Abstract
Four minimal (119 - 145 residue) active site fragments of Escherichia coli Class II histidyl-tRNA synthetase were constructed, expressed as maltose-binding protein fusions, and assayed for histidine activation as fusion proteins and after TEV cleavage, using the 32PPi exchange assay. All contain conserved Motifs 1 and 2. Two contain an N-terminal extension of Motif 1 and two contain Motif 3. Five experimental results argue strongly for the authenticity of the observed catalytic activities: (i) active site titration experiments showing high (∼0.1–0.55) fractions of active molecules, (ii) release of cryptic activity by TEV cleavage of the fusion proteins, (iii) reduced activity associated with an active site mutation, (iv) quantitative attribution of increased catalytic activity to the intrinsic effects of Motif 3, the N-terminal extension and their synergistic effect, and (v) significantly altered Km values for both ATP and histidine substrates. It is therefore plausible that neither the insertion domain nor Motif 3 were essential for catalytic activity in the earliest Class II aminoacyl-tRNA synthetases. The mean rate enhancement of all four cleaved constructs is ∼109 times that of the estimated uncatalyzed rate. As observed for the tryptophanyl-tRNA synthetase (TrpRS) Urzyme, these fragments bind ATP tightly but have reduced affinity for cognate amino acids. These fragments thus likely represent Urzymes (Ur = primitive, original, earliest + enzyme) comparable in size and catalytic activity and coded by sequences proposed to be antisense to that coding the previously described Class I TrpRS Urzyme. Their catalytic activities provide metrics for experimental recapitulation of very early evolutionary events.
Keywords: Aminoacyl tRNA Synthesis, Aminoacyl tRNA Synthetase, Enzyme Catalysis, Enzyme Turnover, Evolution, Histidine, Pre-steady-state Kinetics, Protein Motifs, Active Site Titration
Introduction
The complexity of contemporary enzymes makes it almost certain that ancestral forms were simpler polypeptides. These primordial ancestors have, however, long been extinct. It has been impossible to recapitulate, even approximately, how they functioned without modules that enhance the fitness of contemporary enzymes.
Experimental study of ancestral enzymes began with the inference from multiple sequence alignments of the amino acid sequence for a ribonuclease belonging to an ancestral ruminant (1). Construction of this ancestral gene allowed expression and experimental characterization of the corresponding protein. More recently, this approach, known as “ancestral gene resurrection,” has led to several novel insights into how point mutations in ligand-binding domains of nuclear hormone receptors resulted in irreversible adaptation to entirely new ligands (2–4).
Despite such successes, we still know very little about how the earliest protein catalysts evolved to the point where ancestral gene resurrection from extant multiple sequence alignments (5–8) becomes possible. There are two requisites for addressing this problem. First, model peptides expected to mimic primordial enzymes must be constructed based on observations other than sequence phylogenies. Second, models must have measurable activity, to provide an experimental metric (9) for assessing recapitulation of putative evolutionary events.
We chose to study the aminoacyl-tRNA synthetases (aaRS)2 for a variety of important reasons; (i) these enzymes accelerate amino acid activation which, in the absence of catalysts, is the slowest step along the pathway of protein synthesis3; perhaps 107 times slower than peptide bond formation uncatalyzed (11). For this reason aaRS were therefore likely among the first primitive polypeptide catalysts to evolve; (ii) the twenty canonical aaRS divide evenly into two quite distinct superfamilies whose structures are apparently unrelated; (iii) there is precedent for the catalytic activity of fragments derived from both Class I (12) and Class II (13, 14) aminoacyl-tRNA synthetases. Histidyl tRNA synthetase (HisRS) presents a particularly good system because the steady-state kinetic properties of a 320-residue N-terminal fragment containing the intact catalytic domain have been extensively characterized (14).
We used three-dimensional structural superposition, rather than multiple sequence alignments to identify a peptide scaffold that positions the active site residues for a bacterial Class I tryptophanyl-tRNA synthetase, TrpRS. That 130-residue protein accelerates amino acid activation by ∼109, relative to the estimated uncatalyzed rate of 8.3 × 10−9/mol/s (15). Moreover, steady-state kinetic parameters showed that it binds ATP about as well as the full-length, native enzyme but has ∼2000-fold weaker affinity for tryptophan (16). Because it was catalytically active and appeared to approach a limiting size necessary to position the conserved catalytic residues, we called that construct an Urzyme. On the basis of that work, we hoped to be able to document similar catalytic activity for Class II Urzymes.
We report here the construction and catalytic activity of a series of constructs created from the contemporary Class II histidyl-tRNA synthetase, HisRS. Class II aaRS are defined by three highly conserved signature peptides, referred to as Motifs 1–3. Motif 2 comprises the ATP binding site and provides a partial platform for amino acid binding.
The four constructs described here were extracted from the gene for contemporary Escherichia coli HisRS. No use was made of protein design to compensate for the deletion of ∼300 amino acids. Despite the absence of a systematic approach to redesign a stable fold, active site titration experiments demonstrate that significant fractions of the molecules contribute to the observed activities. We then show that the activity is cryptic in maltose-binding protein fusions, that it is significantly enhanced upon TEV cleavage and that for both fusion proteins and cleaved products, a six-residue N-terminal extension preceding the Motif 1 helix and a 20-residue C-terminal extension containing Motif 3 both contribute consistently to the overall rate acceleration. An active site mutation alters the observed activity of one construct. Finally, Michaelis parameters for ATP and histidine for one construct differ markedly from those of native HisRS.
