Abstract
The ubiquitously distributed MAP1S is a homologue of the exclusively neuronal distributed microtubule-associated protein 1A and 1B (MAP1A/B). They give rise to multiple isoforms through similar post-translational modification. Isoforms of MAP1S have been implicated in microtubule dynamics and mitotic abnormalities and mitotic cell death. Here we show that ablation of the Map1s gene in mice caused reduction in the B-cell CLL/lymphoma 2 or xL (Bcl-2/xL) and cyclin-dependent kinase inhibitor 1B (P27) protein levels, accumulation of defective mitochondria, and severe defects in response to nutritive stress, suggesting defects in autophagosomal biogenesis and clearance. Furthermore, MAP1S isoforms interacted with the autophagosome-associated light chain 3 of MAP1A/B (LC3), a homologue of yeast autophagy-related gene 8 (ATG8), and recruited it to stable microtubules in a MAP1S and LC3 isoform-dependent mode. In addition, MAP1S interacted with mitochondrion-associated leucine-rich PPR-motif containing protein (LRPPRC) that interacts with the mitophagy initiator and Parkinson disease-related protein Parkin. The three-way interactions of MAP1S isoforms with LC3 and microtubules as well as the interaction of MAP1S with LRPPRC suggest that MAP1S isoforms may play positive roles in integration of autophagic components with microtubules and mitochondria in both autophagosomal biogenesis and degradation. For the first time, our results clarify roles of MAP1S in bridging microtubules and mitochondria with autophagic and mitophagic initiation, maturation, trafficking, and lysosomal clearance. Defects in the MAP1S-regulated autophagy may impact heart disease, cancers, neurodegenerative diseases, and a wide range of other diseases.
Keywords: Autophagy, Microtubules, Mitochondria, Mouse, Protein Degradation, C19ORF5, LC3, LRPPRC, RASSF1A, Mitophagy
Introduction
Autophagy, or self-digestion, is a process that begins with the formation of isolation membranes that engulf substrates to form autophagosomes. Then autophagosomes fuse with lysosomes to generate autolysosomes in which substrates are degraded (1). The initiation of autophagy is regulated by the mammalian target of rapamycin (mTOR) pathway. Autophagy will be shut down through either the phosphatidylinositol 3-kinase (PI3K)-v-akt murine thymoma viral oncogene (AKT)-mTOR pathway in response to signals from growth factors or the serine/threonine kinase 11 (LKB1)-AMP-activated protein kinase (AMPK)2-mTOR pathway in response to signals from nutrients and metabolites. The anti-apoptotic protein Bcl-2 and Bcl-xL exhibit opposite functions in autophagy initiation. They inhibit autophagy initiation through the PI3K-AKT-mTOR pathway by sequestering the BCL2 interacting protein (Beclin 1) or activate autophagy initiation through the LKB1-AMPK-mTOR pathway by increasing P27 levels (2, 3).
After initiation, the metabolism of LC3 has emerged as a key biochemical marker for tracking of autophagy and autophagosomes (4, 5). The 22-kDa full-length LC3 (4) is proteolytically modified into LC3I form by ATG4, resulting in exposure of a C-terminal glycine (5). LC3I is first conjugated with the ubiquitin activating enzyme (E1)-like enzyme ATG7 and then transferred to the ubiquitin-conjugating enzyme (E2)-like enzyme ATG3 (5). The LC3I-ATG3 conjugates appear at autophagosomal membrane initiation sites where the C-terminal glycine of LC3I is covalently conjugated with the membrane-associated phosphatidylethanolamine to form LC3II in the assistance of ATG5·ATG12·ATG16 complex (6). The conversion of LC3I to membrane-anchored LC3II is an indicator of initiation of the autophagic process and promotes the development and maturation of autophagosomes (4, 5). Displayed on both sides of isolation membranes, LC3II serves as either a receptor for target cargo to be enveloped (7) or a linker with mature cargo-containing autophagosomes for trafficking. Thus, in addition to autophagosomal biogenesis indicated by the conversion of LC3I to LC3II, LC3II is also a marker for autophagosomal trafficking and degradation by lysosomes.
Autophagosomes are associated with microtubules (8). Microtubules have been implicated in initiation and maturation of autophagosomes (8, 9) and serve as the tracks for movement of mature autophagosomes and bring autophagosomes and lysosomes together for fusion and disposal of autophagosomal cargo (8, 9). Thus, microtubules provide the initiating focal points and coordinate the spatial arrangements among immature LC3II-containing isolation membranes, cargo for isolation, mature autophagosomes, and lysosomes. Different sets of microtubular filaments coexist, with the acetylated microtubules concentrating in the perinuclear region. The regular non-acetylated microtubules are involved in autophagosomal biogenesis, whereas acetylated microtubules are required for the fusion of mature autophagosomes with lysosomes to form autolysosomes (10).
Because of roles of mitochondria in bioenergetics and cell death, their quality control and recycling through the selective autophagic process called mitophagy is critical for cell life and death (1). Mitochondria are in a dynamic state of fission to smaller units and fusion of smaller units into larger units that organize into extended filaments, networks, or clusters for pooling electrical potential, energy, and other mitochondrial functions for specialized intracellular needs and sites (11). Although mitochondrial aggregates are resistant to clearance by mitophagy, they undergo asymmetric fission to generate daughter mitochondria with uneven membrane potentials. Daughter mitochondria with normal membrane potential fuse back into reticular networks and those with low potential are targeted for removal and recycling by mitophagy (12). A blockade in mitophagy causes accumulation of large mitochondria with low membrane potential that, in turn, cause mitochondria-triggered cell death (13).
MAP1S was originally described and named as chromosome 19 open reading frame 5 (C19ORF5) product (14, 15). It also carried the names such as variably charged protein Y2 (VCY2) interacting protein 1 (VCY2IP-1) (16), basic protein on Y chromosome 2 (BPY2) interacting protein 1 (BPY2IP1) (16), ras association domain family 1 (RASSF1)-binding protein 1 (RABP1) (17), and microtubule-associated protein 8 (MAP8) (18). It is a widely distributed homologue of neuron-specific MAP1A and MAP1B (15, 19, 20). MAP1S is an interactive partner of mitochondrion-associated LRPPRC and RASSF1 as well as NADH dehydrogenase subunit 1 (ND1) and cytochrome c oxidase I (COX-I) (14, 15, 20). It associates specifically with the microtubule-stabilizing and the tumor-suppressing RASSF1A on microtubules (17, 21, 22). MAP1S appears on the microtubular spindle during mitosis and reversibly accumulates during mitotic arrest (23). Similar to MAP1A/B, full-length MAP1S (MAP1SFL) gives rise to multiple post-translationally modified isoforms including heavy chain (HC) and light chain (LC) whose functions are unclear. Prolonged mitotic arrest or inhibition of the 26 S proteasome causes accumulation of an otherwise highly labile short chain (SC) of MAP1S (23) (Fig. 1A). Unlike other isoforms, the MAP1SSC associates with mitochondria in addition to microtubules and causes irreversible aggregation of dysfunctional mitochondria resulting in cell death (23). Depletion of either RASSF1A or MAP1S causes mitotic abnormalities (17, 21). The RASSF1A-MAP1S partnership has been proposed to play a role in tumor suppression by preventing propagation of chromosomal abnormalities through mitochondria-mediated mitotic cell death (22, 23). Neuron-specific microtubule-associated proteins MAP1A/B interact with LC3 and facilitate its association with microtubules (4, 24). MAP1B has been implicated in regulation of autophagosomal trafficking in neurons under normal and pathological conditions (25, 26). Therefore, we reasoned that MAP1S may play a general role in autophagy regulation in all tissues.
