Abstract
EpsE is an ATPase that powers transport of cholera toxin and hydrolytic enzymes through the Type II secretion (T2S) apparatus in the Gram-negative bacterium, Vibrio cholerae. On the basis of structures of homologous Type II/IV secretion ATPases and our biochemical data, we believe that EpsE is active as an oligomer, likely a hexamer, and the binding, hydrolysis, and release of nucleotide cause EpsE to undergo dynamic structural changes, thus converting chemical energy to mechanical work, ultimately resulting in extracellular secretion. The conformational changes that occur as a consequence of nucleotide binding would realign conserved arginines (Arg210, Arg225, Arg320, Arg324, Arg336, and Arg369) from adjoining domains and subunits to complete the active site around the bound nucleotide. Our data suggest that these arginines are essential for ATP hydrolysis, although their roles in shaping the active site of EpsE are varied. Specifically, we have shown that replacements of these arginine residues abrogate the T2S process due to a reduction of ATPase activity yet do not have any measurable effect on nucleotide binding or oligomerization of EpsE. We have further demonstrated that point mutations in the EpsE intersubunit interface also reduce ATPase activity without disrupting oligomerization, strengthening the idea that residues from multiple subunits must precisely interact in order for EpsE to be sufficiently active to support T2S. Our findings suggest that the action of EpsE is similar to that of other Type II/IV secretion ATPase family members, and thus these results may be widely applicable to the family as a whole.
Keywords: ATPases, Bacteria, Cholera Toxin, Protein Secretion, Protein Structure, Vibrio cholerae, Hexamer, Type II Secretion
Introduction
The T2S2 system is composed of at least 12 gene products that assemble into a large protein complex spanning the entire cell envelope of Gram-negative bacteria, including Vibrio cholerae (1, 2). Following transport through the cytoplasmic membrane and folding in the periplasm, toxins and hydrolytic enzymes are targeted to the T2S system and secreted in their native form through a pore in the outer membrane formed by the secretin EpsD. This transport is thought to occur through a piston-like mechanism due to the polymerization of a pseudopilus made up of proteins EpsG–K (3–5). The process is energy-dependent, requiring the cytoplasmic ATPase, EpsE (6). EpsE associates with the T2S system through interactions with EpsL, a protein found in an inner membrane complex also composed of proteins EpsC, EpsF, and EpsM (7–11). The use of an ATPase to provide the energy for assembly and/or substrate translocation is an important feature of not just the T2S machinery but of several molecular trafficking systems (3).
EpsE is a member of the large family of Type II/IV secretion ATPases (12, 13), which includes ATPases involved with T2S, Type IV pilus biogenesis, and Type IV secretion (T4S) systems and participates in diverse processes such as protein transport, DNA uptake and export, archaeal flagella assembly, and pilus extension or retraction (14, 15). EpsE and the other T2S ATPases display the greatest level of homology to the Type IV pilus assembly ATPases (12). A lower level of homology is observed with the Type IV pilus retraction ATPases, as shown for Pseudomonas aeruginosa PilT, whereas the lowest degree of homology is observed with the T4S ATPases including Hp0525 from Helicobacter pylori (12) (supplemental Fig. S1).
The structurally characterized members of this family have two distinct domains: the carboxyl-terminal domain (CTD) and the amino-terminal domain (NTD), which are connected through a short flexible linker. Although they have less than 35% overall sequence identity, these proteins share highly homologous sequence motifs in the RecA-like core structure found in the CTD, including a Walker A box encompassing a P-loop structure and a less well defined Walker B box. Additionally, the CTD includes Asp and His boxes, which taken together with the Walker A/B boxes make up the nucleotide binding site (13, 16, 17) (supplemental Fig. S1). There is greater size and sequence variation among the NTDs than among the CTDs. These differences likely impart system and species specificity for the different subgroups. Interactions through the NTD could serve to localize the ATPase to a membrane-bound complex, as was shown for the N-terminal subdomain N1 of the EpsE NTD, which directly interacts with the cytoplasmic membrane protein EpsL within the T2S system (7, 11). Alternatively, NTD interactions with other proteins may help regulate ATPase activity, as was seen for Hp0525 in complex with an inhibitory protein, Hp1451 (18). Regardless of its interaction with other proteins, it should be noted that the NTD may also contribute directly to the enzymatic activity of these ATPases by providing highly conserved residues to complete the active site (13, 19–21) (supplemental Fig. S1).
Structural analyses indicate that significant conformational changes occur in Type II/IV secretion ATPases as they bind nucleotides (13, 19–24). Early work on the T4S ATPase from H. pylori, Hp0525, suggested that it may function as a gating molecule at the bacterial inner membrane and regulate its opening and closing by nucleotide binding, hydrolysis, and release (24). Further analysis indicated that Hp0525 may function as a dynamic hexamer capable of forming a compact, locked structure when in a nucleotide-bound conformation (23). More recently, the structure of AfGspE, an ATPase from Archaeoglobus fulgidus, showed that the NTD domain moves by 10° with respect to the CTD in the presence of nucleotide, thus allowing for two conserved arginine residues from the NTD to clamp around the nucleotide (20).
The identification of two different structural forms of the Type IV pilus retraction ATPase PilT from Aquifex aeolicus (AaPilT) provided additional insight into domain movements and subunit realignments that can occur within hexamers of this family of ATPases (21). In both the ATP-bound and the ADP-bound forms, AaPilT was observed as hexamers with C6 symmetry. Another crystal structure of AaPilT, at 4.2 Å resolution, revealed a hexamer with approximate C2 symmetry, which contains AaPilT subunits in several different conformations, ranging from a more closed conformation than seen in the AaPilT hexamer with C6 symmetry to a wide open conformation. In the latter subunits, a large domain rotation of ∼65° when compared with the closed form was observed (21).
More recently, structures of a PilT homolog from P. aeruginosa (PaPilT) showed that hexamers with a C2 symmetry are formed both in the presence and in the absence of nucleotide (19). Although the differences between these hexamers are not as drastic as those between the AaPilT hexamer with perfect C6 symmetry and quasi-C2 symmetry, the subunit of PaPilT still appears to change from open to closed conformations depending on nucleotide binding. This movement is most clearly demonstrated by looking at the changing positions of two NTD arginines and two “His box” histidines within the three different subunit conformations observed in the hexamer with C2 symmetry. Taken together, the conformational changes associated with ATP binding and hydrolysis proposed for Hp0525, AfGspE, AaPilT, and PaPilT could have a substantial effect on the neighboring subunits of the hexamer as well as on interactions between the hexamer and the greater protein complex with which they interact (19–24).