Additional motivation for these studies comes from our desire to test the unusual hypothesis of Rodin and Ohno (17). According to their hypothesis, the two classes of aaRS descended from a single ancestral gene, one strand of which coded for an ancestral Class I, whereas the other strand coded for the ancestral Class II synthetase. Catalytic activity of a Class II construct composed of the amino acids hypothesized to lie opposite the ancestral Class I enzyme would provide substantial, complementary new evidence for sense/antisense ancestry.
Together with our previous work on the TrpRS Urzyme, these results document that comparable cognate amino acid activation activities arise from both Class I and Class II Urzymes of similar length (∼120–130 residues) in which the conserved signature peptides of the two classes can be arranged approximately antiparallel and opposite one another in keeping with the Rodin-Ohno hypothesis. Moreover, their steady-state kinetic parameters appear appropriate to activate, relatively nonspecifically, the two types of amino acids required for binary encoding of the earliest ancestral globular proteins (18–20).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmid Construction
Restriction enzymes and other molecular biology reagents were obtained from New England Biolabs. Genes for HisRS-1 (residues 16–134) and HisRS-2 (residues 10–134), were amplified by PCR from Ncat_HisRS (PQE-30; (14)). Genes for HisRS-3/4 were extended from HisRS-1/2 sequentially by PCR until they contained coding sequences for residues 301–320, which comprise Motif 3. This procedure formed a peptide bond between residues Gly134 and Pro301 in HisRS 3/4. An EcoRI restriction site, TEV protease cleavage site, and FLAG tag were included in the primers for the 5′ end, and a HindIII restriction site was added at the 3′ end of the fragments. Following the double digestion with EcoRI and HindIII, the HisRS-1–4 constructs were cloned to E. coli expression plasmid vector pMAL-c2X (New England Biolabs). All constructs were confirmed by DNA sequencing. Bacterial strain BL21(DE3)pLysS (Novagen) was used for the expression of all HisRS constructs.
Protein Expression and Purification
The MBP-HisRS fusion proteins were expressed and purified using amylose resin according to the manufacturer's instructions (New England Biolabs). Overnight cultures of E. coli transformed with recombinant plasmid (pMal-c2X/HisRS1–4 Urgenes) were diluted in fresh LB medium containing 100 μg/ml of ampicillin to A600 = 0.5. Then, IPTG was added to a final concentration of 0.3 mm to the culture broth. After 3 h of induction, the cells were harvested by centrifugation at 12,000 × g at 4 °C for 20 min. Cells were resuspended in 1/10 volume of chilled column buffer (200 mm NaCl and 1 mm EDTA in 20 mm Tris-HCl buffer, pH 7.4) and disrupted by sonication on ice and centrifuged at 12,000 × g at 4 °C for 30 min. The crude extract was incubated with amylose resin overnight at 4 °C with shaking, and then poured into a Kontes Flex-Column. The column was washed with 12 volumes of column buffer. Fusion proteins were eluted with the same buffer containing 10 mm maltose, and the eluted fractions were pooled.
TEV Protease Cleavage and Separation of HisRS Constructs
The purified MBP-HisRS fusion proteins were concentrated to at least 1 mg/ml in an Amicon Centricon (Millipore Corp. Bedford, MA). To cleave the fusion proteins, TEV protease was added to the MBP-HisRS fusion proteins (1:30) and incubated at 4 °C for 24 h. The cleavage mixtures were stored at 4 °C for the activity assay and further purification. HisRS constructs could be purified from cleavage mixtures using anti-FLAG M2 affinity gel according to the manufacturer's protocols (Sigma).
Active Site Titration
The concentrations of active sites, compared with the total concentrations of HisRS fusion proteins were determined using the active site titration procedure as described (21) using 10 mm γ-32P-labeled ATP in excess over the total estimated enzyme concentration. Reactions were quenched with SDS at 14 time intervals between 1 and 50 min, and aliquots were examined by thin-layer chromatography on polyethyleneimine cellulose. Plates were scanned after development using a Typhoon phosphoimaging device. Amounts of labeled ATP and PPi were estimated from densitometry and fitted to relation (Equation 1) using JMP (22).
The active fraction, n, is obtained from the fitted parameters (23) as in Equation 2.
 |
Enzymatic Assays
Samples were assayed in 96-well format, using the traditional [32P]pyrophosphate exchange in the presence of the cognate histidine substrate, as described (24, 25). A 3-fold reduction of background 32P counts was achieved by collecting the charcoal containing labeled ATP on disposable filter plates (Whatman Microplate unifilter devices), washing, and eluting the bound ATP with 50 μl of pyridine at 37 °C (15). A unit of activity is 1 μm [32P]ATP formed per minute. Controls with calibrated amounts of labeled ATP were performed to determine the elution efficiency, which was used to correct the experimental results. Four replicates were performed for each measurement. Michaelis-Menten experiments were carried out for one of the four constructs (HisRS3) by varying the ATP and histidine concentrations. Concentration-dependent rates were fitted using JMP (22).