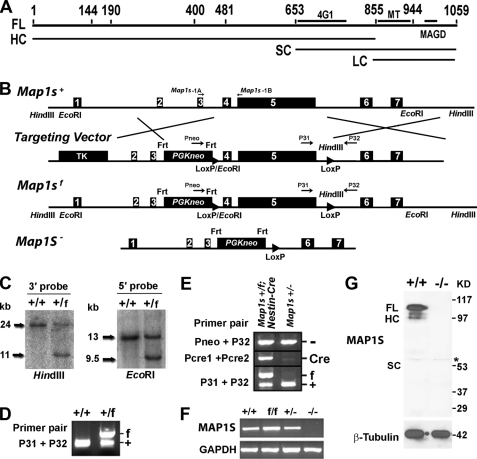
FIGURE 1.
Generation of the Map1s−/− mouse. A, post-translational isoforms of MAP1S are shown. Numbers indicate amino acid residues. FL, 1059-residue full-length translation product; HC, heavy chain; SC, short chain; LC, light chain; 4G1, epitope of the 4G1 antibody; MT, microtubule binding domain; MAGD, mitochondria aggregation and genome destruction domain. B, a schematic of the wild-type mouse Map1s gene (Map1s+), the engineered gene targeting vector, the floxed allele (Map1sf), and the null allele (Map1s−) is shown. Boxed numbers represent the seven exons of the Map1s gene. HindIII and EcoRI restriction enzyme sites are indicated. The indicated TK and PGKneo cassettes carry thymidine kinase and neomycin resistance genes for negative and positive selection, respectively. Frt and LoxP are the target sequences for Flippase or Cre enzymes, respectively. C19-1A, C19-1B, Pneo, P31, and P32 refer to primers for PCR genotyping (supplemental Table 1). C, Southern blot confirmation of Map1sf alleles in ES cells is shown. Genomic DNA from ES clones with wild-type (+/+) and flox allele (+/f) was digested with EcoRI or HindIII and hybridized with the 5′ or 3′ probe, respectively. D, confirmation of Map1sf allele in the mouse genome is shown. E, confirmation of the presence of Map1s− allele in the Nestin-Cre transgenic mice is shown. F, the expression levels of Map1s mRNA in the brains of mice with different genotypes are shown. The mRNA was assessed by RT-PCR with primers C19-1A and C19-1B. G, the expression levels of MAP1S in hearts of mice with different genotypes are shown. MAP1S was assessed by immunoblot with antibody 4G1. The asterisk represents a nonspecific band caused by contaminating mouse antibodies from blood that cross-react with anti-mouse secondary antibody.
We show here that ablation of the Map1s gene in mice causes impairment in both basal autophagy for clearance of abnormal mitochondria and nutritive stress-induced autophagy for nutrient recycling via the Bcl-2/xL/P27 pathway. MAP1S isoforms interact with and specifically recruit the autophagy-associated proteins LC3I and LC3II to stable microtubules in an isoform-specific mode. The profile of three-way interactions of MAP1S isoforms with LC3 isoforms and microtubules as well as the interaction of MAP1S with mitochondrion-associated LRPPRC that interacts with Parkinson disease-related mitophagy initiator Parkin (27) suggests that MAP1S isoforms may differentially underlie the positive role of Map1s gene in autophagosomal biogenesis, maturation, and lysosomal clearance.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Except antibodies against human LC3 (Novus Biologicals, LLC, NB 100–2331) and MAP1S (4G1) (Precision AntibodyTM, AG10006), other antibodies including control IgG antibody (sc-2025), primary antibodies against β-actin (sc-47778), hemagglutinin (HA) (Y-11) (sc-805), ATG5 (sc-33210), Bcl-2 (sc-7382), Bcl-xL (sc-8392), P27 (sc-776), Beclin 1 (sc-11427), phosphatase and tensin homolog (PTEN) (sc-7974), green fluorescent protein (GFP) (sc-8334), and rhodamine-conjugated secondary antibodies (sc-2091 and sc-2300) were from Santa Cruz Biotechnology, Inc. The 4G1 anti-MAP1S monoclonal antibody has been rigorously tested for specificity in immunoblot and immunofluorescence staining of diverse recombinant fragments of MAP1S (19). Lipofectamine (18324-012) was from Invitrogen. MG-132 (474790) was from Calbiochem. The protein G-Sepharose (17-0618-01) was from GE Healthcare. Tubulins (TL238) were from Cytoskeleton, Inc.
Generation of Map1s−/− Mice
Animal protocols were approved by the Institutional Animal Care and Use Committee, Institute of Biosciences and Technology, Texas A&M Health Science Center. Genomic DNA fragments of the Map1s gene were amplified by PCR from genomic DNA purified from the 129S4 mouse-generated AK7 embryo stem (ES) cells with high fidelity enzymes from Roche Applied Science. Briefly, a 2.1-kb fragment for the 5′ homology recombination arm spanning exon 2 and 3 of Map1s (Fig. 1B) in the targeting vector (Fig. 1B) was generated by ligation of two PCR-amplified fragments 1a and 1b. A 3.0-kb floxed fragment flanking the entire exon 4 and 5 was generated by ligation of two PCR fragments flox-a and flox-b. A 5.0-kb fragment for the 3′ homology recombination arm spanning exon 6 and 7 in the targeting vector was generated by ligation of three PCR fragments 3a, 3b, and 3c. The primers used to amplify the fragments are listed in supplemental Table 1. The sequences of all fragments were verified by DNA sequencing before cloning into the targeting vector. Sequences across the cloning sites in the targeting vector were also verified before linearization with Pme1 for electroporation.
The ES cells were transfected with the linearized targeting vector by electroporation and selected in G418-containing medium as described (28). The genotypes of selected clones were analyzed by Southern blot using the 5′ external probe with EcoR1-digested DNAs or 3′ external probe with HindIII-digested DNAs. The 5′ and 3′ probes were PCR-amplified from the genomic DNA purified from the ES cells with the indicated primers (supplemental Table 1). Both fragments were cloned in pBluescript SK vector. The positive clones were further verified by Southern blot using the same probes to the EcoRI- or HindIII-digested DNAs. Two positive clones were selected and microinjected into blastocysts derived from C57BL/6 mice. The chimeric males were bred to C57BL/6 females, and F1 agouti offspring were analyzed by Southern blot or PCR for the presence of the Map1sf allele (Fig. 1, C and D).
Male Map1sf/+ heterozygous mice were crossed with female Nestin-Cre transgenic mice to generate the Map1sf/+;Nestin-Cre mice that express Cre recombinase in the germ line to produce sperm and eggs carrying the Map1s− allele. Genomic DNAs from the tails of progeny of Map1sf/+;Nestin-Cre mice were PCR-analyzed to identify the pups with Map1s− but without the Nestin-Cre allele. Heterozygous mutant mice were out-bred with C57BL/6 mice and inbred to obtain homozygous mutant mice. The presence of Map1sf, Map1s−, Map1s+, or the Cre allele was determined by PCR using the listed primers (supplemental Table 1 and Fig. 1E), and expression of Map1s mRNA (Fig. 1F) and MAP1S protein (Fig. 1G) was further confirmed by immunoblot. The resulting homozygous mice suppressed the expression of exons 4, 5, 6, and 7 that represent 88.9% of the coding region and cover all of the putative functional domains that include the microtubule-binding domain, MAGD (mitochondria aggregation and genomic destruction) domain, and actin binding domain (19, 20).
Generation of GFP-LC3 × Map1s−/− Hybrid Mice
GFP-LC3 transgenic C57BL/6 mice (29) were crossed with C57BL/6 wild-type Map1s+/+ and Map1s−/− mice to generate transgenic wild-type (GFP-LC3+/−Map1s+/+) and Map1s knock-out mice (GFP-LC3+/−Map1s−/−).
Establishment of Mouse Embryonic Fibroblasts
Mouse embryonic fibroblasts were prepared from wild-type (Map1s+/+) and Map1s knock-out mice (Map1s−/−) as described (30). Briefly, embryos were collected at E14.5 and minced in PBS and incubated with 0.25% trypsin, Dulbecco's modified Eagle's medium (DMEM) at 37 °C for 30 min. The separated cells were then harvested with centrifugation and maintained in DMEM with 10% fetal bovine serum.
