regulation, and (−/+) denotes dual regulation. The table was compiled from references in the text and the EcoCyc Database (35).
| Operon | Product | -Fold change in abundance relative to the anaerobic (0% aerobiosis) steady state |
Relevant regulatory proteins | |||
|---|---|---|---|---|---|---|
| 31% | 56% | 85% | 115% | |||
| gadA | Glutamate decarboxylase | 0.9 | 0.2 | 0.1 | 0.2 | CRP (−), Fis (−), GadE (+), GadW (−), GadX (+), RcsB (+) |
| gadBC | Glutamate decarboxylase, Glutamate: 4-aminobutyrate antiporter | 1.0 | 0.2 | 0.1 | 0.2 | CRP (−), Fis (−), GadE (+), GadX (+), RcsB (+) |
| gadE | Regulator of pH homeostasis | 0.8 | 0.2 | 0.1 | 0.2 | CRP (−), EvgA (+), GadE (+), GadW (+), GadX (+), YdeO (+) |
| gadX | Regulator of pH homeostasis | 0.7 | 0.5 | 0.4 | 0.4 | ArcA (+), CRP (−), H-NS (−), Fis (−), FNR (−), GadE (+), GadW (−), RcsB (−), RutR (−), TorR (−) |
| hdeAB-yhiD | Periplasmic acid-resistance chaperones | 0.7 | 0.9 | 0.3 | 0.3 | GadE (+), GadW (−/+), GadX (−/+), H-NS (−), Lrp (−), MarA (−), PhoP (+), TorR (+) |
To gain insight into the changes in the activity of the major oxygen-responsive two-component system ArcBA (49) across the aerobiosis scale, the transcriptomic data sets were analyzed using the probabilistic model TFInfer (22). The model is a simple modification of the dynamical model described by Sanguinetti et al. (50), retaining the same log-linear model for gene expression but replacing the Markovian prior on transcription factor activities with independent priors. The code used is freely available in the open-source C# package TFInfer.
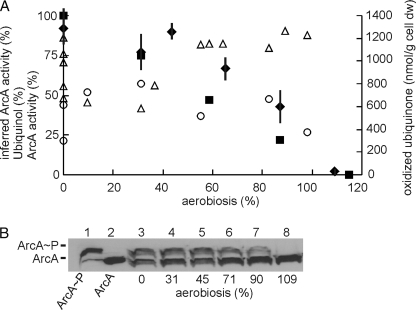
The predicted ArcA activity profile suggested a linear decrease in activity with increasing aerobiosis, a response similar to that of qacetate, which is the basis of the aerobiosis scale (Fig. 3A, closed squares). ArcA is the regulator in the ArcBA two-component system. Under conditions of low oxygen availability, the sensor, ArcB, is autophosphorylated and activates ArcA by transfer of the phospho-group (12). This process is inhibited by ubiquinone (18, 19). Phosphorylated ArcA is dephosphorylated (inactivated) by the phosphatase activity of ArcB. ArcB phosphatase activity is inhibited by fermentation products (20). Therefore, the redox state of the ubiquinone/ubiquinol pool was compared with the inferred ArcA activity profile. At low aerobiosis values (≤31%), the amount of ubiquinol (as a percentage of the total ubiquinone/ubiquinol pool) settled at ∼50%, but with significant variation at 0% aerobiosis (Fig. 3A, open triangles), which was also evident in the measurements of the amount of cytochrome bo′ (Fig. 1B). At higher aerobiosis values (≥31%), the percentage of ubiquinol increased to ∼80% of the total ubiquinone/ubiquinol pool (Fig. 3A). Furthermore, the total amount of oxidized ubiquinone was similar across the micro-aerobic range (Fig. 3A, open circles). These observations, like those of Bekker et al. (21), are not consistent with ubiquinone-mediated inhibition of ArcB autokinase activity being the major determinant of ArcA activity. This could indicate that the inferred ArcA activities are incorrect or that the redox state of the ubiquinone/ubiquinol pool has only a minor influence on ArcA activity in vivo. Therefore, direct experimental verification of the inferred ArcA activity was sought by measuring the phosphorylation state of the protein at different points on the aerobiosis scale. To measure the proportion of phosphorylated ArcA as a percentage of the total ArcA protein, Phos-tagTM-acrylamide gel electrophoresis was used, followed by Western blotting (Fig. 3B). The total amount of ArcA protein per E. coli cell remained relatively constant across the aerobiosis range (∼8000 molecules/cell, estimated by calibrating the Western blots with known amounts of ArcA). At 0% aerobiosis, the proportion of phosphorylated ArcA was 46 ± 5.5%, consistent with the observation that active ArcA is an octamer of ArcA and ArcA∼P at a 1:1 ratio (17). Furthermore, the data obtained show that the proportion of phosphorylated ArcA correlated well with the inferred ArcA activity profile (Fig. 3A, compare closed diamonds and closed squares), with maximum phosphorylation and activity at 0% aerobiosis, consistent with the observation that ArcB-mediated phosphorylation of ArcA is maximal under anaerobic conditions (51).