Although full-length EpsE was not amenable to crystallization, the crystal structure was solved of a truncated form of EpsE lacking a part of the N-terminal domain that interacts with EpsL (13). In contrast to the ATPases discussed above, EpsE was not crystallized as a hexamer; instead, the subunits were found to form a helical filament, a phenomenon that has also been observed with other hexameric ATPases (25, 26). The helical nature of EpsE does not likely represent the quaternary structure of EpsE; however, as subsequent analysis showed that EpsE is able to form active oligomers, which are likely hexamers, in the presence of the cytoplasmic domain of EpsL and acidic phospholipids (27). A correlation between oligomer formation and increased ATPase activity was demonstrated for EpsE (6, 27), suggesting that the communication between adjoining monomers may be key to coupling nucleotide hydrolysis to substrate trafficking for EpsE as well.
In this study, we have modeled EpsE as a hexameric ring with C2 symmetry based on the PaPilT-AMPPCP structure (19). Based on this model, we have designed experiments to probe the importance of putative active site residues and interface residues of EpsE to determine whether side chains from different domains and subunits contribute to the active site and whether the intersubunit interface is similar to those observed in hexameric structures of Type II/IV secretion ATPase homologs. In the EpsE model, three pairs of conserved arginines, one pair from the NTD, one from the CTD, and one from the CTD of a neighboring subunit (CTD′), appear to realign once nucleotide binds to facilitate ATP hydrolysis. The possible involvement of these arginines in ATP binding and hydrolysis was originally proposed by Forest and colleagues (19, 21), but only a few point mutants were tested for twitching motility in vivo, and no mutant protein was purified for in vitro analyses. To determine the role of these arginines in the function of EpsE, we have replaced each of the arginines with alanine. Our results indicate that substitution of each individual arginine abrogates both ATP hydrolysis and the T2S process. Additional mutations within the intersubunit interface similarly disrupts the function of EpsE, providing support for the hexamer model and indicating that a precise arrangement of residues from different subunits around the active site is essential for the action of EpsE and other T2S ATPases.
EXPERIMENTAL PROCEDURES
A Hexameric Ring Model of EpsE
A model was generated using the crystal structure of EpsE (13) and the available C2 hexameric structure of the homologous PaPilT (19–21). The structurally conserved core of the EpsE-CTD was superimposed onto CTDs in this hexameric structure (Protein Data Bank (PDB) ID: 3JVV). The EpsE-NTDs were added next to the hexameric model to maintain the extensive NTD-CTD′ interface observed in the EpsE crystal structure (13).
Cloning and Expression
EpsE point mutations R156A, R156D, R210A, R225A, R320A, R324A, D326R, D328R, R336A, and R369A were introduced into the previously constructed plasmid, pET21dEpsE(2–503)-EpsL(1–253)His6 (27), using the QuikChange site-directed mutagenesis kit (Stratagene) as directed. The fragments of epsE containing the various mutations were cloned into pMMB384 (28) by exchange of an MfeI/BamHI fragment to create the individual pMMBEpsE point mutant plasmids. The pMMB plasmids were then introduced to the epsE− and wild type V. cholerae strains through conjugation.
Purification of Protein
The above constructed EpsE(2–503)-EpsL(1–253)His6 point mutants, with the exception of the one containing D328R, were expressed in Escherichia coli BL21(DE3) under isopropyl-1-thio-β-d-galactopyranoside inducing conditions and purified using metal affinity chromatography and gel filtration as described in Refs. 7 and 29.
Phospholipids
E. coli-derived cardiolipin was prepared in 20 mm Tris, pH 8.0, 150 mm NaCl, 1 mm DTT as described in Ref. 27.
Protease Secretion Assay
pMMB plasmids containing the point mutations of epsE were expressed in wild type V. cholerae, or the V. cholerae epsE− strain in the presence of 10 μm isopropyl-1-thio-β-d-galactopyranoside and tested for secretion of a protease using a modified fluorescence-based assay as described in Ref. 30. Supernatants from LB-grown overnight cultures were separated from cells by two centrifugation steps and assayed for proteolytic cleavage of N-tert-butoxy-carbonyl-Gln-Ala-Arg-7-amido-4-methyl-coumarin (Sigma) as described in Refs. 30 and 31 using excitation and emission wavelengths of 385 and 440 nm, respectively. The protease activity was determined by measuring the change in fluorescence over time and comparing to a methylcoumarin standard. The amount of methylcoumarin generated per minute was normalized by A600 with error reported as S.E. of at least three separate samples assayed in triplicate.
ATPase Activity Assays
ATPase activities of purified proteins were measured using a kinetic assay containing 0.5 μm protein, 5 mm ATP, 5 mm MgCl2, and 125 μm cardiolipin in buffer A (100 mm HEPES, pH 8.5, 65 mm NaCl, 5% glycerol). Reactions were incubated at 37 °C for 0, 15, 30, 45, and 60 min for EL1 and for 0, 120, 180, 240, and 300 min for R210A, R225A, R320A, R324A, R336A, R369A, R156D, R156A, D326R, and BSA and assayed for the release of inorganic phosphate using the BIOMOL Green reagent as directed. The amount of phosphate was determined by comparing sample absorbance with those of a phosphate standard curve with error reported as S.E. of at least three separate samples assayed in duplicate.
Liposome Floatation Assay
Reaction mixtures containing 1 μm protein, 4 mm AMPPNP, 4 mm MgCl2, and 250 μm cardiolipin in buffer A were incubated for 30 min at 37 °C and then mixed with 0.8 ml of 20 mm HEPES, pH 8.5, containing 85.5% sucrose and layered on the bottom of a 5-ml centrifuge tube. 3 ml of 20 mm HEPES buffer containing 65% sucrose was layered on top followed by a 1-ml layer of 20 mm HEPES buffer containing 10% sucrose. Samples were centrifuged at 115,000 × g (35,000 rpm) for 16 h at 4 °C using a Sorvall AH650 rotor. Five 1-ml fractions were collected from the top of each tube, analyzed by SDS-PAGE using the NuPAGE system (Invitrogen), and immunoblotted against EpsE. The top two fractions (fractions 4 and 5) represent the cardiolipin-bound protein.
Cross-linking
Samples containing 125 μm cardiolipin, 5 mm ATP, 5 mm MgCl2, and 1 μm purified proteins were incubated in buffer A and subjected to the chemical cross-linker dithiobis(succinimidyl propionate) (DSP) at concentrations of 0, 10, 50, and 100 μm as described in Ref. 27.