Transmission Electron Microscopy
Mouse heart tissues were fixed for 1 h with a solution containing 3% glutaraldehyde, 2% paraformaldehyde, and 0.1 m cacodylate buffer (pH 7.3). After fixation, the samples were washed and treated with 0.1% Millipore-filtered cacodylate-buffered tannic acid, postfixed with 1% buffered osmium tetroxide for 30 min, and stained en bloc with 1% Millipore-filtered uranyl acetate. The samples were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium followed by polymerization in a 70 °C oven for 2 days. Ultrathin sections were cut in a Leica Ultracut microtome (Leica), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL, USA, Inc.) at an accelerating voltage of 80 kV. Digital images were obtained using the AMT Imaging System (Advanced Microscopy Techniques Corp).
Stress-induced Autophagy by Neonatal Starvation or Adult Fasting
15 wild-type and 15 Map1s knock-out neonatal pups were delivered by Caesarean section from three pairs of congenic wild-type or Map1s knock-out homozygous mothers, respectively. The pups were deprived of feeding in a humidified, thermostat-controlled chamber (30 °C). Pups were monitored for survival over a 30-h period, and the percentage of survived pups of wild-type and Map1s knock-out homozygous mice at different time points were plotted by method of Kaplan-Meier (31) or used to collect tissues for analysis of LC3I and LC3II at times indicated in Fig. 4A. For adult fasting experiments, 1-month-old mice were deprived of food with free access to water for the indicated times.
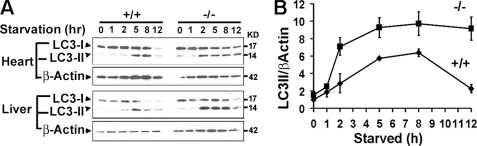
FIGURE 4.
The MAP1S deficiency leads to early accumulation of autophagosomes in neonatal mice under nutritive stress. A, reduced clearance of LC3II in cardiomyocytes and liver tissues of starved MAP1S-deficient neonatal mice is shown. B, intensities of bands in cardiomyocytes revealed by anti-LC3 and β-actin antibodies were measured, and relative intensity to β-actin standard was plotted. Data are the mean ± S.D. of three independent starvation experiments.
Assessment of Mitochondrial Mass by Citrate Synthase Activity
Tissue extracts were prepared with the CelLytic MT Cell Lysis Reagent (C3228) provided with the citrate synthase assay kit from Sigma (CS0720). Protein concentration in cell lysates was assayed by the bicinchoninic acid (BCA) method (19). Procedures for assay of citrate synthase activity were generally carried out according to manufacturer's instructions. Mitochondrial mass was proportional to the citrate synthase activity.
Cell Transfection, Immunoprecipitation, and Immunoblot Analysis
Construction and utility of expression plasmids for GFP-LC3, HA-RASSF1A, HA-MAP1SFL, HA-MAP1SHC, HA-MAP1SSC, and CMYC-MAP1SSC have been described (10, 19, 22, 23). Plasmids coding for MAP1S isoforms fused with HA were constructed by ligation of linearized pCIS vector carrying an HA tag cut with both SalI and NotI enzymes with a fragment coding for the respective isoform, which was first amplified by PCR with the mouse Map1s full-length cDNA as template and the indicated forward and reverse primers (supplemental Table 2) followed by a subsequent cut with both SalI and NotI enzymes. The DNA sequence covering the HA tag and fused fragments in all plasmid products was verified by sequencing.
Vectors coding for MAP1S isoforms fused with HA or GFP-LC3 were transfected into HEK293 cells by the CaCl2 method optimized to achieve high expression levels. Cell lysates were prepared, and immunoprecipitation from them and immunoblot analyses were performed as described (22). Plasmids carrying HA-RASSF1A, cMYC-MAP1SSC HA-MAP1SSC, HA-MAP1SFL, HA-MAP1SHC, or HA-MAP1SSC were transfected alone or cotransfected with each other into HeLa cells or HeLa cells stably expressing GFP-LC3 (23, 32). LC3I, a protein product with a calculated molecular mass of 14,129 Da, migrates at an apparent molecular mass of 16–17 kDa on a 15% gel during SDS-PAGE. LC3II is the conjugated product of LC3I with phosphatidylethanolamine (744 Da) but migrates faster than LC3I as a 14-kDa band (4). Preliminary analyses on 10% gels under normal electrophoresis conditions was unable to routinely separate the GFP-LC3I and GFP-LC3II bands that are expected to be displayed as 43 and 41 kDa, respectively (supplemental Fig. 1). Analysis of the samples on gels of 15% instead of 10% for periods of 5 h instead of 1.5 h clearly separated the bands but at the expanse of a slight distortion of migration patterns for both GFP-LC3I and GFP-LC3II in the immunoprecipitates. Data presented in the text for immunoblot or immunohistochemical analysis (described below) are representative of at least three independent cell transfections, extraction, and analysis. Immunoblot results were developed on x-ray film and scanned into image files, and relative band intensities were determined with the ImageJ software.
Fluorescence Microscopy
Immunofluorescence staining were performed, and images were captured by laser scanning confocal microscope as described (23, 32). Scale bars in the images in the text indicate 10 μm unless otherwise noted. Heart tissues were harvested, quickly frozen, and cryosectioned. The GFP-LC3 distribution in heart tissues was similarly recorded by confocal microscopy. The acquired images were converted to 8-bit binary files, and the number, size, and total occupied area of GFP-LC3 punctate foci with a diameter greater than four pixels on each image were calculated by ImageJ software according to the manufacturer's manual.
Microtubule Interaction in Vitro
Plasmids coding for HA-MAP1SFL, HA-MAP1SHC, HA-MAP1SSC, and HA-MAP1SLC were transfected into HEK293 cells for 48 h. Cells were harvested by scraping into the G-PEM buffer (19) and broken by sonication, and lysates were clarified by centrifugation at 10,000 × g for 10 min. Portions of concentrated lysates containing the indicated HA-MAP1S isoforms (0, 2, or 4 μl), 12 μl of GFP-LC3-containing lysate, 10 μg of tubulins, and 1 μl of 100 μm paclitaxel were mixed and adjusted to a 20-μl final volume with G-PEM buffer. Microtubules were assembled at 37 °C for 30 min and separated from the supernatants by centrifugation at 10,000 × g for 20 min at room temperature (19). Preliminary in vitro microtubule assembly experiments revealed that the majorities of HA-tagged FL and SC proteins were co-precipitated with the assembled microtubular pellet and the majority of HA-tagged HC remained in the supernatant, but HA-LC was not enriched sufficient to include in the analysis (supplemental Fig. 2). Subsequent experiments did not include the LC isoform but clearly demonstrated that both GFP-LC3I and GFP-LC3II were pulled down in the microtubular pellets in the presence of either FL or SC but not HC (supplemental Fig. 3). The high levels of GFP-LC3I relative to GFP-LC3II in the lysates triggered us to suspect that the GFP-LC3I in the microtubular pellet was simply of contaminate. To prevent domination of the assays by GFP-LC3I, HeLa cells stably expressing GFP-LC3 were treated with 20 mm NH4Cl to block degradation of GFP-LC3II by lysosomes, thereby adjusting GFP-LC3I and GFP-LC3II to similar levels for comparative analysis. Lysates were further concentrated three times by centrifugation in MicroconTM tubes (Ultracel YM-30, Millipore) to test the roles of MAP1S isoforms at their saturated concentrations (supplemental Fig. 4).