RESULTS
Structural Design Considerations
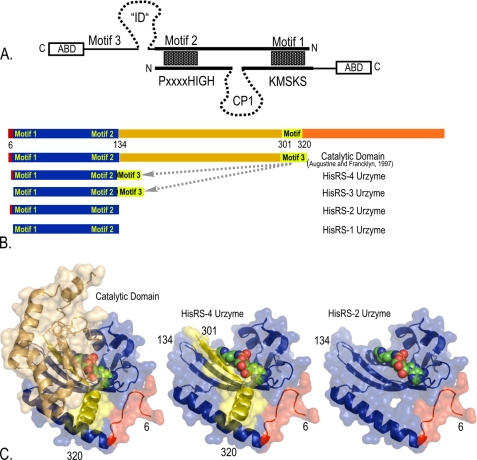
Rodin and Ohno (17) suggested two serious problems associated with their attempt to align the Class I PxxxHIGH and KMSKS sequences sense and antisense with those of Class II Motifs 2 and 1 (Fig. 1A). First, the long and variable CP1 (connecting peptide) insertion precludes simultaneous alignment of both Class I signatures onto Motifs 2 and 1, which are always separated by ∼100 amino acids in Class II aaRS (15). Furthermore, the active site in full-length Class II aaRS includes what appear to be class-defining contributions from Motif 3, which is separated from the C-terminal end of the Motif 2 loop by a long and variable insertion domain (26) that is ∼170 amino acids long in E. coli HisRS, and hence has no N-terminal (antisense) counterpart in many Class I aaRS. Thus, to complement our previous demonstration that Class I catalytic activity does not require the presence of the CP1 insertion, we wished to show here that Motif 3 and the insertion domain also can eliminated without abolishing catalytic activity from comparable Class II constructs.
FIGURE 1.
Motivation and construction of HisRS Urzymes. A, the Rodin-Ohno sense/antisense ancestry hypothesis. Class I and II aaRS proteins are aligned in opposite directions with Class II Motifs 2 and 1 opposite Class I PxxxxHIGH and KMSKS signature peptides. Coding sequences for these four peptides have statistically significant reverse complement homology ((17), filled rectangles). Ancestry of the boldface lines connecting them is incompatible with the Class I CP1 insertion (dashed line). As many Class I aaRS begin within ∼10 residues of the PxxxxHIGH signature, sense/antisense ancestry is also incompatible with the presence of the insertion domain (ID, dashed line) in primordial Class II aaRS. B, schematic maps of HisRS1–4. Motifs 1–3 are conserved peptides that define Class II aaRS (39). The N-terminal catalytic domain was described previously (14). The four Urzymes, HisRS1–4, were constructed in this work. C, three-dimensional structures derived from the 2EL9 crystal structure. Colors match those in B.
Four HisRS polypeptides derived from the HisRS Ncat domain were produced by PCR-based subcloning of the appropriate fragments of the E. coli hisS gene into an expression vector. The catalytic apparatus in Class II aaRS is often described in terms of an antiparallel, seven-stranded β-sheet with flanking helices (27). Our effort here is motivated by the desire to identify much smaller segments of the contemporary enzyme that retain measureable catalytic activity for amino acid activation. For this purpose, we identified an active site built around Motif 2, which forms the ATP-binding site, and the β-strand and α-helix immediately preceding the Motif 2 loop, which comprise much of the amino acid binding site.
Residues Gly134 and Pro301 are ideally positioned to form a β-turn (Fig. 2A) between Motifs 2 and 3. This enabled us to connect the two Motifs directly, without applying the Loop algorithm in Rosetta as we did when removing the internal CP1 peptide in the TrpRS Urzyme (16, 28). None of the conserved residues defining Motif 1 occur before residue Gly37, which is at the C terminus of what has become known as the “Motif 1 α-helix”. We nevertheless elected to include the entire length of this helix in all constructs. Moreover, an N-terminal extension to this helix forms a number of contacts with the back of the Motif 2 loop (Fig. 1C). We reasoned that this connection might afford a detectable functional advantage and thus included variation of this structural feature in the experimental design. The resulting constructs, summarized in Fig. 1, B and C, are as follows: HisRS1, residues 16–134; HisRS2, residues 10–134; HisRS3, residues 16–134 and 301–320; and HisRS4, residues 10–134 and 301–320.
FIGURE 2.
Rationale for the factorial construction design. A, the C-terminal (C) β-strand of Motif 2 (blue) and N-terminal (N) β-strand of Motif 3 (yellow) can be connected by a β-turn between residues Gly134 and Pro301. B, the N-terminal Motif 1 extension (red) makes extensive interactions with the back side of the active site. C, Motif 3 (yellow) provides additional connections to the Motif 1 helix.
Purification of HisRS-3 from TEV Cleavage Mixture Does Not Significantly Alter Its Specific Activity
Purification of the four constructs is illustrated in Fig. 3, together with the results of TEV cleavage. The MBP and HisRS fragments are of sufficiently different sizes that we expected to be able to separate them by gel filtration. We attempted this separation on a Shodex KW-803 column. However, the broad elution profile and the absence of a clear separation (data not shown) implied that the cleaved mixture was aggregated. This aggregation also precluded any clear answer as to the quaternary structure of the active Urzyme.
FIGURE 3.
Purified HisRS1–4 constructs. Lanes are labeled across the top in pairs of fusion protein constructs and their TEV cleavage products. Extra bands present in the cleavage products arise from TEV protease preparation.
