Abstract
The thermophilic lactic acid bacterium Streptococcus thermophilus is widely and traditionally used in the dairy industry. Despite the vast level of consumption of S. thermophilus through yogurt or probiotic functional food, very few data are available about its physiology in the gastrointestinal tract (GIT). The objective of the present work was to explore both the metabolic activity and host response of S. thermophilus in vivo. Our study profiles the protein expression of S. thermophilus after its adaptation to the GIT of gnotobiotic rats and describes the impact of S. thermophilus colonization on the colonic epithelium. S. thermophilus colonized progressively the GIT of germ-free rats to reach a stable population in 30 days (108 cfu/g of feces). This progressive colonization suggested that S. thermophilus undergoes an adaptation process within GIT. Indeed, we showed that the main response of S. thermophilus in the rat's GIT was the massive induction of the glycolysis pathway, leading to formation of lactate in the cecum. At the level of the colonic epithelium, the abundance of monocarboxylic acid transporter mRNAs (SLC16A1 and SLC5A8) and a protein involved in the cell cycle arrest (p27kip1) increased in the presence of S. thermophilus compared with germ-free rats. Based on different mono-associated rats harboring two different strains of S. thermophilus (LMD-9 or LMG18311) or weak lactate-producing commensal bacteria (Bacteroides thetaiotaomicron and Ruminococcus gnavus), we propose that lactate could be a signal produced by S. thermophilus and modulating the colon epithelium.
Keywords: Bacterial Metabolism, Epithelial Cell, Intestine, Lactic Acid, Proteomics, SLC Transporters, Streptococcus thermophilus, Microbiota, p27kip1
Introduction
Streptoccocus thermophilus belongs to the group of the thermophilic lactic acid bacteria and is traditionally and widely used as a starter in manufacturing dairy products (Emmental, Gruyere, Parmigiano, mozarella, yogurt, etc.). Yogurt, which results from the fermentation of milk by S. thermophilus and Lactobacillus delbrueckii sp. bulgaricus (L. bulgaricus), fulfills the current specifications required to be recognized as a probiotic product (1). The health beneficial effect of yogurt consumption is linked to the metabolic properties of S. thermophilus and L. bulgaricus. As such, it improves lactose digestion in the gastrointestinal tract (GIT)3 through their lactose-hydrolyzing activity present in yogurt and in the GIT, thus reducing symptoms of lactose intolerance (2, 3).
Yogurt cultures have been shown to induce other health benefits, such as reduction of diarrhea or allergic disorders as well as modulation of the immune system (1, 4). S. thermophilus is also present at high concentration in VSL#3, a probiotic mixture of eight different bacterial strains that possesses beneficial effects in several intestinal conditions (5, 6).
Recent data indicate that strains related to S. thermophilus LMD-9 are among the 57 bacteria species found in the intestinal microbiota of 90% of 124 European individuals (7). In comparison with the overall human intestinal microbiota, S. thermophilus is a numerically nondominant species with variable levels (7–10). At birth, the Streptococcus genus, with in some studies a precision at the level of S. thermophilus species, is among the first colonizers of the GIT because it has been detected in infant feces and breast milk (11–13). Thus, Streptococcus, as pioneer bacteria colonizing a yet immature GIT, may impact the maturation and homeostasis of the intestinal epithelium after birth.
Convergent data show that intestinal bacteria modulate proliferation and differentiation processes of the intestinal epithelium (14–16), which is one of the most dynamic tissues of the whole organism. Moreover, we have recently demonstrated that microbiota increases colonic epithelium crypt depth in concordance with a well orchestrated modulation of the level of proteins involved in life cycle steps like PCNA, Bcl2, and p21cip1, p27kip1 (markers of proliferation, antiapoptotic pathway, and cell cycle arrest, respectively) (17). Despite its large utilization and consumption, its probiotic-associated traits, and its presence in the intestinal microbiota, the role of S. thermophilus in the gut is still largely unknown. A previous study (18) showed that, in mono-associated animals, S. thermophilus produced an active β-galactosidase in the GIT, which is responsible for lactose hydrolysis. Therefore, the aim of the present study was to expand our view of the metabolic activity of S. thermophilus in the intestinal environment and to investigate its relationship with the host. To do this, we assessed the behavior of two sequenced strains of S. thermophilus, LMD-9 and LMG18311, in gnotobiotic rats. We established the first global protein profile of S. thermophilus after its survival in GIT in mono-associated rats. We also described how S. thermophilus influences colonic epithelium, focusing on monocarboxylic acid transporters and cell cycle arrest proteins. In view of our findings using different mono-associated models, we propose that lactate resulting from the adaptive metabolic activity of S. thermophilus may serve as a biological signal to communicate with host epithelium.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Media, and Culture Conditions
The strains S. thermophilus LMD-9 (ATCC BAA-491), S. thermophilus LMG18311 (Belgian Coordinated Collections of Micro-organisms collection), L. bulgaricus ATCC11842, Bacteroides thetaiotaomicron (BtII8), and Ruminococcus gnavus (FRE1) were used.
Stock cultures of S. thermophilus LMD-9, S. thermophilus LMG18311, and L. bulgaricus ATCC11842 were prepared in reconstituted 10% (w/v) Nilac skim milk (NIZO, Ede, The Netherlands) as described previously (19). S. thermophilus monocultures and S. thermophilus/L. bulgaricus co-cultures were obtained by inoculating Nilac milk with 106 cfu/ml stock cultures of each species and incubated at 42 °C until pH 5.4–5.5. One ml of culture was used for rat gavage, and the remaining was liquid nitrogen-frozen and stored at −20 °C until protein extraction. The cultures were enumerated a posteriori by plating appropriate dilutions on M17 agar lactose (10 g/liter) for S. thermophilus or on acidified MRS agar lactose (20 g/liter) (pH 5.2) for L. bulgaricus. After 16 h (S. thermophilus) or 24 h (L. bulgaricus), incubation at 42 °C under anaerobiosis (Anaerocult A, Merck), colonies were counted.
Animals and Experimental Design
All procedures were carried out according to European and French guidelines for the care and use of laboratory animals (permission 78-123, dedicated to M.T.).