RESULTS
Depletion of the Map1s Gene in Mice Results in Reduction of Autophagy Regulator Bcl-2, Bcl-XL, and P27
Similar to neuronal homologues MAP1A and MAP1B, products of the Map1s gene give rise to multiple isoforms that can be distinguished electrophoretically and immunochemically (Fig. 1A) (19, 20, 23). Cell populations predominantly in interphase exhibit the MAP1SFL, HC, and LC isoforms (19, 23). The MAP1SSC form appears in mitosis and becomes prominent after a paclitaxel-induced mitotic blockade (19, 23). When mouse embryonic fibroblast cells were treated with a general inhibitor of the 26 S proteasome, MAP1SSC specifically accumulated (Fig. 2A). Therefore, all isoforms of MAP1S expressed at high levels but maintained with different stabilities.
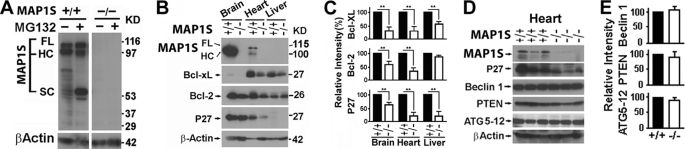
FIGURE 2.
Depressed expression of Bcl-2, Bc-xL, and p27 in MAP1S-deficient mice. A, isoforms of MAP1S in the absence or presence of 26 S proteasome inhibitor MG-132 are shown. Mouse embryo fibroblasts from wild-type (+/+) and Map1s knock-out mice (−/−) (Fig. 1) were treated with 10 μm MG-132 overnight and analyzed with antibody 4G1 (Fig. 1A). B–E, levels of Bcl-2, Bc-xL, and p27 (B and C) and other autophagy regulators (D and E) in mouse tissues were examined. Expression levels of the indicated markers of autophagy in tissues from 3 pairs of unstressed wild-type and Map1s−/− littermates were compared by immunoblot. The relative intensity is the mean ± S.D. of 3 pairs of littermates. Significance was determined by Student's t test. **, p ≤ 0.01. Actual protein levels are dramatically different in different organs and tissues, and low levels of Bcl-XL in brain and P27 in liver were detected in different blots enhanced with increase of protein input and antibody concentration and extended exposure time, and their relative levels were compared.
To test for impact of collective Map1s gene products on autophagy in vivo, we ablated the Map1s gene in mice (Fig. 1). The Map1s knock-out mice exhibited no obvious abnormalities in development, reproduction, or behavior under normal conditions.
The products of the MAP1S gene are mainly the HC in brains and FL and HC in hearts. However, no product was detectable in livers (Fig. 2B). Although the expression profiles of MAP1S in different organs were dramatically different (Fig. 2B), MAP1S depletion caused a similar depression in autophagy-associated markers Bcl-2, Bcl-xL, and P27 in brain, heart, and liver (Fig. 2, B and C). Because MAP1S isoforms are instable, the undetectable levels of MAP1S in livers may suggest a faster turnover rather than a zero expression of MAP1S proteins (23). It is not surprising to observe a similar effect among brains, hearts, and livers upon MAP1S depletion. In cardiomyocytes, no change was observed in Beclin 1 (33) and PTEN (1), which are considered key regulators of initiation of the Beclin 1-dependent PI3K-AKT-mTOR pathway (Fig. 2, D and E). Similarly, no change was observed in ATG5-ATG12 that controls LC3I conjugation with phosphatidylethanolamine to form LC3II (6) (Fig. 2, D and E). Reduction of Bcl-2/XL will potentially have opposite effects; that is, either activating autophagy initiation through the PI3K-AKT-mTOR pathway by releasing more Beclin 1 or inhibiting autophagy initiation through the LKB1-AMPK-mTOR pathway by decreasing the P27 levels (2, 3). The results indicate that MAP1S may either positively or negatively trigger the initiation of basal autophagy.
Accumulation of Dysfunctional Mitochondria in Cardiomyocytes of MAP1S-deficient Mice Indicates a Positive Role of MAP1S in Autophagosomal Biogenesis
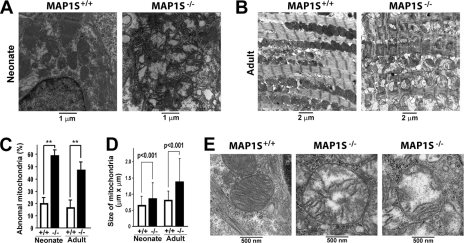
To investigate the actual impact of MAP1S in autophagy initiation under unstressed conditions, we examined the morphology of mitochondria, one of the main substrates of basal autophagy (32), in the cardiomyocytes of MAP1S-deficient mice under transmission electron microscope. Compared with wild type, cardiomyocytes from both neonate and adult MAP1S-deficient mice exhibited a 3-fold increase in abnormal mitochondria that reached 50–60% of the total (Fig. 3, A–C). Those mitochondria were characterized by increased size (Fig. 3D), frequent rupture in the outer membrane, and a clear matrix due to cristolysis (breakdown of internal membranous folds and disorganized cristae) but without isolation membrane-like structures (Fig. 3E). Normally such abnormal mitochondria are present at 20% or less (Fig. 3C) as a part of normal turnover in both cardiomyocytes of neonates and adults. Similar to cardiomyocytes with perturbed autophagy induced by deficiency of ATG5 (34), sarcomeres were also structurally disorganized and misaligned with mitochondria in the MAP1S-deficient cardiomyocytes (Fig. 3B). Because mitophagy is closely coupled with mitochondria fusion/fission dynamics (12), the increased presence of swollen mitochondria in the absence of immature or mature mitophagosomes indicates that the MAP1S deficiency contributes to a reduced ability of the autophagic machinery to either recognize defective mitochondria or initiate mitophagy to isolate and dispose of them. Therefore, MAP1S may enhance P27 levels and positively trigger the initiation of basal autophagy and mitophagy via the LKB1-AMPK-mTOR pathway.
FIGURE 3.
Accumulation of dysfunctional mitochondria in cardiomyocytes of MAP1S-deficient mice. A and B, sections were prepared and analyzed by transmission electron microscopy from hearts of normal unstressed wild-type and MAP1S-deficient neonatal (A) and adult mice (B) at the indicated magnifications. C and D, quantification of abnormal mitochondria in MAP1S-deficient cardiomyocytes is shown. The percentage of abnormal mitochondria was scored directly from a total of 100–300 mitochondria from 15 images acquired from the three pairs of littermates (C). Data are the mean ± S.D. of three littermates, and difference was statistically determined by Student's t test. **, p ≤ 0.01. The average size of mitochondria (±S.D.) from the same samples was calculated from the length and width of individual mitochondria measured on proportional printouts of TEM images (D). Significance of the differences was assessed by the Student's t test. E, the mitochondrial abnormalities are shown in high resolution images.
MAP1S-deficient Mice Exhibit Early Accumulation of Autophagosomes in Response to Nutritive Stress
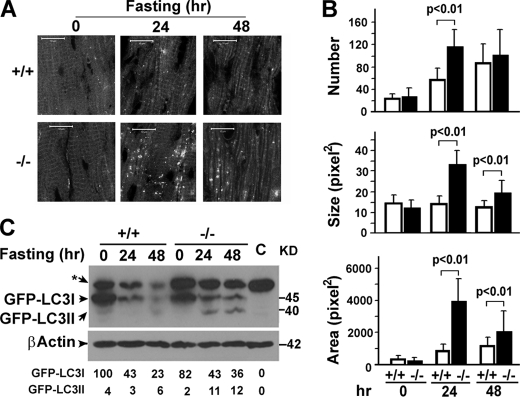
As a prosurvival mechanism, autophagy is activated to degrade subcellular components for nutrient recycling under nutritive stress. To test for a role of MAP1S in the general autophagic response to nutritive stress, neonatal mice delivered by Caesarean section to prevent feeding were separated from their dams and maintained under nutrient deprivation (31). Alterations in LC3I and LC3II contents in hearts and livers (Fig. 4, A and B) under nutritive stress were observed in both the wild-type and MAP1S-deficient pups. Normally wild-type neonatal pups begin to accumulate LC3II at 5 h after Caesarean section (Fig. 4A), which reflected the rate of autophagosomal biogenesis that exceeds the rate of degradation (31). The decline of both LC3I and LC3II by 12 h is indicative of complete conversion of LC3I to autophagosome-associated LC3II and clearance of LC3II-associated autophagosomes via lysosomal degradation for pro-survival nutritive recycling (31). In contrast, the MAP1S-deficient mice exhibited an accumulation of LC3II, indicating an accumulation of autophagosomes as early as 2 h and did not exhibit decrease in LC3I and LC3II at 12 h (Fig. 4, A and B).