However, we isolated sufficient HisRS-3 fusion protein to separate the HisRS construct from both MBP and from the TEV enzyme itself using anti-FLAG antibody. The HisRS-3-specific activities before and after this purification were 1.7 and 1.3 units/mg. Given the similarity of these two values and the high cost of purification, which entails substantial loss of material, we characterized the remaining HisRS constructs in their respective TEV cleavage mixtures.
Active Site Titration of HisRS MBP Fusion Proteins Confirms That Significant Fractions of Total Number of Molecules Contribute to Observed Catalytic Activities
We previously used active site titration (21, 23) to show that the observed activity of the TrpRS Urzyme derived from significant fractions of the total Urzyme concentration (16). As with the TrpRS Urzyme, the HisRS1–4 MBP fusion proteins are substantially less active than are their TEV cleavage products. Furthermore, despite their increased activity, the cleaved products of the HisRS fusion proteins could not be concentrated in sufficient quantity after TEV cleavage to increase the signal to noise in the active site titration assays. We therefore titrated only the fusion proteins themselves. Although there is variation between different preparations, the titration experiments show convincingly that substantial bursts occur (Fig. 4). Thus, as with the TrpRS Urzyme, HisRS 1–4 all retain the histidyl-adenylate intermediate, and turnover is limited, either by product release or by the slow decomposition of the enzyme-bound adenylate.
FIGURE 4.
Active site titration of the HisRS1–4 MBP (A–D, respectively) fusion proteins. All fusion proteins display pre-steady-state bursts with significant amplitudes (lower left corner). The y axis ([ATP]_n, rel) is the fraction of the initial value recorded in the absence of catalyst corrected for dilution.
Catalytic Activity Varies Consistently with Each of Three Design Variables
The experimental design comprises a three-factor, two-level factorial plan testing the effect on the activity of three structural elements: TEV cleavage of the fusion protein, the presence of Motif 3, and the presence of the N-terminal extension to the Motif 1 helix. The fusion purified proteins (Fig. 3) were thus cleaved with TEV protease, and both the cleaved and uncleaved preparations were tested for histidine activation activity using the 32PPi exchange assay (29) as modified previously to enhance sensitivity (15).
Significant evidence that activities presented here represent authentic catalysis by the HisRS constructs and do not arise from contaminating full-length enzyme derives from the systematic variation of activity with the precise characteristics of the different constructs. Neither MBP nor TEV protease by itself has histidine activation activity. Specific activities of the HisRS1–4 constructs, corrected for the fraction of active sites in the respective fusion proteins, are compared in Fig. 5. There are significant differences between the average activities of the three groups of four experiments according to the factorial design: four fusion proteins versus four cleaved products (Fig. 5A); four with versus four without Motif 3 (Fig. 5B), and four with versus four without the N-terminal extension (Fig. 5C).
FIGURE 5.
Specific activities of the four HisRS constructs. HisRS-1, Motifs 1 and 2 only; HisRS-2, core + N-terminal (N-term) extension only; HisRS-3, core + Motif 3 only; HisRS-4, core + N-terminal extension + Motif 3). Specific activities, corrected for the active fractions of the respective fusion proteins (Fig. 4), have been converted to relative free energies by ΔG = −0.597 × ln(Sp_Act). Error bars denote the S.D. for triplicate or quadruplicate measurements of each activity. The three factorial variables TEV cleavage, the N-terminal extension (HisRS-2 and -4) and the presence of Motif 3 (HisRS-3 and -4), all have clearly significant effects. However, the experimental signal-to-noise is comparable for the fusion proteins and their cleaved products. In all panels, the observed activities are shaded according to the comparisons made in fitting the regression model (see Equation 3). Far left panel, activities of the fusion proteins (light gray bars) are suppressed, relative to activities released on TEV cleavage (dark gray bars). Middle panel, activities of all four constructs containing Motif 3 (dark gray bars) are elevated, relative to those that lack Motif 3 (light gray bars). Far right panel, activities of the four constructs containing the N-terminal extension are elevated (dark gray bars), relative to those that lack it. A quantitative assessment is given in Table 2.
To assess the significance of these differences quantitatively, we converted the specific activities to ΔG values and fitted values for each of the eight constructs, ΔGi (Table 1), to a linear multiple regression model, involving four effects (Equation 3) to obtain least-squares estimates for the catalytic contributions,
where Δ(ΔG)Tev, Δ(ΔG)Nterm,M3 and Δ(ΔGint)Nterm×M3 are coefficients estimated by fitting. TEV, N-term, and M3 are matrix elements from the corresponding columns in Table 1. The coefficients represent the free energy contribution in kcal/mol to transition state stabilization from TEV cleavage, the N-terminal extension, Motif 3, and the synergy between the N-terminal extension and TEV cleavage, respectively.
TABLE 1.