At the age of 2 months, germ-free (GF) rats (male, Fisher 344) were inoculated either with S. thermophilus LMD-9 (Ino-LMD9, n = 11), with S. thermophilus LMG18311 (Ino-LMG18311, n = 4), or with a mix of S. thermophilus and L. bulgaricus (Ino-LMD9+Lb, n = 5) according to the following protocol. 1 ml of a culture of S. thermophilus in Nilac milk (5 × 108 S. thermophilus/ml) or 1 ml of a co-culture with S. thermophilus (2 × 109 S. thermophilus/ml) and L. bulgaricus (8 × 107 L. bulgaricus/ml) were transferred to GF rat by oral gavage. As a control, GF rats were also inoculated with 1 ml of sterile Nilac milk (without bacteria). GF, mono-associated, and di-associated rats were housed in sterile Plexiglas isolators (Ingénia, Vitry-sur-Seine, France). Rats mono-associated with either B. thetaiotaomicron (Ino-Bt) or R. gnavus (Ino-Rg) were described previously (17). All groups of rats received the same standard diet (UAR, Villemoisson, France) which was sterilized by γ-irradiation. Throughout the experiment and two times a week, S. thermophilus (and L. bulgaricus in the case of Ino-LMD9+Lb) was enumerated by plating serial dilutions of the feces. All rats were euthanized at 3 months of age (i.e. 30 days after inoculation).
At 9:00 a.m., rats were anesthetized with isoflurane, and tissues were recovered. The colon was immediately used either for cell isolation or for histological procedures. The luminal content of jejunum, ileum, cecum, and feces was diluted in 10 volumes of M17 and vigorously vortexed with sterile glass beads (3.5-mm diameter), and bacteria were enumerated. All results were expressed as log10 (cfu/g).
HT29 Cell Line
HT29 cells were cultivated as described previously (20) and incubated with 0, 20, or 50 mm l-lactate for 18 h.
Scanning Electron Microscopy
Scanning electron microscopy (21) analyses were performed on the MIMA2 platform (available on the World Wide Web) with 0.2 g of cecal samples being suspended in 1 ml of Tris buffer (0.1 m, pH 7.5) and mixed. Debris were removed by centrifugation at 1000 × g for 5 min at 4 °C. Supernatant of cecal samples was centrifuged to recover bacteria, and the pellets were suspended in 0.1 m Tris buffer (pH 7.5) and washed twice in the same buffer by centrifugation at 5000 × g for 5 min at 4 °C. The bacterial pellets were suspended and fixed in 200 μl of glutaraldehyde, 3% ruthenium red and stored at 4 °C. Scanning electron microscopy was performed as reported previously (22).
Protein Extraction
Bacteria were isolated as follows. 500 ml of frozen milk cultures (three replicates) were homogenized with an Ultra-turrax (Bioblock, Paris, France). The suspension was centrifuged at 8000 × g for 10 min at 4 °C and washed three times in sodium phosphate buffer (5 mm) containing 1 mm EDTA, pH 7; bacteria were suspended in 2 ml of sodium phosphate buffer (20 mm, pH 7). Frozen feces from three Ino-LMD9 rats were pooled, and 12 g of feces were suspended in 25 ml of sodium phosphate buffer (20 mm, pH 6.4), homogenized with an Ultra-turrax, and fecal debris was removed by two successive centrifugations at 400 × g for 5 min at 4 °C. The bacteria were then collected by centrifugation at 5000 × g for 10 min at 4 °C and washed three times with sodium phosphate buffer (20 mm, pH 6.4), and the pellets were suspended in 2 ml of this buffer. Bacterial pellets from milk cultures and feces were then submitted to a high speed centrifugation on a Nycodenz density gradient as described previously (23). After ultracentrifugation, the bacterial suspensions were washed with 14 ml of sodium phosphate buffer (20 mm, pH 6.4), and the bacteria were recovered by centrifugation at 5000 × g for 10 min at 4 °C.
The bacterial pellets were suspended in 0.5 ml of sodium phosphate buffer (20 mm, pH 6.4) containing protease inhibitor mixture (Sigma-Aldrich), 40 units/ml catalase, and 10 mm tributylphosphine (Applied Biosystems). Cells were mechanically disrupted with an FP120 FastPrep cell disruptor (Bio 101 Systems, Qbiogen, Irvine, Canada) by two 30-s cycles of homogenization at maximum speed (6.5) at a 1-min interval at 4 °C. Supernatants were collected by centrifugation at 5000 × g, 15 min at 4 °C, and centrifuged (200,000 × g, 30 min, 4 °C); they corresponded to cytoplasmic proteins, whereas the pellet was a fraction enriched with envelope-associated proteins. The protein concentrations were determined by the Coomassie protein assay reagent (Pierce), using bovine serum albumin as a standard.
One-dimensional Electrophoresis Coupled to Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
Proteins of the cell envelope-enriched fraction (4.7 μg) from fecal sample were separated and identified by one-dimensional electrophoresis (SDS-PAGE) coupled with liquid chromatography-tandem mass spectrometry as described previously (24).
Comparative Two-dimensional Protein Analysis and Image Analysis
A volume of cytosolic fraction corresponding to 250 μg of proteins was treated as described previously (25). Fractions of three independent cultures in milk were analyzed. Fractions of two fecal batches were used, each batch resulting from the pool of feces of three rats. Isoelectric focusing was performed with a 24-cm pH 4–7 immobilized pH gradient strip, and 12% SDS-PAGE was used for the second dimension. The gels were stained with BioSafe colloidal Coomassie blue (Bio-Rad) for 1 h and rinsed out with three successive washes in deionized water. Gels were digitized using an Epson Expression 1640XL scanner set (at 256 gray levels) controlled by the Silver Fast software and analyzed using the Progenesis SameSpot software (Nonlinear Dynamics). The relative volume of each spot was obtained from its intensity and normalized with the intensity of all spots. Only spot volume differences of at least 2-fold between the milk and fecal samples were further analyzed. 276 different protein spots on pH 4–7 two-dimensional electrophoresis gels were detected. MS analyses were performed using a Voyager-DE-STR (Applied Biosystems, Framingham, MA) on the PAPSSO proteomic platform (available on the World Wide Web). The proteins were identified using MS-FIT (available on the World Wide Web).
Western Blot Analysis
Colonic epithelial cells were isolated from the whole colon according to the method described by Cherbuy et al. (26). Cell pellet from whole colon or from HT29 cells was immediately used for protein extraction. Western blot analysis was performed as described previously (17) by using a denaturing (SDS)-polyacrylamide gel. Proteins were analyzed using anti-PCNA (GeneTex; diluted 1:1000), anti-p21cip1 (Oncogene (diluted 1:200) or Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) (diluted 1:200)), anti-p27kip1 (Santa Cruz Biotechnology, Inc.; diluted 1:500), or anti-Bcl2 (Santa Cruz Biotechnology, Inc.; diluted 1:400). Several loading controls were used, either GAPDH, Skp1, or cullin1 (17). Signals imprinted on autoradiography films were quantified by scanning densitometry of the autoradiograph using Biovision 1000 and logiciel bio1D (Vilber Lourmat, France).