FIGURE 3.
The redox state of the ubiquinone/ubiquinol pool is not the major determinant of ArcA activity. A, the inferred activity of ArcA from the transcriptomic data assuming 100% activity at 0% aerobiosis (■), the measured percentage of ubiquinol (△), the total amount of oxidized ubiquinone (○), and the calculated ArcA activity assuming that the maximum is reached when ArcA is 50% phosphorylated (17) (♦; bars indicate S.D., n = 4) plotted against the aerobiosis scale. B, a typical Western blot developed using ArcA polyclonal antiserum for steady-state cultures grown at different points on the aerobiosis scale. Lane 1, purified ArcA (15 ng) phosphorylated in vitro using carbamoyl phosphate; lane 2, dephosphorylated ArcA (15 ng); lanes 3–8, extracts from cultures grown at 0, 31, 45, 71, 90, and 109% aerobiosis, respectively. Extracts from an arcA mutant did not produce a signal (not shown). dw, dry weight.
Thus, it is concluded that ArcA activity inferred from analysis of transcript profiles corresponds to the cellular content of phosphorylated ArcA. However, ArcA activity did not correlate with changes in the total amount of oxidized ubiquinone or with the redox state of the ubiquinone/ubiquinol pool. The absence of a correlation between ArcA activity, estimated indirectly using a modified cydA-lacZ fusion, and the redox state of the ubiquinone/ubiquinol pool was also observed previously using a different E. coli strain (MC4100) and growth conditions (40 mm glucose, 0.15-h−1 dilution rate) (21). Furthermore, analysis of ubiC (unable to synthesize ubiquinone) and menB (unable to synthesize dimethylmenaquinone) mutants suggested that menaquinone plays the more important role in ArcBA activation under the conditions used by Bekker et al. (21). However, the ArcA activity profiles reported here differ from those reported by Bekker et al. (21). In the latter, a complex ArcA activity profile was suggested with peaks of β-galactosidase activity at 0 and 60–80% aerobiosis and minimum ArcA activity when the percentage of ubiquinol was maximal (21). It is possible that the different strains and growth conditions used in the two studies might contribute to differences in ArcA activity and ubiquinone/ubiquinol redox profiles. However, it is also possible that the output from the cydA-lacZ reporter was influenced by additional regulators, and thus, the direct determination of ArcA phosphorylation reported here is likely to be a more reliable and accurate measure of ArcA activity. The good correlation between inferred ArcA activity, ArcA phosphorylation, and aerobiosis (i.e. qacetate) implies that fermentation products influence ArcA activity, presumably by inhibition of ArcB phosphatase activity as shown by Iuchi (20). It is perhaps not surprising that, as a regulator of genes that have fundamental roles in bacterial physiology, ArcB should integrate the response to multiple signals (e.g. ubiquinone, menaquinone, acetate, and other fermentation products); however, the inverse linear correlation between ArcA activity and aerobiosis suggests that, at least under the conditions used here, the rate of fermentation product synthesis (qacetate) exerts a greater influence on ArcA activity than the redox state of the quinone pools.
Acknowledgments
We thank Michael Ederer and Ronny Feuer (Institute for System Dynamics, University of Stuttgart) for assistance with statistical analysis of cytochrome numbers, Arthur Moir (University of Sheffield) for N-terminal amino acid sequencing, and all the other members of the SysMO-SUMO consortium (K. Bettenbrock, M. Ederer, E. D. Gilles, S. Henkel, M. Holcombe, S. Maleki-Dizaji, T. Sauter, O. S awodny, S. Stagge, and S. Steinsiek) for many useful discussions.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council UK through SysMO and Project Grant BB/E019943/1.