To directly visualize and quantify autophagosomes, both wild-type and Map1s knock-out mice were crossed with transgenic mice expressing GFP-LC3 (29). Cardiomyocytes from mice on a normal diet exhibited occasional GFP-LC3-labeled punctate foci representing autophagosomes (Fig. 5A). Fasting caused a near 2-fold increase in GFP-LC3-labeled autophagosomes (Fig. 5B), similarly as reported (29) but without a concurrent accumulation of GFP-LC3II (Fig. 5C). The MAP1S-deficient mice exhibited increases in the number and size of GFP-LC3 punctate foci and area occupied by the foci (Fig. 5, A and B) concurrent with an increase in GFP-LC3II levels (Fig. 5C). Similarly, the increase in autophagosomes indicates either increased autophagosomal biogenesis or decreased autophagosomal degradation.
FIGURE 5.
The MAP1S deficiency leads to accumulation of autophagosomes in adult mouse cardiomyocytes under nutritive stress. A, MAP1S depletion causes accumulation of GFP-labeled autophagosomes (white dots) in fasted mouse heart tissue. +/+, wild-type mice expressing the transgenic GFP-LC3 marker; −/−, MAP1S-deficient mice expressing GFP-LC3. Images are representative of samples from three pairs of mice. B, quantification of GFP-LC3 labeled autophagosomes is shown. The number, average size, and total area occupied of GFP-LC3 punctate foci in heart tissue sections is the average and S.D. of 10 randomly selected images in a field of 512 pixel × 512 pixels. The significance of differences was determined by Student's t test. C, nutritionally stressed MAP1S-deficient mice accumulate both LC3I and II. GFP-LC3I and II was assessed with anti-LC3 antibody. Bands were quantified and expressed as percent of the GFP-LC3I intensity in unstressed wild-type mouse. The nonspecific band labeled by an asterisk appears in both GFP-LC3-expressing mice and wild-type mice that do not express GFP-LC3 (C, control). Lysates were prepared from the heart tissues of the same mice shown in A.
Accumulation of Autophagosomes in MAP1S-deficient Mice under Nutritive Stress Indicates an Impairment of Autophagosomal Degradation
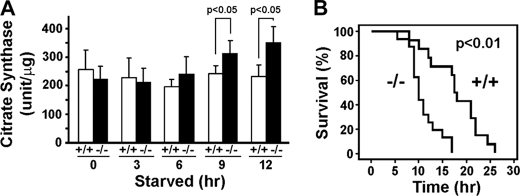
The increases in LC3II levels in the MAP1S-deficient neonates 2 h after starvation suggests an early accumulation of autophagosomes, which is likely an early indicator of either reduced rate of clearance of LC3II-associated autophagosomes or increased conversion of LC3I to LC3II. The maintenance of LC3I levels in addition to LC3II 12 h after starvation (Fig. 4A) suggests that the MAP1S deficiency more likely leads to impairment of autophagosomal degradation, because LC3I levels will decrease if conversion of LC3I to LC3II increases. To further confirm the nature of the early accumulation of autophagosomes in the MAP1S-deficient mice, we compared the mitochondrial mass between the wild-type and MAP1S-deficient mice. An abnormal increase in mitochondrial mass monitored by mitochondrial citrate synthase activity (32) was observed at 9–2 h post-Caesarean section and starvation (Fig. 6A). This confirmed that the autophagic clearance of mitochondria is impaired in the nutritionally stressed MAP1S-deficient neonate hearts.
FIGURE 6.
Increase of mitochondria mass in cardiomyocytes and decrease of survival time caused by MAP1S depletion suggest an impairment of autophagosomal degradation. A, increased mitochondrial mass in stressed MAP1S-deficient cardiomyocytes is shown. Data are the mean ± S.D. of three pairs of littermates. Significance was determined by Student's t test. B, reduced survival in starved MAP1S-deficient neonates is shown. The indicated significance of difference between survival curves for wild-type and knock-out mice was determined by Kaplan-Meier analysis.
Postnatal induction of autophagy is essential for recycling of nutrients and survival of neonates between interruption of trans-placental support at birth and feeding by nursing (31). Because of the impairment of autophagosomal degradation, the survival time for half of the starved neonates was significantly reduced from 18 h in wild-type neonates to 10 h in the MAP1S-deficient mice (Fig. 6B). Thus, products of Map1s gene collectively play a positive role in regulation of autophagy induced by nutrient deprivation in both neonatal and adult mice as well as in the regulation of basal autophagy.
Isoform-dependent Interaction of MAP1S and LC3 in the Absence of Microtubules
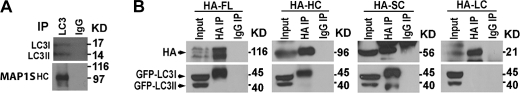
Autophagosomal biogenesis and degradation are closely associated with microtubules, and the truncation of LC3 precursor to LC3I and conversion of LC3I to LC3II regulate the entire process of autophagy (8, 9, 25, 35). To understand why the MAP1S depletion in mice led to impairment of autophagosomal biogenesis and degradation, we started to investigate the interaction of MAP1S isoforms with LC3 and LC3-associated autophagic machinery in addition to the known interaction with the mitochondrion-associated LRPPRC (15, 36). HeLa cells were cultured in media without paclitaxel and MG-132 so that FL and HC were the main isoforms of MAP1S expressed in cells. Cell lysates were prepared in cold lysate buffer in which no microtubule was able to be assembled. Co-immunoprecipitation with an anti-LC3 antibody revealed that, similar to MAP1A/B, at least MAP1SHC interacted with LC3, although it was not clear which LC3 isoform pulled down the HC. MAP1SFL was not pulled down by anti-LC3 antibody, suggesting that either the FL interacted weakly with LC3 or the FL protein itself did not survive the co-immunoprecipitation procedure because of instability (Fig. 7A).
FIGURE 7.
MAP1S interacts with LC3 in an isoform-dependent mode. A, interaction of MAP1S with LC3 is shown. Immunoprecipitates (IP) from HeLa cell lysates were isolated with anti-LC3 antibody or pre-immune IgG and then analyzed by immunoblot with antibody against LC3 or MAP1S (4G1). Ab, antibody. B, interaction profile among both MAP1S and LC3 isoforms is shown. Lysates (Input) from HEK 293 cells co-expressing GFP-LC3 with the indicated HA-tagged MAP1S isoforms were immunoprecipitated with anti-HA antibody (HA IP) or control IgG (IgG IP) and then subjected to immunoblot analysis with antibodies against the HA tag (top panel) or LC3 (lower panel). The GFP-LC3I bands exhibit lower mobility from analyses of immunoprecipitates than Input because of the concentration-dependent influence of proximal bands during the prolonged electrophoretic analysis. Total extract consists of low concentration bands distributed throughout the gel, whereas immunoprecipitates are primarily excess IgG bands. The leading edge of the GFP-LC3I overlaps sufficiently with the tighter band of GFP-LC3I in the Input enough to suggest the band is GFP-LC3I in both cases.