Dosage of d- and l-Lactates
d- and l-lactates were measured in cecal contents and feces of GF and mono-associated rats by 2× diluting of 50–100-mg samples in 0.1 m triethanolamine buffer (pH 9.15). Samples were centrifuged at 13,000 × g for 5 min at 4 °C, and the supernatants were precipitated with trichloroacetic acid (10%) and centrifuged at 4500 × g for 20 min at 4 °C. The lactates were measured in the supernatants with Biosentec d/l-lactic acid enzymatic kits according to the manufacturer's instructions (Biosentec, Toulouse, France).
Histology and Immunohistochemistry
Colon samples were cut into 2-cm sections, fixed in 4% paraformaldehyde (4 h, room temperature), dehydrated, and embedded in paraffin according to standard histological protocols. 4–5-μm sections were mounted on SuperFrost® Plus slides. Slides were stained with hematoxylin and eosin for histological analysis. Crypt depths were analyzed with ImageJ. Only U-shaped longitudinally cut crypts with open lumina along the crypt axis were evaluated. Immunological staining was done with primary antibody anti-PCNA (GeneTex; dilution 1:5000) and with the Envision+ system-HRP (Dako, France) according to the recommendations of the manufacturer. Antigen retrieval was performed by boiling slides for 40 min in 0.01 mol/liter sodium citrate, pH 6.0. Results were expressed as a percentage of PCNA-positive cells relative to total colonic crypt cells. Results were the mean obtained by analysis at least 20 crypts/rat (n = 2).
RNA Isolation and Quantitative RT-PCR Analysis
Total RNAs were extracted from colon epithelial cells of GF and mono-associated with S. thermophilus LMD-9 strain by the guanidinium thiocyanate method (27). The RNA yield was quantified by spectrophotometer analysis, and the RNA purity was determined by an Agilent 2100 Bioanalyzer analysis using the RNA 6000 nanoassay kit. Reverse transcription was performed with 1 μg of total rat RNA using the iScriptTM cDNA synthesis kit (Bio-Rad).
For the p27kip1 gene, quantitative RT-PCR was performed on the Opticon Monitor 3 (Bio-Rad) using specific primers and Master Mix (2×) Universal (KAPA SYBR® FAST qPCR Kits, Kapa Biosystems). Primers were 5′-GGCGGCAAGATGTCAAACGTG-3′ and 5′-GGGCCGAAGAGGTTTCTG-3′. Primers were validated to ensure efficient amplification of a single product at 60 °C (annealing temperature). Comparisons were made by using GAPDH as the reference gene and the p27kip1 gene as the target gene.
For SLC16A1, SLC16A2, and SLC5A8 genes, real-time quantitative PCR was performed on cDNAs in an ABI PRISM 7000 sequence detection system. Rn00562332-m1, Rn00596041-m1, and Rn01503812-m1 TaqMan references were used for SLC16A1, SLC16A2, and SLC5A8, respectively (Applied Biosystems). 18 S rRNA was used as a reference gene and measured with an Hs99999901-s1 TaqMan gene expression assay. Results obtained on each transporter were normalized to 18 S rRNA (reference gene) and compared with the mean target gene expression of GF rats as calibrator sample. At least four rats were used for each group of rats (GF and Ino-LMD9). The formula used was the equation, -fold change = 2−ΔΔCt, where ΔΔCt = (Cttarget − Ctreference)sample − (Cttarget − Ctreference)calibrator sample.
Statistical Analysis
Results are presented as means ± S.E. for the number of animals indicated. Comparisons of group data between different batches of rats were performed using one-way analysis of variance, followed by Tukey's Student range test where appropriate. Significance was a p value lower than 0.05. Statistical analysis was performed using the JMP® software (version 7, SAS Institute, Inc.).
RESULTS
Progressive Adaptation of S. thermophilus to the GIT
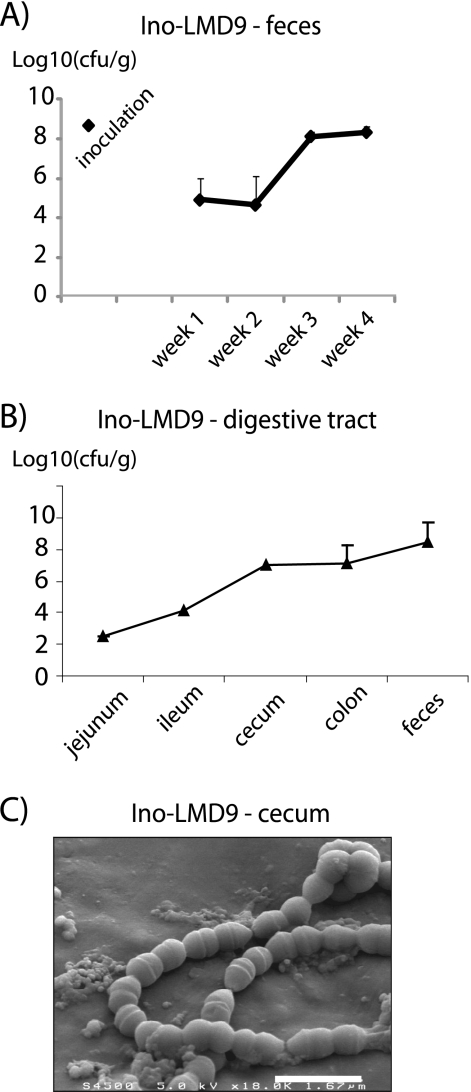
After a single gavage, S. thermophilus LMD-9 progressively colonized the GIT of GF rats, leading to Ino-LMD9 mono-associated rats (Fig. 1A). The implantation of S. thermophilus LMD-9 occurred in a three-step way: 1) initiation phase with implantation of 105 cfu/g of feces; 2) growth period between weeks 2 and 3, where it reached a population of 108 cfu/g of feces; and 3) maintenance phase (post-third week). Thus, 4 weeks were necessary to reach a level of 108 cfu/g of feces. At day 30, the population of S. thermophilus LMD-9 was higher in the distal digestive compartment with an increasing gradient of population from jejunum (1.6 × 103 cfu/ml) to colon (9.3 × 108 cfu/ml) (Fig. 1B). The progressive implantation curve was also observed with another strain of S. thermophilus, LMG18311 (Ino-LMG18311 rats). In Ino-LMG18311 rats, the final implantation of LGM18311 was similar to that obtained with LMD-9, with 2 × 108 cfu/g (n = 4). To mimic the levels present in yogurt, GF rats were inoculated with a co-culture containing 109 cfu/ml S. thermophilus LMD-9 and 107 cfu/ml L. bulgaricus, in accordance with the relative proportion of both strains in yogurt. 24 h after gavage, L. bulgaricus was undetectable in feces, whereas the implantation curve of S. thermophilus LMD-9 was similar to that observed with Ino-LMD9 (data not shown). The progressive implantation of S. thermophilus suggested that an adaptation of S. thermophilus occurred in the GIT.