REFERENCES
- 1. Guest J. R., Green A. S., Irvine, Spiro S. (1996) in Regulation of Gene Expression in Escherichia coli (Linn E. C. C., Lynch A. S. eds) pp. 317–342, R. G. Landes & Co., Austin, TX [Google Scholar]
- 2. Sawers G. (1999) Curr. Opin. Microbiol. 2, 181–187 [DOI] [PubMed] [Google Scholar]
- 3. Taylor C. T., Colgan S. P. (2007) J. Mol. Med. 85, 1295–1300 [DOI] [PubMed] [Google Scholar]
- 4. Garcia-Ochoa F., Gomez E. (2009) Biotechnol. Adv. 27, 153–176 [DOI] [PubMed] [Google Scholar]
- 5. Alexeeva S., Hellingwerf K. J., Teixeira de Mattos M. J. (2002) J. Bacteriol. 184, 1402–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pirt S. J. (1975) Principles of Microbe and Cell Cultivation, pp. 81–106, Blackwell Scientific Publications, Oxford, UK [Google Scholar]
- 7. Constantinidou C., Hobman J. L., Griffiths L., Patel M. D., Penn C. W., Cole J. A., Overton T. W. (2006) J. Biol. Chem. 281, 4802–4815 [DOI] [PubMed] [Google Scholar]
- 8. Kang Y., Weber K. D., Qiu Y., Kiley P. J., Blattner F. R. (2005) J. Bacteriol. 187, 1135–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Partridge J. D., Scott C., Tang Y., Poole R. K., Green J. (2006) J. Biol. Chem. 281, 27806–27815 [DOI] [PubMed] [Google Scholar]
- 10. Partridge J. D., Sanguinetti G., Dibden D. P., Roberts R. E., Poole R. K., Green J. (2007) J. Biol. Chem. 282, 11230–11237 [DOI] [PubMed] [Google Scholar]
- 11. Salmon K., Hung S. P., Mekjian K., Baldi P., Hatfield G. W., Gunsalus R. P. (2003) J. Biol. Chem. 278, 29837–29855 [DOI] [PubMed] [Google Scholar]
- 12. Green J., Paget M. S. (2004) Nat. Rev. Microbiol. 2, 954–966 [DOI] [PubMed] [Google Scholar]
- 13. Lazazzera B. A., Beinert H., Khoroshilova N., Kennedy M. C., Kiley P. J. (1996) J. Biol. Chem. 271, 2762–2768 [DOI] [PubMed] [Google Scholar]
- 14. Crack J. C., Green J., Cheesman M. R., Le Brun N. E., Thomson A. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2092–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jervis A. J., Crack J. C., White G., Artymiuk P. J., Cheesman M. R., Thomson A. J., Le Brun N. E., Green J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4659–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khoroshilova N., Popescu C., Münck E., Beinert H., Kiley P. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6087–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeon Y., Lee Y. S., Han J. S., Kim J. B., Hwang D. S. (2001) J. Biol. Chem. 276, 40873–40879 [DOI] [PubMed] [Google Scholar]
- 18. Georgellis D., Kwon O., Lin E. C. (2001) Science 292, 2314–2316 [DOI] [PubMed] [Google Scholar]
- 19. Malpica R., Franco B., Rodriguez C., Kwon O., Georgellis D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iuchi S. (1993) J. Biol. Chem. 268, 23972–23980 [PubMed] [Google Scholar]
- 21. Bekker M., Alexeeva S., Laan W., Sawers G., Teixeira de Mattos J., Hellingwerf K. (2010) J. Bacteriol. 192, 746–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Asif H. M., Rolfe M. D., Green J., Lawrence N. D., Rattray M., Sanguinetti G. (2010) Bioinformatics 26, 2635–2636 [DOI] [PubMed] [Google Scholar]
- 23. Blattner F. R., Plunkett G., 3rd, Bloch C. A., Perna N. T., Burland V., Riley M., Collado-Vides J., Glasner J. D., Rode C. K., Mayhew G. F., Gregor J., Davis N. W., Kirkpatrick H. A., Goeden M. A., Rose D. J., Mau B., Shao Y. (1997) Science 277, 1453–1462 [DOI] [PubMed] [Google Scholar]
- 24. Datsenko K. A., Wanner B. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Evans C. G., Herbert D., Tempest D. W. (1970) Methods Microbiol. 2, 278–327 [Google Scholar]