Quantification of Immunoblots
Densitometry analysis was performed on a Typhoon TRIO (Amersham Biosciences) using ImageQuant TL software. For the liposome binding assay, the percentage of EpsE floated represents the amount of EpsE in F4+F5 divided by the total amount of EpsE in F1, F2, F4, and F5. For quantification of EpsE cross-linking, the amount of monomer remaining in the 100 μm DSP lane was divided by the amount of monomer in the 0 μm DSP lane to obtain the percentage of uncross-linked monomer.
Statistical Analysis
A Student's t test was used for all statistical analyses. Values were considered significant at p < 0.05.
Thermal Melts
The thermal stability of proteins was measured by fluorescence using Sypro Orange fluorophore as described in Ref. 32, either without additives or in the presence of 5 mm ATP and 5 mm MgCl2.
RESULTS
Modeling EpsE as an Asymmetric Hexamer
The crystal structure of EpsE without its N-terminal 90 residues was previously solved in space group P6122 where the individual subunits were found in a helical filament; however, based on its structural similarity with the T4S ATPase Hp0525, then the only known hexameric structure of the family, EpsE was modeled as a hexamer (13). In view of the fact that EpsE is more closely related to PilT than to Hp0525 (supplemental Fig. S1) (12), we have updated our working model of the functional hexameric ring of EpsE based on the structure of PaPilT.
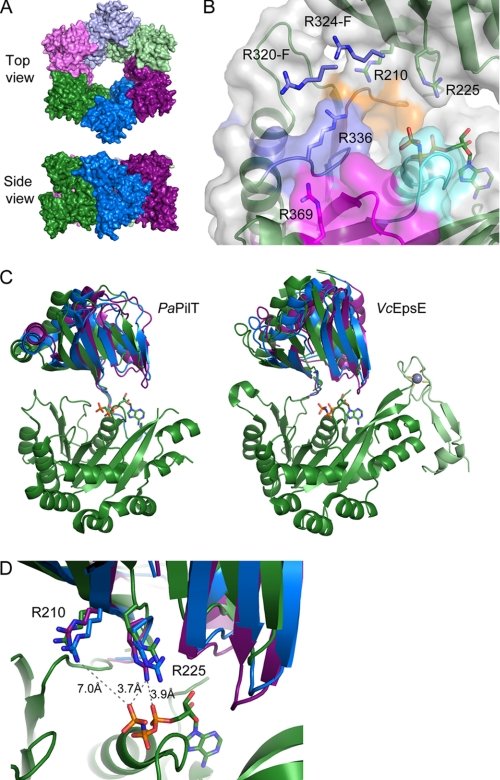
To identify residues outside the immediate nucleotide binding site that are important for ATP hydrolysis and to test the hypothesis originally put forth by Forest and colleagues (21) that there are conserved arginines that move in to complete the active site once nucleotides bind, we have constructed a C2 hexamer model of EpsE using the structure of the related PaPilT as a template (Fig. 1A). The positions of six highly conserved arginines (supplemental Fig. S1) are conserved in the new EpsE hexameric model with C2 symmetry, which emphasizes the idea of a universal function for these residues in the activity of these ATPases. A closer assessment of the two conserved NTD arginines reveals a similar movement toward the bound nucleotide as seen for both AfGspE and PaPilT, further substantiating the hypothesis that these residues may be required for EpsE to hydrolyze ATP (Fig. 1B). When examining the four invariant CTD arginine residues relative to the bound nucleotide, we see that their placement within the conserved motifs would likely allow them to contribute to the completion of the active site, if not directly participate in nucleotide hydrolysis (Fig. 1, C and D). To determine the functional role of the identified arginines in vivo and in vitro, we constructed EpsE variants with the following mutations: R210A, R225A, R320A, R324A, R336A, and R369A.
FIGURE 1.
Hexameric model of EpsE. A, EpsE modeled after the PaPilT (PDB ID: 3JVV (19)) structure, which forms hexameric rings with C2 symmetry, i.e. with three different subunit conformations (19). Subunits A, B, and C are shown in dark green, blue, and purple, respectively. Subunits D, E, and F are shown in light green, blue, and purple, respectively, to indicate that subunits A and D, B and E, and C and F have identical structures due to the C2 symmetry of the PaPilT hexamer. B, the nucleotide binding site of EpsE in the hexamer model formed by subunits A and F. The nucleotide shown is AMPPNP bound to the CTD of EpsE (13). Residues corresponding to the Walker A, Walker B, Asp, and His boxes are highlighted in cyan, blue, orange, and magenta, respectively. The side chains of functionally important arginine residues are shown in stick representation. Arg210 and Arg225 belong to the NTD, and Arg336 and Arg369 belong to the CTD of subunit A, whereas R320-F and R324-F belong to CTD′ of the neighboring subunit F. C, the EpsE hexamer model predicts NTD-CTD motions similar to PaPilT structure. Left, subunits A, B, and C of PaPilT structure superimposed using CTD only. NTDs of subunits A, B, and C are shown in green, blue, and purple, respectively; only the CTD of subunit A is shown for clarity. Right, a superposition of subunits A, B, and C of the EpsE hexamer model. A zinc binding CM domain of EpsE is highlighted in light green with the Zn2+ ion shown as a gray sphere and the side chains of coordinating cysteines shown as sticks. D, a close-up view of nucleotide binding sites in different EpsE subunits shown in C. Side chains of Arg210 and Arg225 are shown as sticks.
Replacement of Conserved Arginine Residues Disrupts Type II Secretion
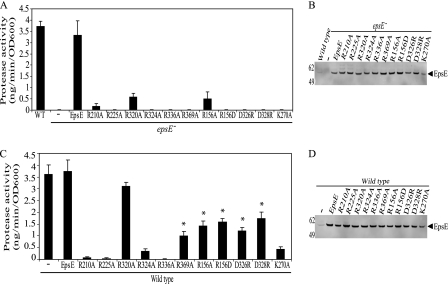
To test the importance of these arginine residues, first, the plasmid-encoded EpsE mutant proteins were analyzed for their ability to complement the secretion defect observed in an epsE− mutant strain of V. cholerae (Fig. 2A). Wild type EpsE was able to restore secretion; however, none of the six individual point mutants (including R320A) were able to complement the epsE− mutant to a statistically significant level over the vector only control, as shown in Fig. 2A. Subsequent immunoblotting analysis indicated that the mutant proteins were expressed at levels similar to that of wild type EpsE (Fig. 2B), suggesting that the mutations do not affect the stability of EpsE but rather its function.