FIGURE 1.
S. thermophilus in the digestive tract. Shown is enumeration of viable S. thermophilus isolated from feces (A) and different rat digestive tract sections (B) of Ino-LMD9 rats. Fecal or luminal samples were diluted and immediately plated on culture medium M17 + lactose. Colonies were numbered after a 24-h anaerobic culture. Results are expressed on a log scale. n, number of rats used. C, snapshot of S. thermophilus isolated from cecum of Ino-LMD9 rats obtained by scanning electron microscopy. Error bars, S.E.
It has been previously shown that the morphology of Lactobacillus sakei changed after its survival through the GIT (28). Thus, the global morphology of S. thermophilus recovered from cecum of Ino-LMD9 rats was observed by scanning electron microscopy (Fig. 1C). In cecum, S. thermophilus exhibited the expected ovococcus-shaped chains and was present as dividing cells with visible septa.
Proteomic Profiles of S. thermophilus in the Digestive Tract
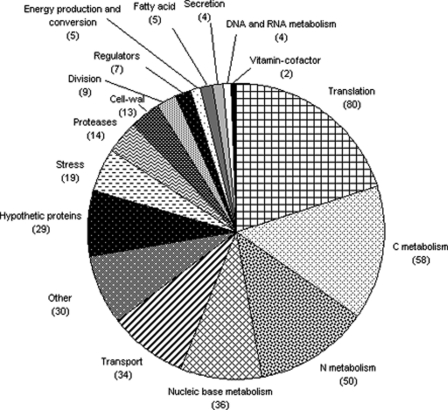
In the feces of Ino-LMD9 rats, proteins of S. thermophilus were analyzed by LC-MS/MS, listed (see supplemental Table 1), and classified into different functional categories (Fig. 2). This result showed that the main S. thermophilus LMD-9 proteins expressed in the GIT are involved in staple cellular functions, such as translation, carbon metabolism, nitrogen metabolism, nucleic base metabolism, or transport. This indicated that S. thermophilus LMD-9 was metabolically active in the rat GIT 30 days after inoculation.
FIGURE 2.
Functional distribution of the proteins of S. thermophilus LMD-9 isolated from rat feces. Proteins from the envelope-enriched fraction were separated and identified by LC-MS/MS. Transport and Regulators, proteins involved in transport and regulators, other than those involved in carbon metabolism, nitrogen metabolism, and nucleic base metabolism. The numbers of proteins are shown in parentheses.
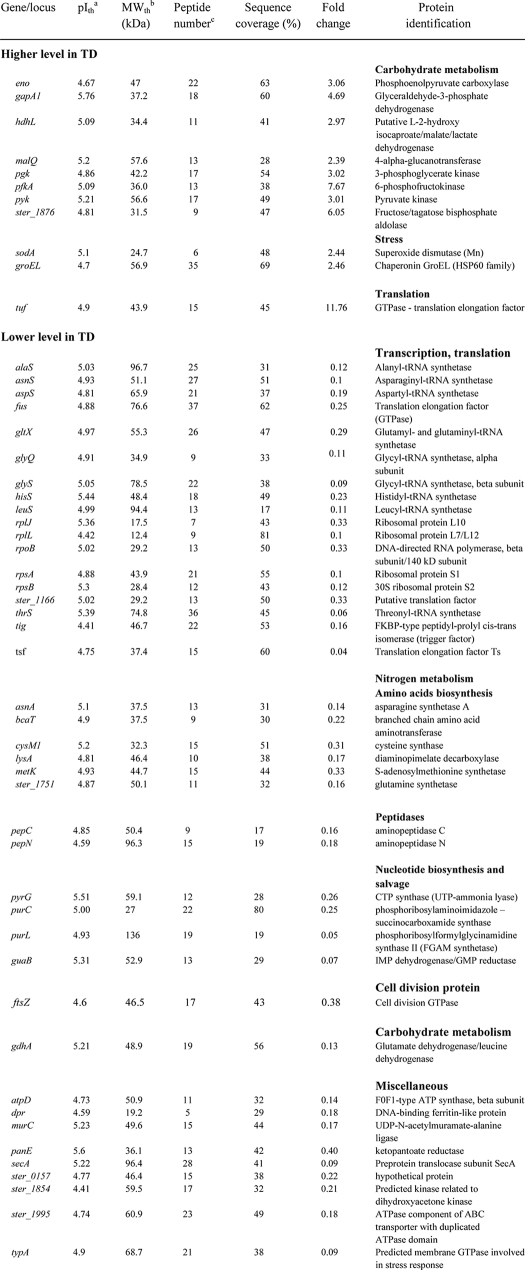
Using a comparative proteomic analysis, we then identified the proteins that were differentially produced when S. thermophilus LMD-9 was grown in milk (inoculum) or when it was recovered from feces of Ino-LMD9 rats (day 30). In both conditions, bacterial populations were similar (i.e. 1–5 × 108 cfu/ml) (Fig. 1). 52 proteins displayed different abundances between the two conditions; 11 were up-regulated (induction from 2- to 11-fold), and 41 were down-regulated (repression from 3- to 25-fold) in feces compared with milk (Table 1).
TABLE 1.
-Fold changes in protein abundance (two-dimensional electrophoresis) of S. thermophilus LMD-9 between feces and late growth phase in milk
a Theoretical isoelectric point.
b Theoretical molecular weight.
c Number of peptides identified.
Of the 11 up-regulated proteins, three are related to environmental signal responses: SodA (superoxide dismutase) and GroEL, which are stress proteins, and EF-Tu (elongation factor Tuf), which is a translation/adhesion-related protein. The remaining up-regulated eight proteins in the gut played a role in carbon metabolism, specifically in the glycolytic pathway. We have also verified that these proteins involved in glycolysis were more abundant in feces of Ino-LMG18311 than in milk (data not shown). Thus, proteins involved in glycolysis that are already expressed at a high level in milk (25) were overexpressed after survival in GIT. The proteins down-regulated in feces compared with milk were involved in protein synthesis, cell division (FtsZ), nucleotide biosynthesis and salvage (PyrG, PurC, PurL, and GuaB), energy provision (F0-F1-ATP synthase AtpD), and iron metabolism (Dpr bacterioferritin). Overall, the proteins of the two strains of S. thermophilus that were overexpressed in the GIT compared with milk are devoted to the glycolytic pathway.