The demonstration of an endogenous HC-LC3 complex triggered us to investigate in detail the interaction of different isoforms of MAP1S with different isoforms of LC3 in HEK 293 cells that were able to express recombinant proteins tagged with either HA or GFP at high levels. Immunoprecipitation analysis using microtubule-free cell lysates revealed that the MAP1SFL, HC, and SC forms interacted with LC3I, MAP1SSC uniquely interacted with both LC3I and LC3II, and MAP1SLC failed to interact with either form of LC3 (Fig. 7B). Because the LC isoform constantly associates with microtubules (23) and did not react with LC3, it was not included in subsequent analyses. The isoform-specific interaction between MAP1S and LC3 in the absence of microtubules also suggests MAP1S isoforms may play microtubule-independent roles through either LC3I or LC3II when the LC3 isoforms are not associated with microtubules.
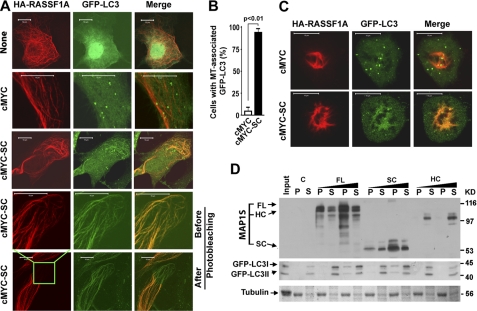
MAP1S Isoforms Differentially Facilitate the Association of LC3 Isoforms with Microtubules
RASSF1A stabilized and completely associated with microtubules so that microtubules can be visualized by RASSF1A staining (19, 22, 23). RASSF1A alone did not recruit GFP-LC3 on the stabilized microtubules (Fig. 8A). Because microtubules stabilized by either RASSF1A or paclitaxel recruit and accumulate MAP1SSC (19, 22, 23), GFP-LC3 colocalized with the RASSF1A-induced microtubular bundles in cells co-expressing RASSF1A and MAP1SSC (Fig. 8, A and B). Although there were much fewer mitotic cells expressing RASSF1A and MAP1SSC, the same results were obtained in mitotic cells blocked in premetaphase due to the RASSF1A-induced stabilization of spindle microtubules (Fig. 8C). A similar microtubular association of LC3 was not observed in cells overexpressing MAP1SFL due to lower levels of MAP1SFL relative to MAP1SSC (23). Based on the fluorescence microscopy data shown above, we cannot differentiate whether the microtubule-associated GFP-LC3 is GFP-LC3I or GFP-LC3II.
FIGURE 8.
MAP1S recruits LC3 to microtubules in an isoform-dependent mode. A, MAP1S recruits LC3 to stabilized microtubules. Association of LC3 (green) with microtubules was monitored in HeLa cells expressing recombinant GFP-LC3 and microtubule stabilizer HA-RASSF1A (red) in the presence or absence of MAP1SSC introduced by transient transfection. A control without cMYC vector (None) was included to test the effect of the cMYC vector. Transfection efficiency driven by the CMYC vector was monitored separately by immunostaining. Yellow indicates overlap of microtubules decorated with RASSF1A and LC3. Images in the fourth panels were captured from the boxed area in the bottom panel that was then subjected to photobleaching under the red laser to demonstrate that the GFP-LC3-associated fibril structures were not caused by leaking through of the red fluorescent signal. B, percentage of cells exhibiting microtubular association of GFP-LC3 in cells expressing both HA-RASSF1A and GFP-LC3 was determined by direct count. Data are the mean ± S.D. of 3 transfection experiments with 100 randomly selected transfected cells including both interphase and mitotic cells counted for each transfection. The p value was determined by Student's t test. C, MAP1S recruits LC3 to microtubules in mitotic cells. Mitotic cells blocked in premetaphase due to RASSF1A were located morphologically from cultures in A. Fibril structures were confirmed to be mitotic spindles made of microtubules by us and others (47, 48). D, a specific profile of interactions among isoforms of MAP1S, LC3I, LC3II, and microtubules is shown. Pelleted microtubules (P) were resuspended in an equal volume of reaction buffer to the supernatants (S). Both fractions were analyzed with antibodies against MAP1S (4G1) and LC3. Tubulin was visualized with Coomassie Blue. Input, reaction mixture in the absence of MAP1S lysates before microtubule assembly; C, control mixture, the same as Input but after assembly and separation of microtubules. A representative of at least three repeats is shown here.
We further analyzed the association of LC3 isoforms with paclitaxel-stabilized microtubules assembled from purified tubulins in the presence of different MAP1S isoforms in vitro. In the absence of MAP1S, neither LC3I nor LC3II associated with the paclitaxel-stabilized microtubules (Fig. 8D). MAP1SSC that interacted with both LC3I and II independent of microtubule (Fig. 7B) also recruited both LC3I and LC3II to stabilized microtubules (Fig. 8D). Because MAP1SHC only interacted with LC3I in solution (Fig. 7B) and it does not associate with microtubules on its own (23), it induced neither LC3I nor LC3II to co-assemble with microtubules (Fig. 8D). Although MAP1SFL only interacted with LC3I in the absence of microtubule (Fig. 7B), it facilitated association of both LC3I and LC3II on stabilized microtubules (Fig. 8D). This potentially indicates a self-inhibitory effect from MAP1SFL on its interaction with LC3II had been released upon its association with stabilized microtubules. The amounts of GFP-LC3I and GFP-LC3II associated with microtubules depended on the concentration of FL or SC (Fig. 8D) and reached saturated levels when concentrated FL- or SC-containing lysate was added in the assembly mixtures (supplemental Fig. 4).
Taken together, these interaction studies show that the interaction of different isoforms of MAP1S with microtubules is contingent on the dynamic state of microtubules (23). Different isoforms of MAP1S exhibit distinct interactions with LC3. The MAP1SLC isoform that is constitutively associated with microtubules independent of dynamic state (23) does not interact with LC3. Both MAP1SFL and HC interact with LC3I and may exhibit potential roles when LC3I is disassociated with microtubules. MAP1SSC uniquely and independently forms complexes with LC3I or LC3II. Both FL and SC isoforms have affinity for stabilized microtubules and bridge both LC3I and LC3II to them. The association of either LC3I or II isoform with microtubules requires the presence of either FL or SC of MAP1S. The isoform-dependent interaction of MAP1S and LC3 and recruitment to stabilized microtubules may implicate roles of MAP1S in autophagosomal biogenesis and degradation.
DISCUSSION
To maintain cellular homeostasis, robust basal levels of autophagy persist during the entire cell cycle including both interphase and mitosis (32). Normal cell functions are critically dependent on ATP from mitochondria. Detection and removal of defective mitochondria are critical for cell life and death, and defective mitochondria are one of the main substrates of autophagy (32). A balance of autophagosomal biogenesis and degradation is essential to maintain normal autophagic homeostasis. Acute induction of autophagy under nutritive stress results in autophagosomal biogenesis exceeding the capacity of degradation. Consistent with numerous reported results, our data demonstrate that levels of LC3II are elevated upon starvation (4).
LC3 was originally identified as an interactive partner of microtubule-associated proteins MAP1A and MAP1B and suggested to regulate their microtubule binding activity (24). It has attracted attention as the first mammalian protein identified that specifically associates with autophagosome membranes (4, 37). Overexpression of MAP1B in mice has been reported to impact autophagy in the neuronal system where it is selectively expressed (26). Here we have shown for the first time with a knock-out mouse model that MAP1S, the ubiquitously distributed homologue of neuronal MAP1A and MAP1B, positively regulates both autophagosomal biogenesis and degradation.
To further understand the mechanism by which MAP1S regulates autophagy and mitophagy, we employed in vitro biochemical and cell biological experiments to show that isoforms of MAP1S interact not only with microtubules but also with LC3I or LC3II variants of the autophagosome-associated protein LC3 (ATG8) in an isoform-specific manner. The profile of three-way interactions of specific MAP1S isoforms with LC3I and LC3II and microtubules strongly support the idea that MAP1S through its isoforms participates in microtubular coordination of the autophagic process from autophagosomal biogenesis to degradation. In addition, it appears that MAP1S may regulate autophagy in multiple ways.