Factorial design matrix for HisRS1–4 Urzyme constructs
| Variant | TEV | N terminus | M3 | Rate | ΔG‡ |
|---|---|---|---|---|---|
| s−1 | kcal/mol | ||||
| HisRS-1 | 0 | 0 | 0 | 0.018 | 2.404 |
| HisRS-1+TEV | 1 | 0 | 0 | 1.070 | −0.040 |
| HisRS-2 | 0 | 1 | 0 | 0.017 | 2.430 |
| HisRS-2+TEV | 1 | 1 | 0 | 0.879 | 0.077 |
| HisRS-3 | 0 | 0 | 1 | 0.015 | 2.501 |
| HisRS-3+TEV | 1 | 0 | 1 | 1.067 | −0.038 |
| HisRS-4 | 0 | 1 | 1 | 0.297 | 0.725 |
| HisRS-4 | 0 | 1 | 1 | 0.283 | 0.753 |
| HisRS-4 | 0 | 1 | 1 | 0.311 | 0.697 |
| HisRS-4+TEV | 1 | 1 | 1 | 8.351 | −1.267 |
The units of specific activity, mol/s/mol protein, are those of kcat. Values reported here, however, are lower bounds for kcat, as we have not titrated the fraction of active sites for the cleaved HisRS constructs. The Student's t test probabilities indicate the statistical significance of the coefficients and hence of the quantitative catalytic contributions of each of the four predictors. These values are summarized, together with analysis of variance statistics, in Table 2. Consistent activity differences between constructs with respect to each of the three design variables would likely not be observed unless the HisRS constructs were the authentic source of the observed catalytic activity.
TABLE 2.
Regression modeling of HisRS Urzyme activities
Analysis of variance statistics were as follows: N(obs) = 9; R2 = 0.993; F = 22.1, and p > F, 0.0002.
| Term | Estimate | S.E. | t Ratio | p > |t| |
|---|---|---|---|---|
| Intercept | 2.84 | 0.115 | 24.58 | < 0.0001 |
| TEV | −2.31 | 0.110 | −20.92 | < 0.0001 |
| N-terminal extension | −0.83 | 0.110 | −7.57 | 0.0016 |
| M3 | −0.85 | 0.110 | −7.78 | 0.0015 |
| N-terminal extension × M3 | −1.63 | 0.220 | −7.38 | 0.0018 |
Negative coefficients imply that the presence of the factor reduces ΔG‡, thus accelerating the rate of histidine activation. TEV cleavage, the N-terminal extension, and Motif 3 each accelerate the rate, with effects of different magnitude. TEV cleavage lowers ΔG‡ by −2.31 kcal/mol, or about three times the extent afforded by Motif 3 and by the N-terminal extension (0.8 kcal/mol).
The effect of Motif 3 differs, depending on whether or not the construct containing it also has the N-terminal extension. This effect is reflected in the sign of the interaction term (N-term extension × TEV) = −1.63 kcal/mol. It changes the ΔG‡ by only −0.83 kcal/mol in HisRS-3, which has only M3, but by −1.46 kcal/mol (−0.83 − 0.19 × 1.63) kcal/mol in HisRS-4, which has both.4 Thus, the contribution of both the N-terminal extension or Motif 3 are enhanced if the other is present, due to their synergistic interaction term.
Active Site Mutation R113A in HisRS-4 Reduces Activity ∼4-Fold
Arginine 113 is a strictly conserved basic residue in the Motif 2 catalytic sequence. Density functional theory calculations suggest that it is critical to transition-state stabilization of acyl transfer by HisRS transition state stabilization (30). We therefore constructed the R113A mutant in HisRS-4, the most active of the four constructs. Replicated assays of the WT and R113A HisRS-4 MBP fusion proteins (Table 3) show that the mutation reduces activity 4-fold, by an amount that is 9.8 times the estimated S.D. of the measurements. This experiment provides orthogonal evidence that the observed activity resides in the HisRS-4 construct and not another source of adventitious contamination. We return in the discussion to the unexpectedly small magnitude of this mutational effect.
TABLE 3.
Specific Activities of HisRS4 R113A versus wild type HisRS 4_MBP fusion proteins
The Student's t test probability (p) that the WT and R113A have the same activities is 0.0052.
| Specific acitivities | S.D. | |
|---|---|---|
| mm/min/mg × 103 | ||
| HisRS-4, WT | 1.38 | 0.09 |
| HisRS4_R113A | 0.33 | 0.06 |
HisRS-3 Binds ATP Far More Tightly and Histidine Less Strongly Than Native, Full-length HisRS or Its Intact Catalytic Domain
Complementary evidence that the HisRS1–4 constructs are responsible for observed activities comes from the fact that Michaelis-Menten Km values of HisRS-3 for ATP and histidine are distinctly different from those of the full-length enzyme or the Ncat catalytic domain (14). The activities of HisRS-1–4 are roughly comparable, and the constructs are difficult to purify in large amounts. The HisRS-3 preparation provided a surplus of fusion protein, which we used to determine Michaelis-Menten kinetic parameters for 32PPi exchange (Fig. 6).
FIGURE 6.
Michaelis-Menten kinetics for HisRS-3. Error bars denote the root mean square deviation for quadruplicate measurements at each concentration. A, histidine dependence of 32PPi exchange. B, ATP-dependence of 32PPi exchange. Chisq is the goodness of fit, and R is the correlation coefficient between observed and calculated values.