Production of Lactate by S. thermophilus in the Digestive Tract
Considering the preponderant abundance of enzymes involved in glycolysis, l-lactate concentration was measured in the cecum of gnotobiotic rats. Although no lactate (neither d- nor l-lactate) was found in GF rats, 13.6 ± 0.9 and 9.8 ± 2.5 mm l-lactate was detected, respectively, in cecum and feces of Ino-LMD9 rats. Note that only the l-form was detected in cecum of Ino-LMD9 rats, as expected due to the well known capacity of S. thermophilus to produce this isoform. This production of l-lactate did not induce any acidification of the cecal content of Ino-LMD9 rats (mean pH 7.7 ± 0.08, n = 11; versus pH 7.7 ± 0.2, n = 6 GF). Acetate, propionate, and butyrate levels in Ino-LMD9 (1.8 ± 0.7, <0.1, and <0.1 mm respectively) were not statistically different from that obtained with GF rats (1.7 ± 1, <0.1, and <0.1 mm). Similar cecal concentration of lactate was measured in Ino-LMG18311 rats (10.02 ± 3.0 mm) and in Ino-LMD9+Lb rats (9.91 ± 0.99 mm); thus, lactate was found in GIT of mono-associated rats with two different S. thermophilus strains.
Cross-talk between S. thermophilus and the Colonic Epithelium
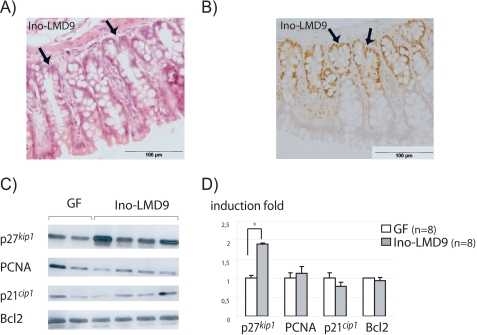
In the intestine, epithelial cells expressed a set of carriers involved in the transport of luminal metabolites, such as short chain fatty acids (29). Thus, we quantified at the mRNA level in GF and Ino-LMD9 rats three transporters that are able to shuttle monocarboxylic acid as lactate (SLC16A1, SLC16A2, and SLC5A8). In both lots of rats, the three RNAs were differently abundant, with the following hierarchy of expression levels: SLC16A1 > SLC5A8 > SLC16A2 (data not shown). The SLC16A1 mRNA amount was statistically different (p < 0.05) between GF (mean ΔCt = 14.9 ± 0.1, n = 6) and Ino-LMD9 rats (ΔCt = 13.8 ± 0.1, n = 8), that represented a 2.2-fold stimulation in the presence of LMD-9. SLC5A8 was significantly 1.7-fold more expressed in Ino-LMD9 (n = 8) than in GF rats (n = 5), whereas SLC16A2 tended to be more expressed in Ino-LMD9, but it was not statistically different. This observation suggests that lactate produced in lumen by LMD-9 may be transported inside the colonic cells. Because of the pivotal role of bacterial metabolites in colonic tissue homeostasis, we studied in Ino-LMD9 and GF rats the global structure of colonic epithelium and the content of cell cycle-related proteins. The colonic tissue of Ino-LMD9 rats was structured in crypts (Fig. 3A), with a 196 ± 26-μm crypt depth mean and the detection of bifurcating crypts (noted with arrows in Fig. 3, A and B); the global histological trait of colonic epithelium in Ino-LMD9 rats was thus similar to that we have previously observed in GF rats (17). Because it is well established in colon tissue, the proliferating cells, stained by PCNA (Fig. 3B) or by Ki67 (data not shown), were located at the bottom of crypt in Ino-LMD9 rats. The PCNA-positive cells represented 60 ± 5% of total cells inside a crypt structure (n = 2 rats, 20 measures/rat). This percentage was similar to what we obtained with GF rats (17). When comparing the amount of colon proteins involved in cell cycle between Ino-LMD9 and GF rats, we noticed that PCNA, Bcl2, and p21cip1 were similar between GF and Ino-LMD9 rats (Fig. 3B), whereas p27kip1 abundance was significantly higher (1.8-fold induction) in the presence of S. thermophilus LMD-9. The induction of p27kip1 was also observed in Ino-LMG18311 (2-fold) and Ino-LMD9+Lb (1.5-fold) (Fig. 4). In contrast to the protein level, the p27kip1 mRNA amount was not statistically different between GF (mean ΔCt = 4.8 ± 0.3, n = 3) and Ino-LMD9 rats (ΔCt = 4.7 ± 0.1, n = 4). Thus, the induction of protein p27kip1 was observed in vivo with two different strains of S. thermophilus and did not result from a parallel induction at the p27kip1 mRNA level.
FIGURE 3.
Histological and molecular analysis of the colonic epithelium of Ino-LMD9 rats. A, representative photomicrograph of colonic sections obtained from Ino-LMD9 rats stained with hematoxylin and eosin. The arrows indicate a bifurcating crypt in the colonic epithelium. B, representative photomicrograph of immunohistochemical staining with PCNA in the colon of Ino-LMD9 rats. C, representative autoradiography films of Western blot revealing p27kip1, PCNA, p21cip1, and Bcl2 from colonic epithelial cells of GF (n = 2) and Ino-LMD9 (n = 4) rats. D, quantifications of Western blot by densitometric analysis are means ± S.E. (error bars) obtained with eight Ino-LMD9 rats and eight GF rats. Values are expressed as -fold induction considering the mean of the GF group (n = 8) as 1. *, statistically significant differences (p < 0.05) between groups.
FIGURE 4.
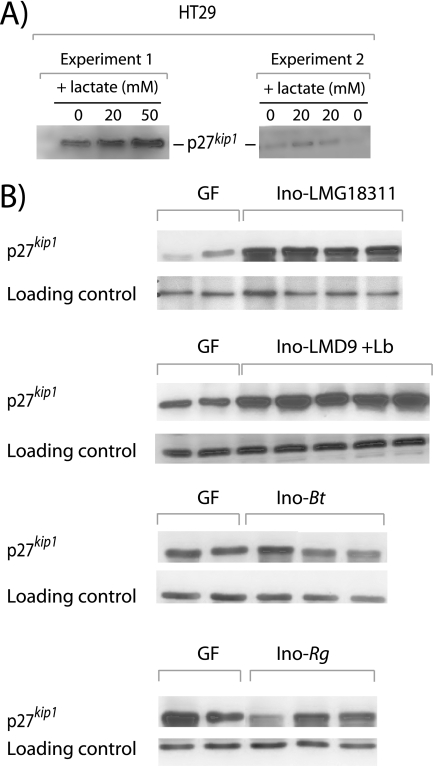
p27kip1 detection in the HT29 cell line and in different mono-associated rats. A, typical autoradiography films of Western blot revealing p27kip1 from HT29 incubated with 0, 20, or 50 mm l-lactate for 18 h (two independent experiments are shown). B, typical autoradiography films of Western blot revealing p27kip1 and loading control from colonic epithelial cells of GF (n = 2) and four different mono-associated lots of rats. Ino-LMG18311 (n = 4), Ino-Bt (n = 3), and Ino-Rg (n = 3), respectively, harbored S. thermophilus strain LMG18311, B. thetaiotaomicron, and R. gnavus. Ino-LMD9+Lb rats (n = 5) have been inoculated with a mixture of S. thermophilus and L. bulgaricus. At the end of the experiment (30 days after the gavage), rats were only mono-associated with S. thermophilus because L. bulgaricus did not efficiently implant.