FIGURE 2.
EpsE point mutations inhibit protease secretion in vivo. epsE− (A and B) or wild type (C and D) strains of V. cholerae containing pMMB67 vector, pMMB EpsE, or the plasmid-encoded EpsE with the following point mutations, R210A, R225A, R320A, R324A, R336A, R369A, R156A, R156D, D326R, D328R, or K270A, were grown overnight in LB + 10 μm isopropyl-1-thio-β-d-galactopyranoside. Culture supernatants and cells were separated by centrifugation. A and C, culture supernatants were analyzed for the presence of extracellular protease activity using a fluorescence-based assay with peptide-conjugated methylcoumarin as a substrate. The amount of methylcoumarin liberated was compared with a standard curve with each sample assayed at least three independent times. The results are presented as nanogram of methylcoumarin hydrolyzed per minute, normalized to the A600 of the culture ± S.E. * indicates a statistically significant difference (p < 0.05) when compared with both wild type and EpsE-K270A. OD, optical density. B and D, cell extracts were subjected to SDS-PAGE and immunoblotted with anti-EpsE. Molecular mass markers are indicated on the left, and the position of EpsE is indicated by the arrow.
To further verify that the mutations are not grossly affecting the folding of EpsE, the mutant proteins were analyzed for their ability to interact with the rest of the T2S apparatus and inhibit secretion in wild type V. cholerae. Their effect on secretion was compared with both plasmid-encoded wild type EpsE as well as the nucleotide binding site mutant, EpsE-K270A, which was previously shown to block secretion in wild type V. cholerae (6, 11). As shown in Fig. 2C, all of the mutant EpsE proteins, with the exception of EpsE-R320A, acted in a dominant negative manner when overexpressed in wild type V. cholerae. Although the R369A mutant did display trans-dominance over wild type, it did not inhibit secretion as efficiently as the K270A mutant (p < 0.05). The dominant phenotype could be due to competition between wild type and mutant EpsE for binding to other T2S components such as EpsL and/or due to the formation of mixed oligomers containing wild type and mutant EpsE.
EpsE Point Mutations Abolish ATPase Activity in Vitro
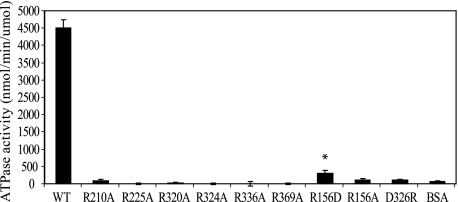
Previously we have demonstrated that purified EpsE, when in the presence of EpsL(1–253)His6 and acidic phospholipids, such as cardiolipin, assembles into larger oligomers and displays ATPase activity (27). Therefore, we next tested whether the above chosen residues are required for hydrolysis of ATP in the presence of EpsL(1–253)His6 and cardiolipin. The point mutations R210A, R225A, R320A, R324A, R336A, and R369A were individually introduced into the previously created EpsE(2–503)-EpsL(1–253)His6 construct, and the mutant proteins were purified by metal affinity chromatography and gel filtration as described (7, 29). The finding that all of these mutants (including EpsE-R320A) could be co-purified with EpsL(1–253)His6 by these methods indicates that the interaction of EpsE and EpsL was not disrupted as a result of these mutations. As Fig. 3 demonstrates, each individual point mutation abrogated the ATPase activity of EpsE in the presence of cardiolipin more than 50-fold when compared with wild type protein. The rates of ATP hydrolysis for the mutant proteins were not statistically different from that of the BSA control, indicating that all six arginine mutants are inactive.
FIGURE 3.
EpsE point mutations block cardiolipin-stimulated ATPase activity in vitro. Cardiolipin-stimulated ATPase activity was measured kinetically for wild type EpsE-EpsL(1–253)His6 over a period of 60 min and the individual EpsE mutants with the following mutations, R210A, R225A, R320A, R324A, R336A, R369A, R156D, R156A, and D326R, or BSA over a period of 300 min in reactions containing Mg-ATP and cardiolipin. Each protein was assayed at least three separate times, and the means ± S.E. are presented. * indicates a statistically significant difference (p < 0.05) when compared with BSA.
EpsE Mutants Retain Nucleotide Binding Capabilities
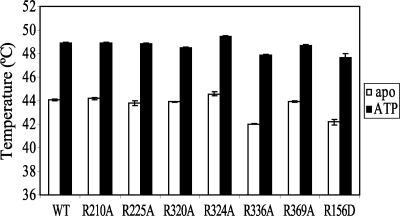
To determine whether the reduced ability to hydrolyze ATP for the mutant EpsE proteins was due to defects in ATP binding, we performed thermal melt assays. We determined the temperature at which wild type EpsE and the individual EpsE point mutants R210A, R225A, R320A, R324A, R336A, and R369A in complex with EpsL(1–253)His6 melted in the presence or absence of Mg-ATP as a measure of nucleotide binding. As seen in Fig. 4, each protein showed a significant increase in melting temperature in the presence of Mg-ATP over their respective apo forms, indicating that nucleotide binding increases the stability of each EpsE variant. Furthermore, the wild type and mutant proteins responded with a very similar thermal melt shift to the presence of nucleotide, demonstrating that these proteins have comparable nucleotide binding capabilities. Another important item to note is the fact that the EpsE mutant proteins melted within 2 °C of the average melting temperature of wild type protein in both the presence and the absence of ATP, suggesting comparable overall stability for these proteins. Taken together with the data from the ATPase assay, the mutations do not appear have a negative effect on ATP binding, but rather on the ability of EpsE to hydrolyze ATP.
FIGURE 4.
EpsE point mutants can bind ATP. Average thermal melt temperatures for wild type protein, and EpsE variants with the following mutations, R210A, R225A, R320A, R324A, R336A, R369A, and R156D, in the absence or presence of ATP are shown. Melting temperatures for each protein were assayed at least eight times on two separate days, and the means ± S.E. are presented.