The histidine-dependent Michaelis-Menten experiment also demonstrates that there is no activity in the absence of histidine. As the exchange condition affords thermodynamic dissociation parameters (31, 32), these experiments provide measures of the affinity of the HisRS-3 Uryzme for both substrates. As is true for the TrpRS Urzyme (16), and in contrast to either full-length enzyme, the HisRS-3 binds ATP (Km = 15 μm) an order of magnitude more tightly than it does histidine (Km = 119 μm). HisRS-3 binds both substrates an order of magnitude more tightly in the ground state complexes than corresponding substrates bind to the TrpRS Urzyme. It is worth noting that the high ATP affinity implies that the HisRS constructs probably use less of the binding energy resulting from interaction with ATP to lower the transition state free energy than is observed for the full-length HisRS. Moreover, the higher Km for histidine is consistent with the fact that all constructs are missing important contact residues, e.g. the HisA loop, residues 257–264 in the insertion domain (33).
DISCUSSION
Reconstruction of extinct enzymes from beyond the era that is supported by a well defined sequence phylogeny is necessarily controversial. It is thus worth summarizing at the outset the logic underlying our proposal that these fragmentary catalysts are legitimate models for ancestral aminoacyl tRNA synthetases, that is that they do indeed do deserve to be called “Urzymes.” There is widespread consensus that structural conservation is evidence for common ancestry. This phylogenetic argument holds that the most highly conserved structures, both sequences and three-dimensional structures, within contemporary enzyme families are most ancient. To this strong prior prejudice that the most widely shared, invariant structures are indeed the most ancestral, we have added essential experimental confirmation; these conserved fragments exhibit ∼9 of the ∼14 orders of magnitude in rate acceleration, i.e. they afford ∼65% of the transition state stabilization associated with the contemporary enzymes.
Substrate-binding affinities reinforce the conclusion that the TrpRS and HisRS Urzymes are plausible models for the early Class I and Class II aaRS. Both Urzymes bind ATP more tightly than amino acid, consistent with the expectation that the primordial function of ancestral aaRS probably was to mobilize ATP for chemical group activation. The posterior probability that these Urzymes are legitimate models for ancestral aaRS is therefore substantially strengthened by demonstrating such significant catalytic activity and relative substrate specificites (16). Viewed another way, we have shown experimentally that significant catalytic activities and appropriate specificities are associated with the most highly conserved, and hence arguably most ancient portions of class I and II synthetases. Thus, it is proper to invoke Ockham's razor (34) in arguing that ancestral synthetases were unlikely to have differed radically from the HisRS and TrpRS Urzymes.
The results presented here complement our previous study of the TrpRS Urzyme (15, 16) in several ways. This is the first study of an Urzyme representing Class II aaRS. All HisRS Urzymes were constructed without using protein design to compensate for structural damage inflicted by deleting large portions of the native enzyme. In retrospect, it will be interesting to assess possible reasons for this relative independence from mutational changes to the contemporary sequence. A work in progress is devoted to a more comprehensive testing of the design parameters and their application to type II HisRS and ProRS Urzymes. The immediate significance of this work, however, arises from our demonstration that the core catalytic activities of both Class I and Class II aminoacyl-tRNA synthetases can be implemented in peptides as short as ∼120–130 amino acids, containing their most highly conserved active site residues.
Relative Rate Enhancements Observed for Successive Fragmentation of Full-length HisRS Enzyme
Specific activities illustrated in Fig. 5 are correlated, roughly, with the fitted kchem (r = 0.48) and kcat (r = 0.72) parameters from the active site titrations. The better correlation, as expected, is for kcat values. The correlations are significantly better between values for the first three constructs (r = 0.97 and 0.99) than for HisRS4, whose corrected activity is substantially higher than would be expected from the active site titrations alone.
It is interesting to compare the relative rate enhancements for a series of successively smaller E. coli HisRS active site fragments (Fig. 7). The N-terminal catalytic domain studied previously (14) represents 76% of the full-length enzyme; its activity is reduced by ∼2000-fold. HisRS 1, 2, 3, and 4 represent, respectively, 29%, 30%, 33%, and 35% of the full-length HisRS. Their activities are reduced by 4–9 × 106-fold. An excellent linear relationship exists between the fractional loss of mass and the loss of ln(enzymatic activity) (R2 = 0.99, P(t) < 0.0001) over nearly seven orders of magnitude.
FIGURE 7.
Relative rate enhancements for successive fragmentation of the full-length E. coli HisRS. Relative rates from specific activity values for the full-length enzyme and the full N-terminal catalytic domain (gray) were taken from (14); those for HisRS-1–4 are from this work.
This observation reinforces the notion that historical improvements in evolutionary fitness were associated with the accumulation of modular blocks of new genetic information. Furthermore, the insertion domains are clearly different protein folds in prokaryotic and eukaryotic HisRS. This implies that the insertion domain was probably the most recent domain addition to the enzyme and suggests that it was acquired subsequently to the HisA loop.
Large Fraction of Catalytically Active Molecules and Consistent Dependence of Urzyme Activities on TEV Cleavage and Presence of N- and C-terminal Extensions Afford Strong Evidence for Authenticity of Observed Catalytic Activity
In our previous study of the TrpRS Urzyme, we outlined six criteria by which we could assess the authenticity of its catalytic activity. The four most compelling of these criteria were as follows: the high percentage (≫1%) of active molecules contributing to the observed activity in active site titrations, the cryptic nature of the activity of the fusion protein, the fact that an active site mutation altered the observed activity, and the unique value of the Km value for tryptophan.