In the epithelial keratinocyte cell line, p27kip1 is induced by lactate in vitro (30). By using intestinal line cells (HT29), we also observed that 20–50 mm l-lactate for 18 h increased 1.5-fold p27kip1 protein amounts (see Fig. 4A). Taken together, our data suggest that lactate accumulates in the GIT of mono-associated rats, is transported by carriers inside colonic cells, and may increase p27kip1 abundance. In order to demonstrate the preponderant role of lactate, we have tried to inactivate in LMD-9 the ldh gene encoding the lactate dehydrogenase responsible for lactate production. We have failed to generate this mutant (data not shown), confirming previous unsuccessful attempts to obtain this probably lethal mutant (31). However, in rats mono-associated with Bacteroides tethaiotaomicron (Ino-Bt) or Ruminococcus gnavus (Ino-Rg) (17) containing 0 and 3.2 ± 0.7 mm of cecal lactate, respectively, no p27kip1 induction was observed in comparison with GF (Fig. 4B).
DISCUSSION
In order to gain a better understanding of the behavior of lactic acid bacteria within the intestinal environment and the complex bacterium/host cross-talk system, here we combined in vivo characterization of S. thermophilus metabolism and the resulting host response. This study led to novel insights into the interplay between S. thermophilus and the host through its major metabolite product, lactate.
The implantation of S. thermophilus in GF rats occurred progressively, and this trend was observed with both strains studied, LMD-9 and LMG18311. These two strains differ in particular by the presence of a cell wall protease, PrtS, and a sortase, A1, present in the LMD-9 strain. This suggests that the latter proteins were probably not essential for the kinetics of implantation of this bacterium. In contrast to S. thermophilus, the implantation was maximal in a few days for Escherichia coli (32), R. gnavus and B. thetaiotaomicron (17), Lactobacillus casei and Bifidobacterium breve (33), and Lactococcus lactis (23). This adaptive response of S. thermophilus was not accompanied by significant morphological changes, contrary to what was observed with L. sakei or E. coli (28, 34). The proteomic analysis of S. thermophilus before (milk inoculum) and after GIT transit (feces) shed light on its adaptive metabolic profile. Indeed, we showed that the main response to the passage of S. thermophilus through the rat GIT was the massive induction, at the maintenance phase, of the glycolysis pathway leading to formation of lactate in cecum. Here, we propose that S. thermophilus modulates p27kip1 through production of lactate. This hypothesis stems from five observations: 1) the main S. thermophilus metabolic pathway enhanced in GIT compared with a milk culture was the glycolysis leading to production of lactate; 2) two transporters of monocarboxylic acids were induced in Ino-LMD9, suggesting that lactate may be shuttled in colonic cells; 3) the increase of p27kip1 level in the colon was observed in the presence of two S. thermophilus strains producing equivalent amounts of lactate; 4) the increase of p27kip1 level in the colon did not appear in the presence of non-lactate-producing or low lactate-producing bacteria (B. thetaiotaomicron and R. gnavus); and 5) p27kip1 was induced in vitro by the presence of lactate in intestinal epithelial cells (Fig. 4A) and in keratinocytes (30). Obviously, our hypothesis does not exclude other underlying mechanisms involved in the interplay between S. thermophilus and the host (see below).
The massive induction of glycolysis in GIT compared with milk was unexpected for S. thermophilus because these proteins are already present at high levels in the milk (25). The capacity for a bacterium to diversify its carbon sources seems to be essential for the colonization in GF rodents (23, 32, 35–37). S. thermophilus is particularly well adapted to milk, in particular via the assimilation of lactose, which is its preferential sugar and is present at high and non-limiting concentrations in milk (4.5 g/liter). However, we showed here that S. thermophilus may develop an efficient ability to use other sources of saccharides present in GF rat (38) because the rat diet did not contain lactose. This flexibility is rather unexpected because S. thermophilus has a small genome with a large proportion of nonfunctional genes (10% of pseudogenes), in particular genes involved in carbon utilization, in line with the paucity of carbon sources in milk (39). Therefore, it would be of interest to determine whether the implantation level and the adaptive responses of S. thermophilus may be improved by using mono-associated rats receiving a diet enriched with lactose, as previously suggested (40).
The use of mono-associated rats is an efficient tool to reveal the activity of bacterial proteins, especially those with dual functions. For example, EF-Tu is an elongation factor playing a key role in protein synthesis but also in the maintenance in the GIT. The up-regulation of EF-Tu was observed in vivo for L. lactis (41) and Bifidobacterium (42) and in vitro for L. plantarum (43). Although S. thermophilus reduced most of its translation and protein synthesis machinery, EF-Tu was the sole protein belonging to protein synthesis pathway that was increased at a high level in digestive tract compared with milk (induction factor 12). We thus propose that in Ino-LMD9 rats, EF-Tu, which displays the characteristics of an adhesion factor (21), is mainly involved in the maintenance of S. thermophilus rather than in the protein synthesis.
We have recently demonstrated that the colonization of GF rats with a complex microbiota leads to an increase in the absorptive surface by deepening crypts and splitting bifurcated crypts by a crypt fission process (17). This trophic effect of microbiota is associated with a modulation of several cell cycle-related proteins. Along the colonization with a complex microbiota, we have previously described a precocious hyperproliferative phase at the level of colonic epithelium that is counterbalanced by an induction of proteins restraining the proliferation and a decrease of antiapoptotic proteins (17). In contrast to what was observed with a complex microbial population, the presence of S. thermophilus did not trigger deeper crypts and the division of bifurcating crypts. Thus, as we reported previously (17), a single strain is not sufficient to switch on proliferation and the associated greater absorptive surface. In contrast, the induction of cell cycle arrest proteins could be triggered by a single bacterium. Indeed, B. thetaiotaomicron and R. gnavus increased p21cip1 (17), whereas S. thermophilus increased p27kip1. p21cip1 and p27kip1 are inhibitors of cyclin-dependent kinases and are preponderant cell cycle regulators in the GIT (44). In particular, p27kip1 is involved in the capacity of epithelium and host to attenuate deleterious effects of environmental stimuli (45). Thus, we have shown that p27kip1 is stabilized in vivo by a complex microbial population (17) and by S. thermophilus (Figs. 3 and 4). The induction of p27kip1 by a complex microbiota or by S. thermophilus may not result from similar mechanisms because no lactate was detected in luminal content of conventional rats (data not shown). It has also been observed that p27kip1 was enhanced by a bacterial cyclomodulin like Cif (cyclin inhibitor factor) (46). Thus, p27kip1 seems to be sensitive to different bacterial effectors.