Interaction with Cardiolipin Is Unaltered in EpsE Mutants
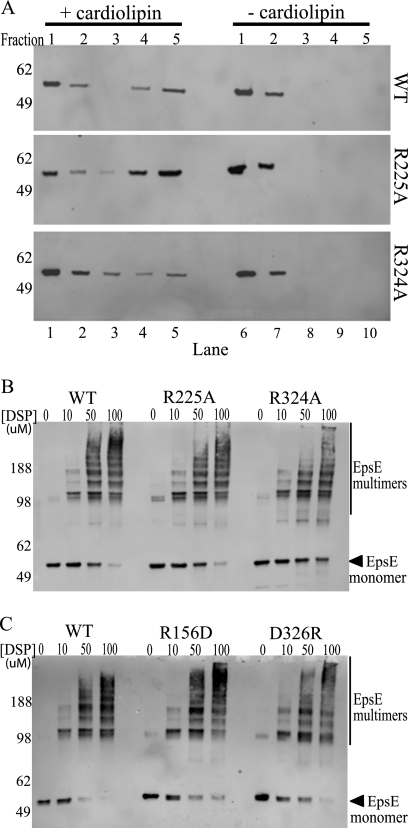
To determine whether the loss of activity of the point mutants is solely due to a defect in forming the complete active site or whether there is another factor indirectly contributing to the reduced ATP hydrolysis such as altered cardiolipin binding, we incubated the purified EpsE-EpsL(1–253)His6 variants at 37 °C in the presence of Mg-AMPPNP with or without cardiolipin. These liposome binding experiments sought to directly measure whether the mutant proteins retained their ability to interact with acidic phospholipids comparable with that of wild type as failure to interact with cardiolipin could account for the loss of ATPase activity of the point mutants. After incubation, the samples were added to an 85.5% sucrose solution in the bottom of a centrifuge tube, and a sucrose gradient was layered on top. After centrifugation, samples were taken from the top of the tube with the highest fractions, representing those that contained the phospholipids and phospholipid-bound protein. Each fraction was analyzed by SDS-PAGE and immunoblotting. All EpsE mutant proteins in complex with EpsL(1–253)His6 were able to float to the highest two fractions in the presence of cardiolipin but were retained in the lowest two fractions in the absence of cardiolipin, similar to that of wild type EpsE-EpsL(1–253)His6 (Fig. 5A and data not shown). When quantified over at least three individual experiments, each point mutant had a comparable percentage of protein floating up to the highest fractions when compared with that of the wild type protein (Table 1). Although the CTD′ arginines 320 and 324 cluster toward the lower end, they were not statistically different from wild type (Table 1). Because EpsL was previously determined to be the main protein involved with the interaction with cardiolipin (27), it was not surprising that mutations in EpsE far away from the EpsE-EpsL interface are still able to bind phospholipids in the presence of EpsL, and this confirms that the reduction in ATPase activity is not due to loss of cardiolipin binding.
FIGURE 5.
EpsE point mutants interact with cardiolipin and can form oligomers. A, protein-cardiolipin interactions were compared using a liposome floatation assay. Reaction mixtures containing wild type, R225A, or R324A protein ± cardiolipin were subjected to membrane floatation. After centrifugation, five 1-ml fractions were collected and analyzed by SDS-PAGE and immunoblotting with anti-EpsE. Molecular mass markers are indicated on the left, fraction numbers are listed across the top, and lane numbers are listed across the bottom. Lanes 4 and 5 are the top fractions in the cardiolipin (+) samples and represent liposome-bound protein, whereas lanes 9 and 10 represent the highest fractions of the cardiolipin (−) samples. B, reactions containing either wild type EpsE-EpsL(1–253)His6 or the point mutants EpsE-R225A or EpsE-R324A protein were cross-linked in the presence of Mg-ATP and cardiolipin using increasing concentrations of DSP (0, 10, 50, or 100 μm). Samples were analyzed by SDS-PAGE and immunoblotted with anti-EpsE. Molecular mass markers are denoted on the left, and the position of monomeric EpsE is indicated with an arrow. C, reactions containing either wild type EpsE-EpsL(1–253)His6 or the point mutant EpsE-R156D or EpsE-D326R proteins were cross-linked in the presence of Mg-ATP and cardiolipin as described in B. Molecular mass markers are denoted on the left, and the position of monomeric EpsE is indicated with an arrow.
TABLE 1.
Quantification of cross-linking and liposome floatation assays
Quantification of at least three independent assays was performed for each protein using the ImageQuantTL software. For cross-linking, the amount of monomer remaining in the highest [DSP] lane was compared with that in the no cross-linker lane (±S.E.). Protein floatation (±S.E.) was determined by dividing the amount of protein in F4+F5 to the total protein from F1, F2, F4, and F5. For both assays, mutant proteins were not statistically different from WT. CL, cardiolipin; nt, not tested.
| Protein | % of uncross-linked monomer | p value | % of total protein associated with CL | p value |
|---|---|---|---|---|
| WT | 25 (8.4) | 63 (18.3) | ||
| R210A | 31 (14) | 0.71 | 77 (5.2) | 0.30 |
| R225A | 35 (17.7) | 0.58 | 80 (6.3) | 0.24 |
| R320A | 25 (10) | 0.99 | 38 (13.5) | 0.21 |
| R324A | 23 (7.5) | 0.87 | 43 (10.7) | 0.25 |
| R336A | 26 (11.2) | 0.44 | 66 (17.9) | 0.89 |
| R369A | 41 (9.7) | 0.25 | 62 (6.3) | 0.91 |
| R156D | 47 (15.5) | 0.21 | 53 (11.4) | 0.54 |
| D326R | 17 (8.6) | 0.60 | nt |
EpsE Mutants Can Still Form Higher Molecular Weight Oligomers
Finally, to determine whether the mutant proteins are still capable of forming higher molecular weight complexes in the presence of EpsL(1–253)His6, ATP, and cardiolipin, we subjected the proteins to chemical cross-linking analysis using DSP (27). The formation of higher molecular weight species was unchanged for the mutant proteins when compared with wild type in the presence of cardiolipin when analyzed by SDS-PAGE and immunoblotting (Fig. 5B and data not shown). The percentage of monomers remaining uncross-linked was calculated for the highest DSP concentration as shown in Table 1. None of the EpsE point mutants have a statistically different amount of uncross-linked monomers remaining when compared with wild type protein, indicating that the point mutations introduced into EpsE do not inhibit its ability to form higher molecular weight species. Taken together with all the above mentioned data, it is likely that the substitutions R210A, R225A, R320A, R324A, R336A, and R369A specifically interfere with the ability of EpsE to hydrolyze ATP.