- 26. Yang Y. H., Speed T. (2002) Nat. Rev. Genet. 3, 579–588 [DOI] [PubMed] [Google Scholar]
- 27. Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004) FEBS Lett. 573, 83–92 [DOI] [PubMed] [Google Scholar]
- 28. Barbieri C. M., Stock A. M. (2008) Anal. Biochem. 376, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalnenieks U., Galinina N., Bringer-Meyer S., Poole R. K. (1998) FEMS Microbiol. Lett. 168, 91–97 [DOI] [PubMed] [Google Scholar]
- 30. Kita K., Konishi K., Anraku Y. (1986) Methods Enzymol. 126, 94–113 [DOI] [PubMed] [Google Scholar]
- 31. Neidhardt F. C., Ingraham J. L., Schaechter M. (1990) Physiology of the Bacterial Cell: A Molecular Approach, Sinauer Associates, Inc. Sunderland, MA [Google Scholar]
- 32. Bekker M., Kramer G., Hartog A. F., Wagner M. J., de Koster C. G., Hellingwerf K. J., de Mattos M. J. (2007) Microbiology 153, 1974–1980 [DOI] [PubMed] [Google Scholar]
- 33. Lange H. C., Eman M., van Zuijlen G., Visser D., van Dam J. C., Frank J., de Mattos M. J., Heijnen J. J. (2001) Biotechnol. Bioeng. 75, 406–415 [DOI] [PubMed] [Google Scholar]
- 34. Chart H. (1994) Methods in Practical Laboratory Bacteriology, pp. 1–10, CRC Press Inc., Boca Raton, FL [Google Scholar]
- 35. Keseler I. M., Bonavides-Martínez C., Collado-Vides J., Gama-Castro S., Gunsalus R. P., Johnson D. A., Krummenacker M., Nolan L. M., Paley S., Paulsen I. T., Peralta-Gil M., Santos-Zavaleta A., Shearer A. G., Karp P. D. (2009) Nucleic Acids Res. 37, D464–D470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh R., Lemire J., Mailloux R. J., Appanna V. D. (2008) PLoS One 3, e2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Storz G., Zheng M. (2000) Bacterial Stress Responses, pp. 47–59, ASM Press, Washington, D. C [Google Scholar]
- 38. Gennis R. B., Stewart V. (1996) Escherichia coli and Salmonella Cellular and Molecular Biology, 2nd Ed., pp. 217–261, ASM Press, Washington, D. C [Google Scholar]
- 39. D'Mello R., Hill S., Poole R. K. (1996) Microbiology 142, 755–763 [DOI] [PubMed] [Google Scholar]
- 40. D'Mello R., Hill S., Poole R. K. (1995) J. Bacteriol. 177, 867–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stolper D. A., Revsbech N. P., Canfield D. E. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18755–18760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johansen J., Rasmussen A. A., Overgaard M., Valentin-Hansen P. (2006) J. Mol. Biol. 364, 1–8 [DOI] [PubMed] [Google Scholar]
- 43. Marshall F. A., Messenger S. L., Wyborn N. R., Guest J. R., Wing H., Busby S. J., Green J. (2001) Mol. Microbiol. 39, 747–753 [DOI] [PubMed] [Google Scholar]
- 44. Haikarainen T., Papageorgiou A. C. (2010) Cell. Mol. Life Sci. 67, 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lacqua A., Wanner O., Colangelo T., Martinotti M. G., Landini P. (2006) Appl. Environ. Microbiol. 72, 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Allen L. A. (2000) J. Exp. Med. 191, 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moreau P. L. (2007) J. Bacteriol. 189, 2249–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kern R., Malki A., Abdallah J., Tagourti J., Richarme G. (2007) J. Bacteriol. 189, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salmon K. A., Hung S. P., Steffen N. R., Krupp R., Baldi P., Hatfield G. W., Gunsalus R. P. (2005) J. Biol. Chem. 280, 15084–15096 [DOI] [PubMed] [Google Scholar]
- 50. Sanguinetti G., Lawrence N. D., Rattray M. (2006) Bioinformatics 22, 2775–2781 [DOI] [PubMed] [Google Scholar]
- 51. Lynch A. S., Lin E. C. (1996) J. Bacteriol. 178, 6238–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]