The expression levels of the two transporters, SLC16A1 and SLC5A8, were stimulated in Ino-LMD9 rats, where luminal lactate was around 10 mm. Our observation is in accord with previous reports suggesting that monocarboxylic acid transporters provide a link between bacteria and the host (47–49). In human healthy adults, no or low (0–2 mm) lactate is detected in fecal samples (50, 51) because the luminal lactate is absorbed by host and is also used as an energy source by other commensal bacteria (52–54). However, increased fecal lactate has been associated with intestinal malabsorption and colitis (55). It could also be of interest to measure fecal lactate in humans with short bowel syndrome because we have shown that their microbiota is deeply enriched in lactobacilli at the expense of other bacterial members (22). Thus, the effect of lactate that we evidenced here by using a model of lactic acid bacteria in an experimental model may have significance in human digestive diseases.
Because S. thermophilus is able to adapt its global metabolism to the gut environment and to induce monocarboxylic acid transporters and p27kip1, this work provides new insights into the functional “panoply” of one of the two yogurt bacteria. Finally, the fact that S. thermophilus emphasizes its carbohydrate metabolism in the digestive tract is in accord with the beneficial role of fermented milk consumption in improving lactose intolerance.
Acknowledgments
We thank Niriaina Rasoava and Catherine Philippe for technical assistance, Didier Chevret for LC-MS/MS analysis, and Thierry Meylheuc for SEM analyses. We thank the team of animal facilities (ANAXEM platform, MICALIS). We acknowledge Monique Zagorec and Mihai Covasa for fruitful discussions and critical reading of the manuscript.
This work was supported by Syndifrais-CNIEL (Paris, France).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- GIT
- gastrointestinal tract
- PCNA
- peripheral cell nuclear antigen
- GF
- germ-free.
REFERENCES
- 1. Guarner F., Perdigon G., Corthier G., Salminen S., Koletzko B., Morelli L. (2005) Br. J. Nutr. 93, 783–786 [DOI] [PubMed] [Google Scholar]
- 2. Lomer M. C., Parkes G. C., Sanderson J. D. (2008) Aliment Pharmacol. Ther. 27, 93–103 [DOI] [PubMed] [Google Scholar]
- 3. Rabot S., Rafter J., Rijkers G. T., Watzl B., Antoine J. M. (2010) J. Nutr. 140, 677S–689S [DOI] [PubMed] [Google Scholar]
- 4. Higashikawa F., Noda M., Awaya T., Nomura K., Oku H., Sugiyama M. (2010) Nutrition 26, 367–374 [DOI] [PubMed] [Google Scholar]
- 5. Pagnini C., Saeed R., Bamias G., Arseneau K. O., Pizarro T. T., Cominelli F. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Preidis G. A., Versalovic J. (2009) Gastroenterology 136, 2015–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D. R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J. M., Hansen T., Le Paslier D., Linneberg A., Nielsen H. B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Bork P., Ehrlich S. D., Wang J. (2010) Nature 464, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elli M., Callegari M. L., Ferrari S., Bessi E., Cattivelli D., Soldi S., Morelli L., Goupil Feuillerat N., Antoine J. M. (2006) Appl. Environ. Microbiol. 72, 5113–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Firmesse O., Alvaro E., Mogenet A., Bresson J. L., Lemée R., Le Ruyet P., Bonhomme C., Lambert D., Andrieux C., Doré J., Corthier G., Furet J. P., Rigottier-Gois L. (2008) Int. J. Food Microbiol. 125, 176–181 [DOI] [PubMed] [Google Scholar]
- 10. Mater D. D., Bretigny L., Firmesse O., Flores M. J., Mogenet A., Bresson J. L., Corthier G. (2005) FEMS Microbiol. Lett. 250, 185–187 [DOI] [PubMed] [Google Scholar]
- 11. Moreau M. C., Thomasson M., Ducluzeau R., Raibaud P. (1986) Reprod. Nutr. Dev. 26, 745–753 [PubMed] [Google Scholar]
- 12. Palmer C., Bik E. M., DiGiulio D. B., Relman D. A., Brown P. O. (2007) PLoS Biol. 5, e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perez P. F., Doré J., Leclerc M., Levenez F., Benyacoub J., Serrant P., Segura-Roggero I., Schiffrin E. J., Donnet-Hughes A. (2007) Pediatrics 119, e724–e732 [DOI] [PubMed] [Google Scholar]
- 14. Alam M., Midtvedt T., Uribe A. (1994) Scand. J. Gastroenterol. 29, 445–451 [DOI] [PubMed] [Google Scholar]
- 15. Nougayrède J. P., Taieb F., De Rycke J., Oswald E. (2005) Trends Microbiol. 13, 103–110 [DOI] [PubMed] [Google Scholar]
- 16. Willing B. P., Van Kessel A. G. (2007) J. Anim. Sci. 85, 3256–3266 [DOI] [PubMed] [Google Scholar]
- 17. Cherbuy C., Honvo-Houeto E., Bruneau A., Bridonneau C., Mayeur C., Duée P. H., Langella P., Thomas M. (2010) Am. J. Physiol. Gastrointest. Liver Physiol. 299, G348–G357 [DOI] [PubMed] [Google Scholar]
- 18. Drouault S., Anba J., Corthier G. (2002) Appl. Environ. Microbiol. 68, 938–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Herve-Jimenez L., Guillouard I., Guedon E., Gautier C., Boudebbouze S., Hols P., Monnet V., Rul F., Maguin E. (2008) Proteomics 8, 4273–4286 [DOI] [PubMed] [Google Scholar]
- 20. Druesne N., Pagniez A., Mayeur C., Thomas M., Cherbuy C., Duée P. H., Martel P., Chaumontet C. (2004) Carcinogenesis 25, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 21. Dallo S. F., Kannan T. R., Blaylock M. W., Baseman J. B. (2002) Mol. Microbiol. 46, 1041–1051 [DOI] [PubMed] [Google Scholar]
- 22. Joly F., Mayeur C., Bruneau A., Noordine M. L., Meylheuc T., Langella P., Messing B., Duée P. H., Cherbuy C., Thomas M. (2010) Biochimie 92, 753–761 [DOI] [PubMed] [Google Scholar]