The most attractive is that MAP1S may regulate autophagy by interaction with LC3I and LC3II and recruiting them to microtubules. Autophagy is a dynamic process during which isolation membranes package substrates to form autophagosomes that later are fused with lysosomes to form autolysosomes for degradation. Before LC3I is conjugated with phosphatidylethanolamine to form LC3II, MAP1S may assist LC3I to associate with microtubules. Different sets of microtubular filaments coexist in cells, with the acetylated microtubules concentrating in the perinuclear region. Depolymerization of both non-acetylated and acetylated microtubules results in impairment of LC3I-LC3II conversion (10). Both FL and SC of MAP1S interact with LC3I and recruit it on stabilized microtubules to regulate the LC3I to II conversion or autophagosomal biogenesis.
It is agreed that the LC3II-associated mature autophagosomes move along microtubular tracks (8, 9). Recently, it has been confirmed that acetylated microtubules are required for fusion of autophagosomes with lysosomes to form autolysosomes (10). Paclitaxel and RASSF1A may similarly enhance tubulin acetylation to stabilize microtubules (10, 22, 38). Microtubule-associated isoforms of MAP1SFL and SC interact with LC3II and facilitate its association with stabilized microtubules. This is consistent with a role in bridging LC3II-associated autophagosomes with relatively stable acetylated microtubule tracks for migration rather than a simple role in regulation of microtubule dynamics per se (19, 23). This is in contrast to the LC isoform that is constitutively associated with microtubules and affects microtubule dynamics but does not interact with LC3. Therefore, impairment of autophagosomal degradation resulting from MAP1S depletion indicates a positive role of Map1s gene products such as the FL and SC.
Besides functions mediated by microtubular association, isoforms of MAP1S may impact autophagy through its interaction with either isoforms of LC3 independent of microtubules. The microtubule-independent interactions of LC3I with FL, HC, and SC indicate potential roles of isoforms before initiation of autophagy when LC3I is not associated with microtubules. When LC3II molecules are inserted into isolation membranes, they may serve as a linker to bridge autophagosomes with the microtubule cytoskeleton if protruding externally, but with cargo receptors, such as P62, if facing internally (4, 37, 39). The microtubule-independent interaction of MAP1SSC with LC3II potentially plays a role in packaging cargos like dysfunctional mitochondria. In previous reports, using the specific MAP1SSC as a prototypic product of the Map1s gene, we showed that it not only is a stabilized microtubule-associated protein but also exhibits a unique and conditional association with mitochondria (19, 23). Microtubule- and mitochondria-associated activities of the SC isoform reside in independent sequence domains (19). Therefore, MAP1SSC may specifically interfere with mitophagy at the stages of deposition of LC3I to mitochondria and conversion of LC3I to LC3II and initiation, maturation, and isolation of mitochondria by LC3II-containing isolation membranes. The MAP1SSC-induced mitochondrial aggregates likely reflect a failure in the process. Progressive, irreversible, and lethal aggregation of mitochondria is the final consequence of the failure (22).
In addition to the microtubule-dependent and -independent roles of MAP1S through interaction with LC3 isoforms, conceivably MAP1S isoforms may also directly affect autophagy or mitophagy. A role for the MAP1SLC isoform in autophagy and general microtubule dynamics beyond its constitutive association with microtubules remains to be assigned. However, interaction of MAP1S with mitochondrion-associated proteins such as LRPPRC (15, 36) may suggest another pathway to affect autophagy because LRPPRC interacts with Parkin (27) that translocates specifically to uncoupled mitochondria to induce their autophagy (40–42). As an E3 ubiquitin ligase, Parkin directly interacts with Bcl-2 and mono-ubiquitinates Bcl-2 to regulate autophagy (43). LRPPRC alone basically acts as a checkpoint for initiation of mitophagy, and its depletion results in autophagy activation.3 An in-depth study on the mechanism by which MAP1S regulates autophagy through LRPPRC is under way.
The MAP1S-deficient mice exhibited alterations in the Bcl-2/Bcl-xL/p27 axis in multiple tissues. The depression of the Bcl-2/xL family is involved in initiation of autophagy through release of sequestered Beclin 1 that activates autophagy through the PI3K-AKT-mTOR pathway (33). Elevated levels of Bcl-2 through an increase in P27 that increases ATG5 (3) can result in Beclin-1-independent activation of autophagy through the LKB1-AMPK-mTOR pathway (2, 44). Cardiomyocytes of the Map1s-depleted mice where alterations of the Bcl-2/xL/P27 axis are prominent exhibit an accumulation of large swollen mitochondria indicative of a potential defect in mitophagy. The results suggest that the effects of MAP1S on the LKB1-AMPK-mTOR pathway dominate relative to the PI3K-AKT-mTOR pathway.
In addition to its role in initiation of autophagy, Bcl-2/xL have been widely implicated in maintenance of mitochondrial membrane integrity and mitochondrial homeostasis (13). Mitochondrial fusion-fission and mitophagy are tightly coupled (12). Mitochondrial tubules that are generally resistant to removal by mitophagy are asymmetrically separated into smaller units that allow selection of the weaker daughter units with respect to membrane potential for clearance by mitophagy. It has been proposed as a mechanism of maintenance of mitochondrial tubules with the highest membrane potential (9). Wild-type dynamin-related protein 1 (Drp1) is essential for mitochondrial fission (45). Depression of Bcl-2/xL contributes to inhibition of fission by reduction of its positive regulation of the GTPase activity of the mitochondrial fission-fusion regulator Drp1 (46). The accumulation of large swollen mitochondria in cardiomyocytes of the Map1s-depleted mice also indicates a potential defect in mitochondrial fission or recognition and/or removal of the defective mitochondria by mitophagy. The deficient mice exhibited severe defects in the autophagic response to nutritional stress that was reflected in the accumulation of mitochondria, reduced clearance of autophagosomes, and reduced survival of starved neonates.
Taken together, these results strongly suggest that products of the Map1s gene positively regulate autophagy and, particularly, mitophagy in vivo. As a multifunctional protein, MAP1S isoforms may act as a linker to bridge autophagy components not only with the microtubular cytoskeleton through interacting with LC3 isoforms but also directly to mitochondria, one of the major substrates of autophagy through the interaction with LRPPRC. Such a profile may extend to MAP1A and -B, the LC3-interactive neuronal homologues of MAP1S with which LC3 was first described to form an interactive complex (24). MAP1S interacts with tumor suppressor RASSF1A (21, 22), autophagy-related protein LC3, and mitochondrion-associated LRPPRC that interacts with mitophagy initiator and Parkinson disease-related Parkin (27) and regulates autophagy. Defects in MAP1S-regulated autophagy may result in heart disease, cancers, neurodegenerative diseases, and a wide range of other diseases.
Acknowledgments
We thank Drs. Le Sun and Joe Corvera (A&G Pharmaceuticals, Inc., Columbia, MD) for anti-MAP1S mouse monoclonal antibody 4G1, which is now sold by Precision AntibodyTM with catalog number AG10006. LC3 cDNA and GFP-LC3 transgenic mice were gifts from Dr. Noboru Mizushima, the Department of Physiology and Cell Biology, Tokyo Medical and Dental University Graduate School and Faculty of Medicine. Transmission electron microscopy was performed by the High Resolution Electron Microscopy Facility in the University of Texas MD Anderson Cancer Center supported by National Institutes of Health Cancer Center Core Grant CA16672.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R01CA142862 (NCI; to L. L.) and NCI P50 CA140388 (NCI; to W. L. M.). This work was also supported by Department of Defense New Investigator Award W81XWH (to L. L.), the Susan Komen Foundation (to W. L. M.), and aid from the John S. Dunn Research Foundation (to W. L. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2 and Figs. S1–S4.
Leyuan Liu, unpublished results.