Similar criteria also establish the authenticity of the HisRS Urzyme catalytic activities. Active site titration reveals significant proportions of active molecules (Fig. 4). As was true for the MBP-TrpRS Urzyme fusion protein, the MBP fusion inactivates the HisRS Urzymes by a factor of ∼50, relative to the activity released upon TEV cleavage. Moreover, addition to the minimal HisRS Urzyme of either the N-terminal extension to Motif 1 or Motif3 enhances activity by reproducible amounts in the fusion proteins and the TEV cleavage products (Fig. 5). The consistent attribution of catalytic effects of Motif 3, the N-terminal extension and their interaction in the regression model in Table 2 also argues strongly in favor of the authenticity of the observed catalytic activity, as does the significant impact of the HisRS-4 R113A active site mutation.
Results shown in Fig. 5 also establish important existence proofs for the ability to manipulate modular structural details related to catalytic activity. The approaches we used here may prove useful in implicating short peptide segments, including individual secondary structure elements, in modulating fitness on a broader scale. Fuller characterization of the steady-state kinetic parameters for HisRS1–4 will be key to progressing further along these lines.
HisRS-3 Urzyme Steady-state Kinetic Parameters Differ Substantially from Those of Native Full-length HisRS (Fig. 6)
The Michaelis-Menten parameters are preliminary, and must eventually be measured for all three constructs. Nevertheless, they are significantly different from those either of the full-length HisRS or the N-terminal catalytic domain (14). From the standpoint of whether or not the observed activity can be attributed reasonably to contaminating full-length enzyme, the important parameters are the half-saturation values, Km, for ATP and histidine. Contamination by wild-type HisRS would be half saturated at [His] = 30 μm and at [ATP] = 890 μm. We observe half saturation instead at [His] = 209 μm and at [ATP] = 19 μm. The Urzymes require seven times the histidine concentration and yet require only ∼0.02 as much ATP as wild type HisRS. These values far exceed the errors of the estimated Km parameters, which are both ∼50%. We conclude from these comparisons that the Urzyme kinetic parameters afford strong, orthogonal evidence for the authenticity of HisRS 1–4 catalytic activities.
HisRS Urzyme Catalysis Likely Illustrates Catalytic Importance of Distributed Active Site-binding Properties
The R113A mutation might have been expected to have a larger impact than the observed 4-fold reduction. The modest observed effect suggests that the detailed electrostatic transition-state stabilization by the arginine residue in the native enzyme is much reduced in the Urzyme. In turn, this suggests that the observed catalysis arises to a greater extent from the distributed substrate binding properties of the active site. We noted a similar and possibly related phenomenon with respect to the effect of the D146A mutation in the TrpRS Urzyme.
Catalytic Activities of HisRS-1 and HisRS-2 Provide Complementary Support for Rodin-Ohno Hypothesis That Ancestral Class I and II aaRS Were Encoded on Opposite Strands of Same Gene
Rodin and Ohno (17) observed that codons for the conserved, Motif 2 and Motif 1 catalytic signatures of Class II aaRS were conspicuously homologous, respectively, to the reverse complements of sequences coding for the corresponding PxxxxHIGH and KMSKS catalytic signatures in the Class I aaRS. This observation led them to propose that the two apparently unrelated superfamilies descended from opposite strands of the same ancestral gene. Our work on the TrpRS Urzyme verified one essential prediction of the sense/antisense origin hypothesis, that Class I catalytic activity did not require the CP1 insertion (15, 16, 28).
As Motif 3 is C-terminal to Motif 2, and as the PxxxxHIGH signature is within ∼10 amino acids of the N terminus in roughly half of the Class I synthetases, a comparable prediction of the Rodin-Ohno hypothesis is that Motif 3 was not essential for the catalytic activity of the progenitor Class II aaRS. We have shown here that HisRS-1 and HisRS-2 catalyze histidine activation at rates similar to those observed for the Class I TrpRS Urzyme (15, 16), confirming a complementary experimental prediction of the Rodin-Ohno hypothesis.
Significantly Different Rate Enhancements by Four HisRS Urzymes Demonstrate That Amino Acid Activation Provides an Experimental Metric for Recapitulating very Early Evolutionary Events
The factorial design of the experiments reported here illustrates that significant experimental differences in fitness can be attributed to new blocks of genetic information. In particular, we have demonstrated improvements in catalytic activity associated with both Motif 3 and the N-terminal extension to Motif 1. The four HisRS Urzymes range from 119 to 145 residues, representing two putative discontinuous evolutionary stages in the generation of modern Class II aaRS. Table 1 emphasizes that the sensitivity of our 32PPi exchange assay suffices to implicate both modules with different contributions to histidine activation, which is certainly an appropriate metric of fitness in the early evolution of translation.
Translation of the genetic code was a signal event in the origin of life, requiring not only the activation of amino acids, considered here, but also the recognition and acyl transfer to cognate tRNAs. Cognate tRNA is required for induced-fit active site assembly in a subset of class I aaRS (35–37) and helps to coordinate the activities of the two monomers in class II aaRS dimers (38). It seems likely that ancestral synthetases developed rudimentary functional integration of enzyme and tRNA. Demonstration of experimental metrics for the HisRS and TrpRS (16) Urzymes provide a realistic hope of unraveling their evolution.