Mutations in the EpsE Subunit Interface Have a Similar Phenotype to the Arginine Mutations
We have thus far demonstrated that substitutions of conserved arginine residues abolish ATPase activity and suggest that these mutations disrupt the formation of the complete active site, resulting in failure to hydrolyze the nucleotide and inactivation of the entire complex. Further, we have shown that the defect is in the hydrolysis step and not due to inefficient nucleotide binding and hypothesize that oligomerization is a key step in obtaining full activity. To strengthen the argument that oligomerization is necessary to form the complete active site needed for efficient hydrolysis, we introduced point mutations within the subunit-subunit interface. We rationalized based on the EpsE hexamer model that a single mutation within this interface would be unlikely to fully disrupt oligomerization but may result in altered ability to communicate nucleotide binding in one subunit, which could further inhibit the movement of the conserved residues necessary to complete the active site. We chose three residues: Arg156 in the NTD as well as Asp326 and Asp328 in the CTD. These three residues form two salt bridges at the edge of the NTD-CTD′ interface of the EpsE filament and also in the C2 hexamer model and are likely important for properly aligning the subunits with respect to each other in the hexameric ring. The salt bridges are also likely preserved between the active, closed form and the neighboring open form as the distance between these residues does not vary between the different hexamers. R156A, R156D, D326R, and D328R mutations were introduced into EpsE, and the mutant proteins were first tested for complementation and dominance in vivo using the protease secretion assay described earlier. As Fig. 2, A and B, demonstrate, all four of the interface mutants failed to complement the epsE-deficient strain when assayed for protease secretion, yet they were stably overexpressed as verified by immunoblotting. The interface mutants also acted in a dominant negative manner when overexpressed in the wild type strain, indicating that the mutants likewise retain their ability to interact with the T2S complex (Fig. 2, C and D). EpsE with the R156A, R156D, D326R, and D328R substitutions showed the same intermediate effect on secretion as the previously discussed R369A, meaning that they were dominant when compared with the plasmid-encoded wild type EpsE (p < 0.05); however, they did not inhibit secretion as efficiently as the K270A mutant (p < 0.05). This may indicate a trend toward a less severe dominance phenotype for mutations in the interface than those directly participating in the active site.
Next we purified EpsE-R156D and EpsE-D326R in complex with EpsL(1–253)His6 and tested them in the ATPase and cross-linking experiments described above. As predicted, both interface mutants were still able to cross-link into higher molecular weight complexes statistically comparable with that of wild type EpsE-EpsL(1–253)His6 (Fig. 5C and Table 1). Neither mutant, however, displayed high levels of ATP hydrolysis in the presence of cardiolipin (Fig. 3). Interestingly, the only point mutant that retained any ATPase activity greater than BSA was EpsE-R156D; however, this was reduced by at least 15-fold when compared with wild type protein and is insufficient to support T2S. Further, the EpsE-R156D mutant was tested in both the liposome binding assay and the thermal melt assay, which showed that it interacts with cardiolipin comparable with that of wild type protein (data not shown and Table 1) as well as that it has a comparable melting temperature with wild type EpsE-EpsL(1–253)His6 in the absence of nucleotide (Fig. 4). Likewise, it also showed an increase in melting temperature over its apo form in the presence of Mg-ATP, which indicates that it is still able to bind nucleotide similar to that of the other proteins tested (Fig. 4). Overall, these data support the hypothesis that a complete active site requires residues from multiple domains and subunits and that the movements of the subunits into their proper position in response to nucleotide binding and hydrolysis require oligomerization of the protein.
DISCUSSION
EpsE and other secretion ATPases participate in diverse processes, and as a consequence, they share low overall sequence homology. What stands out, however, are a few areas of high conservation. Forest and colleagues (19) have mapped the conservation of residues among 27 secretion ATPases and determined that the residues within the active site and at the NTD-CTD′ subunit-subunit interface are the most conserved residues across the entire protein (19). We have shown in this study that mutations in either area of conservation are detrimental to the ability of EpsE to hydrolyze ATP, thus further supporting the functional importance of these areas in the Type II/IV ATPases. Through in vivo methods, we have demonstrated that replacement of each one of six conserved arginines in EpsE inhibits the function of the T2S complex. Additionally, in vitro experiments have demonstrated that alanine replacements of these arginines abolish the ability of EpsE to efficiently hydrolyze ATP but not to: (i) bind nucleotide; (ii) form higher molecular weight oligomers; or (iii) interact with cardiolipin. We have further gone on to show that even with an intact active site, EpsE is nonfunctional if the subunit-subunit interface is altered. The finding that replacement of Arg156 and Asp326 in the subunit interface has an adverse effect on ATPase activity without preventing oligomerization suggests that altering the interactions in this conserved interface may lead to oligomers that are unable to sense and/or move appropriately when nucleotide binds.
Although the arginine residues from the NTD (Arg210, Arg225), CTD (Arg336, Arg369), and CTD′ (Arg320, Arg324) were all found to be required for ATP hydrolysis, not all of them are within hydrogen-bond distance to the nucleotide in our model (Fig. 1B); therefore, the contribution of each arginine to the enzymatic activity of EpsE may differ. Taking structural information into account, the potential roles of the individual arginines in the ATPase activity of EpsE are discussed below.
We have demonstrated in this study that both Arg225 and Arg210 are necessary for function both in vivo and in vitro. These NTD arginines are both highly conserved and likely important for closure of the protein around the bound nucleotide. Modeling of the EpsE hexamer based on the PaPilT structure brings these arginines in the vicinity of the bound nucleotide. As seen in Fig. 1D, the Arg225 subunit C from EpsE moves sufficiently close to the γ-phosphate of the bound nucleotide to directly participate in ATP coordination or hydrolysis. The EpsE-Arg225 equivalent in Xanthomonas campestris XpsE (XpsE-Arg286) has been pinpointed as being an important residue for sensing the bound nucleotide and coordinating the movement of the NTD relative to the CTD (33). The EpsE-Arg225 and Arg210 counterparts in both AfGspE and PaPilT have been shown to come close to the bound nucleotide suggesting a conserved function (20, 33). Interestingly, the equivalent arginines in PaPilT structure show marked conformational differences in the three observed states (19). The guanidinium moiety of PaPilT-Arg97 (the EpsE-Arg225 equivalent) stays at a quite similar distance of 4.3–5.2 Å from the α- and β-phosphate groups in all three states. In contrast, the guanidinium group of PaPilT-Arg82 (EpsE-Arg210) moves dramatically between 3.2 and 9.0 Å distance from the γ-phosphate. It may be argued that Arg225 serves as a static sensor of ATP whereas Arg210 plays a more active role by engaging the γ-phosphate directly and removing it from the active site after hydrolysis. In our model, Arg210 does not come close to the γ-phosphate to coordinate or assist directly in the hydrolysis of ATP (Fig. 1D). This may be attributed to the limitations of our rigid body model. Indeed a change of the arginine side chain conformation could bring the guanidinium group of EpsE-Arg210 in subunit B close to the γ-phosphate. Also, Arg210 is within hydrogen-bond distance to the carboxylate group of Glu296 responsible for Mg2+ ion coordination. Therefore, an alternative or additional function of Arg210 might be to stabilize Glu296, thereby supporting its coordination of Mg2+ ion. Regardless of the immediate roles Arg210 and Arg225 play in ATP hydrolysis, the observations that these two residues are far from the nucleotide in the initial EpsE structure (13) but come near the nucleotide bound in our hexameric EpsE model (Fig. 1), along with our protease secretion and ATPase data (Figs. 2 and 3), suggest that EpsE needs to be hexameric to perform its function.