- AMPK
- AMP-activated protein kinase
- ATG
- autophagy-related gene
- Bcl-2
- B-cell CLL/lymphoma 2
- GFP-LC3
- GFP-fused LC3
- HC
- heavy chain
- LC
- light chain
- LC3
- light chain 3 of MAP1A/B
- MAP1A
- microtubule-associated protein 1A
- MAP1B
- microtubule-associated protein 1B
- MAP1S
- microtubule-associated protein 1S
- PTEN
- phosphatase and tensin homolog
- RASSF1
- ras association domain family 1
- SC
- short chain
- ES
- embryo stem
- LRPPRC
- leucine-rich PPR-motif containing protein
- RASSF1
- ras association domain family 1
- FL
- full-length.
REFERENCES
- 1. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liang J., Shao S. H., Xu Z. X., Hennessy B., Ding Z., Larrea M., Kondo S., Dumont D. J., Gutterman J. U., Walker C. L., Slingerland J. M., Mills G. B. (2007) Nat. Cell Biol. 9, 218–224 [DOI] [PubMed] [Google Scholar]
- 3. Zhang G., Park M. A., Mitchell C., Walker T., Hamed H., Studer E., Graf M., Rahmani M., Gupta S., Hylemon P. B., Fisher P. B., Grant S., Dent P. (2008) J. Biol. Chem. 283, 24343–24358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanida I., Sou Y. S., Ezaki J., Minematsu-Ikeguchi N., Ueno T., Kominami E. (2004) J. Biol. Chem. 279, 36268–36276 [DOI] [PubMed] [Google Scholar]
- 6. Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. (2008) Mol. Biol. Cell 19, 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) Mol. Cell 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 8. Fass E., Shvets E., Degani I., Hirschberg K., Elazar Z. (2006) J. Biol. Chem. 281, 36303–36316 [DOI] [PubMed] [Google Scholar]
- 9. Köchl R., Hu X. W., Chan E. Y., Tooze S. A. (2006) Traffic 7, 129–145 [DOI] [PubMed] [Google Scholar]
- 10. Xie R., Nguyen S., McKeehan W. L., Liu L. (2010) BMC Cell Biol. 11, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skulachev V. P. (2001) Trends Biochem. Sci. 26, 23–29 [DOI] [PubMed] [Google Scholar]
- 12. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuwana T., Newmeyer D. D. (2003) Curr. Opin. Cell Biol. 15, 691–699 [DOI] [PubMed] [Google Scholar]
- 14. Liu L., Amy V., Liu G., McKeehan W. L. (2002) In Vitro Cell Dev. Biol. Anim. 38, 582–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu L., McKeehan W. L. (2002) Genomics 79, 124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong E. Y., Tse J. Y., Yao K. M., Lui V. C., Tam P. C., Yeung W. S. (2004) Biol. Reprod. 70, 775–784 [DOI] [PubMed] [Google Scholar]
- 17. Song M. S., Chang J. S., Song S. J., Yang T. H., Lee H., Lim D. S. (2005) J. Biol. Chem. 280, 3920–3927 [DOI] [PubMed] [Google Scholar]
- 18. Ding J., Valle A., Allen E., Wang W., Nardine T., Zhang Y., Peng L., Yang Y. (2006) Biochem. Biophys. Res. Commun. 339, 172–179 [DOI] [PubMed] [Google Scholar]
- 19. Liu L., Vo A., Liu G., McKeehan W. L. (2005) Cancer Res. 65, 4191–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Orbán-Németh Z., Simader H., Badurek S., Tranciková A., Propst F. (2005) J. Biol. Chem. 280, 2257–2265 [DOI] [PubMed] [Google Scholar]
- 21. Dallol A., Cooper W. N., Al-Mulla F., Agathanggelou A., Maher E. R., Latif F. (2007) Cancer Res. 67, 492–500 [DOI] [PubMed] [Google Scholar]
- 22. Liu L., Vo A., McKeehan W. L. (2005) Cancer Res. 65, 1830–1838 [DOI] [PubMed] [Google Scholar]
- 23. Liu L., Xie R., Yang C., McKeehan W. L. (2009) Cell Oncol. 31, 393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mann S. S., Hammarback J. A. (1994) J. Biol. Chem. 269, 11492–11497 [PubMed] [Google Scholar]
- 25. Monastyrska I., Rieter E., Klionsky D. J., Reggiori F. (2009) Biol. Rev. Camb. Philos. Soc. 84, 431–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Q. J., Ding Y., Kohtz D. S., Kohtz S., Mizushima N., Cristea I. M., Rout M. P., Chait B. T., Zhong Y., Heintz N., Yue Z. (2006) J. Neurosci. 26, 8057–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davison E. J., Pennington K., Hung C. C., Peng J., Rafiq R., Ostareck-Lederer A., Ostareck D. H., Ardley H. C., Banks R. E., Robinson P. A. (2009) Proteomics 9, 4284–4297 [DOI] [PubMed] [Google Scholar]
- 28. Lin Y., Zhang J., Zhang Y., Wang F. (2007) Genesis 45, 554–559 [DOI] [PubMed] [Google Scholar]
- 29. Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van der Weyden L., Tachibana K. K., Gonzalez M. A., Adams D. J., Ng B. L., Petty R., Venkitaraman A. R., Arends M. J., Bradley A. (2005) Mol. Cell. Biol. 25, 8356–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 32. Liu L., Xie R., Nguyen S., Ye M., McKeehan W. L. (2009) Cell Cycle 8, 1616–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levine B., Sinha S., Kroemer G. (2008) Autophagy 4, 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakai A., Yamaguchi O., Takeda T., Higuchi Y., Hikoso S., Taniike M., Omiya S., Mizote I., Matsumura Y., Asahi M., Nishida K., Hori M., Mizushima N., Otsu K. (2007) Nat. Med. 13, 619–624 [DOI] [PubMed] [Google Scholar]
- 35. Jahreiss L., Menzies F. M., Rubinsztein D. C. (2008) Traffic 9, 574–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu L., Vo A., Liu G., McKeehan W. L. (2005) Biochem. Biophys. Res. Commun. 332, 670–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanida I., Ueno T., Kominami E. (2004) Int. J. Biochem. Cell Biol. 36, 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu L., Tommasi S., Lee D. H., Dammann R., Pfeifer G. P. (2003) Oncogene 22, 8125–8136 [DOI] [PubMed] [Google Scholar]
- 39. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 40. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanaka A. (2010) FEBS Lett. 584, 1386–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. (2010) J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen D., Gao F., Li B., Wang H., Xu Y., Zhu C., Wang G. (2010) J. Biol. Chem. 285, 38214–38223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scarlatti F., Maffei R., Beau I., Codogno P., Ghidoni R. (2008) Cell Death Differ. 15, 1318–1329 [DOI] [PubMed] [Google Scholar]
- 45. Frank S., Gaume B., Bergmann-Leitner E. S., Leitner W. W., Robert E. G., Catez F., Smith C. L., Youle R. J. (2001) Dev. Cell 1, 515–525 [DOI] [PubMed] [Google Scholar]
- 46. Li H., Chen Y., Jones A. F., Sanger R. H., Collis L. P., Flannery R., McNay E. C., Yu T., Schwarzenbacher R., Bossy B., Bossy-Wetzel E., Bennett M. V., Pypaert M., Hickman J. A., Smith P. J., Hardwick J. M., Jonas E. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2169–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Song M. S., Song S. J., Ayad N. G., Chang J. S., Lee J. H., Hong H. K., Lee H., Choi N., Kim J., Kim H., Kim J. W., Choi E. J., Kirschner M. W., Lim D. S. (2004) Nat. Cell Biol. 6, 129–137 [DOI] [PubMed] [Google Scholar]
- 48. Rong R., Jin W., Zhang J., Sheikh M. S., Huang Y. (2004) Oncogene 23, 8216–8230 [DOI] [PubMed] [Google Scholar]