- 23. Roy K., Meyrand M., Corthier G., Monnet V., Mistou M. Y. (2008) Proteomics 8, 1661–1676 [DOI] [PubMed] [Google Scholar]
- 24. Gardan R., Besset C., Guillot A., Gitton C., Monnet V. (2009) J. Bacteriol. 191, 4647–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Derzelle S., Bolotin A., Mistou M. Y., Rul F. (2005) Appl. Environ. Microbiol. 71, 8597–8605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cherbuy C., Darcy-Vrillon B., Morel M. T., Pégorier J. P., Duée P. H. (1995) Gastroenterology 109, 1890–1899 [DOI] [PubMed] [Google Scholar]
- 27. Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 28. Chiaramonte F., Blugeon S., Chaillou S., Langella P., Zagorec M. (2009) Appl. Environ. Microbiol. 75, 4498–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thwaites D. T., Anderson C. M. (2007) Exp. Physiol. 92, 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsiao Y. P., Huang H. L., Lai W. W., Chung J. G., Yang J. H. (2009) J. Dermatol. Sci. 54, 175–184 [DOI] [PubMed] [Google Scholar]
- 31. Hols P., Hancy F., Fontaine L., Grossiord B., Prozzi D., Leblond-Bourget N., Decaris B., Bolotin A., Delorme C., Dusko Ehrlich S., Guédon E., Monnet V., Renault P., Kleerebezem M. (2005) FEMS Microbiol. Rev. 29, 435–463 [DOI] [PubMed] [Google Scholar]
- 32. Alpert C., Scheel J., Engst W., Loh G., Blaut M. (2009) Environ. Microbiol. 11, 751–761 [DOI] [PubMed] [Google Scholar]
- 33. Shima T., Fukushima K., Setoyama H., Imaoka A., Matsumoto S., Hara T., Suda K., Umesaki Y. (2008) FEMS Immunol. Med. Microbiol. 52, 69–77 [DOI] [PubMed] [Google Scholar]
- 34. Giraud A., Arous S., De Paepe M., Gaboriau-Routhiau V., Bambou J. C., Rakotobe S., Lindner A. B., Taddei F., Cerf-Bensussan N. (2008) PLoS Genet. 4, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bron P. A., Grangette C., Mercenier A., de Vos W. M., Kleerebezem M. (2004) J. Bacteriol. 186, 5721–5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang D. E., Smalley D. J., Tucker D. L., Leatham M. P., Norris W. E., Stevenson S. J., Anderson A. B., Grissom J. E., Laux D. C., Cohen P. S., Conway T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7427–7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marco M. L., Peters T. H., Bongers R. S., Molenaar D., van Hemert S., Sonnenburg J. L., Gordon J. I., Kleerebezem M. (2009) Environ. Microbiol. 11, 2747–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sonnenburg J. L., Xu J., Leip D. D., Chen C. H., Westover B. P., Weatherford J., Buhler J. D., Gordon J. I. (2005) Science 307, 1955–1959 [DOI] [PubMed] [Google Scholar]
- 39. Bolotin A., Quinquis B., Renault P., Sorokin A., Ehrlich S. D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G. D., Fonstein M., Overbeek R., Kyprides N., Purnelle B., Prozzi D., Ngui K., Masuy D., Hancy F., Burteau S., Boutry M., Delcour J., Goffeau A., Hols P. (2004) Nat. Biotechnol. 22, 1554–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mater D. D., Drouault-Holowacz S., Oozeer R., Langella P., Anba J., Corthier G. (2006) Br. J. Nutr. 96, 177–181 [DOI] [PubMed] [Google Scholar]
- 41. Beganović J., Guillot A., van de Guchte M., Jouan A., Gitton C., Loux V., Roy K., Huet S., Monod H., Monnet V. (2010) J. Proteome Res. 9, 677–688 [DOI] [PubMed] [Google Scholar]
- 42. Yuan J., Wang B., Sun Z., Bo X., Yuan X., He X., Zhao H., Du X., Wang F., Jiang Z., Zhang L., Jia L., Wang Y., Wei K., Wang J., Zhang X., Sun Y., Huang L., Zeng M. (2008) J. Proteome Res. 7, 375–385 [DOI] [PubMed] [Google Scholar]
- 43. Ramiah K., van Reenen C. A., Dicks L. M. (2007) Int. J. Food Microbiol. 116, 405–409 [DOI] [PubMed] [Google Scholar]
- 44. Besson A., Dowdy S. F., Roberts J. M. (2008) Dev. Cell 14, 159–169 [DOI] [PubMed] [Google Scholar]
- 45. Zheng Y., Bie W., Yang R., Perekatt A. O., Poole A. J., Tyner A. L. (2008) Cancer Biol. Ther. 7, 873–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Samba-Louaka A., Nougayrède J. P., Watrin C., Jubelin G., Oswald E., Taieb F. (2008) Cell Microbiol. 10, 2496–2508 [DOI] [PubMed] [Google Scholar]
- 47. Cresci G. A., Thangaraju M., Mellinger J. D., Liu K., Ganapathy V. (2010) J. Gastrointest. Surg. 14, 449–461 [DOI] [PubMed] [Google Scholar]
- 48. Ganapathy V., Thangaraju M., Gopal E., Martin P. M., Itagaki S., Miyauchi S., Prasad P. D. (2008) AAPS J. 10, 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Frank H., Gröger N., Diener M., Becker C., Braun T., Boettger T. (2008) J. Biol. Chem. 283, 24729–24737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hove H., Mortensen P. B. (1995) Dig. Dis. Sci. 40, 320–330 [DOI] [PubMed] [Google Scholar]
- 51. Hove H., Rye Clausen M., Brøbech Mortensen P. (1993) Gut 34, 625–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He T., Venema K., Priebe M. G., Welling G. W., Brummer R. J., Vonk R. J. (2008) Eur J. Clin. Invest. 38, 541–547 [DOI] [PubMed] [Google Scholar]
- 53. Marquet P., Duncan S. H., Chassard C., Bernalier-Donadille A., Flint H. J. (2009) FEMS Microbiol. Lett. 299, 128–134 [DOI] [PubMed] [Google Scholar]
- 54. Veiga P., Gallini C. A., Beal C., Michaud M., Delaney M. L., DuBois A., Khlebnikov A., van Hylckama Vlieg J. E., Punit S., Glickman J. N., Onderdonk A., Glimcher L. H., Garrett W. S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18132–18137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hove H., Mortensen P. B. (1995) Dig. Dis. Sci. 40, 1372–1380 [DOI] [PubMed] [Google Scholar]