The arginines at positions 336 and 369 in the CTD of EpsE are found in the Walker B and His box motifs, respectively, and our data demonstrate that both of these residues are required for function of EpsE. In all three subunit conformations of the EpsE hexamer model, EpsE-Arg336 forms salt bridges with Asp293 and with the catalytic Glu334, a residue essential for ATPase activity (34). Therefore, the coordination of EpsE-Arg336 by both Asp293 and Glu334 is likely crucial for correct positioning of the latter for catalysis. A mutation in either Arg336 or Asp293 would have negative consequences on function as seen both in this study and in previous ones looking at the effects of mutating the EpsE-Asp293 equivalent in PaPilT (34) and PulE (17, 34). Interestingly, the equivalent salt bridges in the PaPilT structure are observed only in active subunit B. In subunits A and C, a simultaneous shift of a loop containing residues 203–206 along with a change in side chain conformation of PaPilT-Glu208 (EpsE-Glu334) brings the carboxylate in contact with His box residue His229 instead. This leads to movement of PaPilT-Arg206 (EpsE-Arg336) toward the interface of neighboring CTD′.
EpsE-Arg369 of the CTD does not appear to make significant contacts with bound nucleotide in our hexamer model, nor in the EpsE crystal structure (13). Instead, the guanidinium group of EpsE-Arg369 forms a hydrogen bond with the main chain carbonyl of EpsE-Arg336 and, therefore, could be important for stabilizing or sensing the conformation of the loop described above. The movement of that loop could be an important element of synchronized structural rearrangements during the ATP hydrolysis cycle. The intrasubunit EpsE-Arg336 interaction with Arg369 has not been observed in the PaPilT structure where the EpsE-Arg369 equivalent (PaPilT-Arg239) instead makes an intersubunit contact with PaPilT-Glu219′. Intriguingly, this salt bridge between PaPilT-Arg239 and PaPilT-Glu219′ is maintained in all three subunits of the PaPilT C2 hexamer despite the changes in side chain conformations and relative orientation of CTD and CTD′; however, PaPilT-Glu219 is not conserved in related ATPases (supplemental Fig. S1). Based on the EpsE hexamer model, residue EpsE-Glu376 from the neighboring CTD′ is a plausible candidate for contacting EpsE-Arg369. This fits with our in vivo secretion data showing that the EpsE-R369A dominant phenotype shares characteristics with the interface mutants, indicating that the primary role of this residue may be to properly align the subunits rather than to directly participate in ATP hydrolysis. Further experiments are needed to establish the precise functional role of Arg369.
Two residues from the CTD′, EpsE-Arg320 and Arg324, both seem to participate in completing the active site of the neighboring subunit and perhaps support the efficient removal of the γ-phosphate (Fig. 1B), although of the two, only Arg324 is strictly conserved. Arg324 of CTD′ forms a salt bridge with Asp195 in the NTD of EpsE; however, the importance of this interaction for the stability of the NTD-CTD′ interface has not been investigated. Although we have demonstrated that Arg320 is necessary to support secretion through the T2S apparatus of V. cholerae, a mutation in the EpsE-Arg320 equivalent in PaPilT showed only a minor effect on twitching motility (21), further substantiating the idea that the importance of this residue may vary across species.
Although both EpsE-R320A and EpsE-R324A are defective in ATP hydrolysis, and thus secretion, only EpsE-R324A has a dominant negative effect on secretion when expressed in a wild type V. cholerae strain, suggesting that the defect in EpsE-R320A may be compensated for in vivo by the wild type EpsE, possibly through the formation of mixed oligomers. In the presence of wild type EpsE, the mutant protein EpsE-R320A may adopt a conformation amenable to ATP hydrolysis. Alternatively, it is possible that not every subunit in the hexamer needs to be active to support ATP hydrolysis and secretion; therefore an EpsE hexamer could function even when it consists of a mixture of wild type EpsE and mutant EpsE-R320A. Interestingly, work investigating the formation of active, mixed hexamers of the AAA+ ATPase ClpX, generated by covalently linking wild type and inactive subunits, concluded that six wild type subunits are not necessary for the ATPase activity of ClpX (35). The observation that the wild type EpsE alleviates the defective EpsE-R320A mutant in vivo further confirms the idea that EpsE is active as an oligomer.
Mutations in the subunit interface were also detrimental to the function of EpsE; however, based on our data, it is possible that somewhat of a hierarchy exists in the ability of EpsE to compensate for mutations either within the active site or within the subunit interface, with mutations of residues directly participating in ATP hydrolysis being more severe. This idea is supported by the results from the protease secretion assays in which the trans-dominance phenotype was not nearly as severe for the interface mutants as seen for mutants within the active site, indicating that these mutants may be more easily compensated for by the wild type protein. Further, the only mutant protein to retain any appreciable ATPase activity in vitro was the interface mutant EpsE-R156D, and although it was not sufficiently active to support secretion on its own in vivo, this mutant was able to hydrolyze ATP at a significant level above the BSA control (p < 0.05).
Based on our findings on the requirement of the conserved arginines for ATP hydrolysis and secretion and on observations published earlier on conformational differences between nucleotide-bound and nucleotide-free Hp0525, PilT, and AfGspE (19–21, 23), we speculate that the binding of nucleotide to one EpsE subunit likely causes drastic conformational changes that can be relayed to not only the adjoining subunits but also to the larger T2S complex through the interaction of EpsE with the inner membrane protein EpsL. The interactions of EpsL with the major pseudopilin EpsG (36) may in turn be essential in the formation of the pseudopilus and/or drive the transport of proteins through the EpsD pore in the outer membrane. In addition, understanding how EpsE powers the transport through the T2S system may provide insight into the function of Type II/IV secretion ATPases as a whole due to the high level of structural homology among members of this protein family.
Acknowledgments
We acknowledge Mark Robien and Jan Abendroth for earlier hexameric models of EpsE.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1AI049294 (to M. S.) and RO1AI34501 (to W. G. J. H.) from the NIAID.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- T2S
- Type II secretion
- T4S
- Type IV secretion
- CTD
- carboxyl-terminal domain
- NTD
- amino-terminal domain
- DSP
- dithiobis(succinimidyl propionate)
- AMPPCP
- adenosine 5′-(β,γ-methylene)triphosphate
- AMPPNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1. Johnson T. L., Abendroth J., Hol W. G., Sandkvist M. (2006) FEMS Microbiol. Lett. 255, 175–186 [DOI] [PubMed] [Google Scholar]
- 2. Cianciotto N. P. (2005) Trends Microbiol. 13, 581–588 [DOI] [PubMed] [Google Scholar]
- 3. Filloux A. (2004) Biochim. Biophys. Acta 1694, 163–179 [DOI] [PubMed] [Google Scholar]
- 4. Hobbs M., Mattick J. S. (1993) Mol. Microbiol. 10, 233–243 [DOI] [PubMed] [Google Scholar]
- 5. Sandkvist M. (2001) Mol. Microbiol. 40, 271–283 [DOI] [PubMed] [Google Scholar]
- 6. Camberg J. L., Sandkvist M. (2005) J. Bacteriol. 187, 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abendroth J., Murphy P., Sandkvist M., Bagdasarian M., Hol W. G. (2005) J. Mol. Biol. 348, 845–855 [DOI] [PubMed] [Google Scholar]
- 8. Py B., Loiseau L., Barras F. (2001) EMBO Rep. 2, 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lybarger S. R., Johnson T. L., Gray M. D., Sikora A. E., Sandkvist M. (2009) J. Bacteriol. 191, 3149–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robert V., Filloux A., Michel G. P. (2005) FEMS Microbiol. Lett. 252, 43–50 [DOI] [PubMed] [Google Scholar]
- 11. Sandkvist M., Bagdasarian M., Howard S. P., DiRita V. J. (1995) EMBO J. 14, 1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Planet P. J., Kachlany S. C., DeSalle R., Figurski D. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2503–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robien M. A., Krumm B. E., Sandkvist M., Hol W. G. (2003) J. Mol. Biol. 333, 657–674 [DOI] [PubMed] [Google Scholar]
- 14. Peabody C. R., Chung Y. J., Yen M. R., Vidal-Ingigliardi D., Pugsley A. P., Saier M. H., Jr. (2003) Microbiology 149, 3051–3072 [DOI] [PubMed] [Google Scholar]
- 15. Craig L., Pique M. E., Tainer J. A. (2004) Nat. Rev. Microbiol. 2, 363–378 [DOI] [PubMed] [Google Scholar]
- 16. Thomsen N. D., Berger J. M. (2008) Mol. Microbiol. 69, 1071–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Possot O., Pugsley A. P. (1994) Mol. Microbiol. 12, 287–299 [DOI] [PubMed] [Google Scholar]
- 18. Hare S., Fischer W., Williams R., Terradot L., Bayliss R., Haas R., Waksman G. (2007) EMBO J. 26, 4926–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Misic A. M., Satyshur K. A., Forest K. T. (2010) J. Mol. Biol. 400, 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamagata A., Tainer J. A. (2007) EMBO J. 26, 878–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Satyshur K. A., Worzalla G. A., Meyer L. S., Heiniger E. K., Aukema K. G., Misic A. M., Forest K. T. (2007) Structure 15, 363–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hare S., Bayliss R., Baron C., Waksman G. (2006) J. Mol. Biol. 360, 56–66 [DOI] [PubMed] [Google Scholar]
- 23. Savvides S. N., Yeo H. J., Beck M. R., Blaesing F., Lurz R., Lanka E., Buhrdorf R., Fischer W., Haas R., Waksman G. (2003) EMBO J. 22, 1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yeo H. J., Savvides S. N., Herr A. B., Lanka E., Waksman G. (2000) Mol. Cell 6, 1461–1472 [DOI] [PubMed] [Google Scholar]
- 25. Story R. M., Weber I. T., Steitz T. A. (1992) Nature 355, 318–325 [DOI] [PubMed] [Google Scholar]
- 26. Sawaya M. R., Guo S., Tabor S., Richardson C. C., Ellenberger T. (1999) Cell 99, 167–177 [DOI] [PubMed] [Google Scholar]
- 27. Camberg J. L., Johnson T. L., Patrick M., Abendroth J., Hol W. G., Sandkvist M. (2007) EMBO J. 26, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sandkvist M., Morales V., Bagdasarian M. (1993) Gene 123, 81–86 [DOI] [PubMed] [Google Scholar]
- 29. Abendroth J., Bagdasarian M., Sandkvist M., Hol W. G. (2004) J. Mol. Biol. 344, 619–633 [DOI] [PubMed] [Google Scholar]
- 30. Sandkvist M., Michel L. O., Hough L. P., Morales V. M., Bagdasarian M., Koomey M., DiRita V. J., Bagdasarian M. (1997) J. Bacteriol. 179, 6994–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson T. L., Scott M. E., Sandkvist M. (2007) J. Bacteriol. 189, 9082–9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lo M. C., Aulabaugh A., Jin G., Cowling R., Bard J., Malamas M., Ellestad G. (2004) Anal. Biochem. 332, 153–159 [DOI] [PubMed] [Google Scholar]
- 33. Shiue S. J., Chien I. L., Chan N. L., Leu W. M., Hu N. T. (2007) Mol. Microbiol. 65, 401–412 [DOI] [PubMed] [Google Scholar]
- 34. Chiang P., Sampaleanu L. M., Ayers M., Pahuta M., Howell P. L., Burrows L. L. (2008) Microbiology 154, 114–126 [DOI] [PubMed] [Google Scholar]
- 35. Martin A., Baker T. A., Sauer R. T. (2005) Nature 437, 1115–1120 [DOI] [PubMed] [Google Scholar]
- 36. Gray M. D., Bagdasarian M., Hol W. G., Sandkvist M. (2011) Mol. Microbiol. 79, 786–798 [DOI] [PMC free article] [PubMed] [Google Scholar]