Acknowledgments
pMAL-c2x vector was a kind gift from A. Sancar. We thank H. Fried for numerous helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM078227.
Aminoacyl adenylates hydrolyze spontaneously in aqueous conditions at a rate of 1.06 × 10−4 /sec (40). Alcohols are intrinsically about an order of magnitude more reactive than water on a molar basis (41). In a naïve active site unable to stabilize electron rearrangements but which retains both tRNA and adenylate, the rate of acyl transfer would be expected to be about the same, ∼105 times, or more, faster than amino acid activation.
The factor of 0.19 arises because JMP calculates the interaction term in terms of (N-terminal − 0.555) × (M3 − 0.555) because the absence and presence of the two variables are coded by 0 and 1 in Table 1 and 0.44 × 0.44 = 0.19.
- aaRS
- aminoacyl tRNA synthetases
- HisRS
- histidyl-tRNA synthetase
- TrpRS
- tryptophanyl-tRNA synthetase
- IPTG
- isopropyl β-D-1-thiogalactopyranoside
- MBP
- maltose-binding protein
- TEV
- Tobacco Etch Virus protease.
REFERENCES
- 1. Stackhouse J., Presnell S. R., McGeehan G. M., Nambiar K. P., Benner S. A. (1990) FEBS Lett. 262, 104–106 [DOI] [PubMed] [Google Scholar]
- 2. Thornton J. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5671–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bridgham J. T., Carroll S. M., Thornton J. W. (2006) Science 312, 97–101 [DOI] [PubMed] [Google Scholar]
- 4. Bridgham J. T., Ortlund E. A., Thornton J. W. (2009) Nature 461, 515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benner S. A., Sassi S. O., Gaucher E. A. (2007) Adv. Enzymol. Rel. Areas Mol. Biol. 75, 9–140 [DOI] [PubMed] [Google Scholar]
- 6. Gaucher E. A., Govindarajan S., Ganesh O. K. (2008) Nature 451, 704–707 [DOI] [PubMed] [Google Scholar]
- 7. Thornton J. W. (2004) Nat. Rev. Genet. 5, 366–375 [DOI] [PubMed] [Google Scholar]
- 8. Thornton J. W., Need E., Crews D. (2003) Science 301, 714–1717 [DOI] [PubMed] [Google Scholar]
- 9. Benner S. A. (1989) Chem. Rev. 89, 789–806 [Google Scholar]
- 10. Deleted in proof.
- 11. Sievers A., Beringer M., Rodnina M. V., Wolfenden R. (2004) Proc. Nat. Acad. Sci., U.S.A. 101, 7897–7901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwob E., Söll D. (1993) EMBO J. 12, 5201–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borel F., Vincent C., Leberman R., Härtlein M. (1994) Nucleic Acids Res. 22, 2963–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Augustine J., Francklyn C. (1997) Biochemistry 36, 3473–3482 [DOI] [PubMed] [Google Scholar]
- 15. Pham Y., Li L., Kim A., Erdogan O., Weinreb V., Butterfoss G. L., Kuhlman B., Carter C. W., Jr. (2007) Mol. Cell 25, 851–862 [DOI] [PubMed] [Google Scholar]
- 16. Pham Y., Kuhlman B., Butterfoss G. L., Hu H., Weinreb V., Carter C. W., Jr. (2010) J. Biol. Chem. 285, 38590–38601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodin S. N., Ohno S. (1995) Orig. Life Evol. Biosph. 25, 565–589 [DOI] [PubMed] [Google Scholar]
- 18. Kamtekar S., Schiffer J. M., Xiong H., Babik J. M., Hecht M. H. (1993) Science 262, 1680–1685 [DOI] [PubMed] [Google Scholar]
- 19. Moffet D. A., Foley J., Hecht M. H. (2003) Biophys. Chem. 105, 231–239 [DOI] [PubMed] [Google Scholar]
- 20. Patel S. C., Bradley L. H., Jinadasa S. P., Hecht M. H. (2009) Protein Sci. 18, 1388–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Francklyn C. S., First E. A., Perona J. J., Hou Y. M. (2008) Methods 44, 100–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. SAS (2006) JMP, version 6.0, The SAS Institute, Cary, NC [Google Scholar]
- 23. Fersht A. R., Ashford J. S., Bruton C. J., Jakes R., Koch G. L., Hartley B. S. (1975) Biochemistry 14, 1–4 [DOI] [PubMed] [Google Scholar]
- 24. Joseph D. R., Muench K. H. (1971) J. Biol. Chem. 246, 7602–7609 [PubMed] [Google Scholar]
- 25. Joseph D. R., Muench K. H. (1971) J. Biol. Chem. 246, 7610–7615 [PubMed] [Google Scholar]
- 26. Arnez J. G., Harris D. C., Mitschler A., Rees B., Francklyn C. S., Moras D. (1995) EMBO J. 14, 4143–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. (1990) Nature 347, 249–255 [DOI] [PubMed] [Google Scholar]
- 28. Pham Y. B. (2009) PhD Thesis in Biochemistry and Biophysics, pp. 95, University of North Carolina at Chapel Hill, Chapel Hill, NC [Google Scholar]
- 29. Fersht A. R. (1974) Proc. Roy. Soc. Lond. B. 187, 397–407 [DOI] [PubMed] [Google Scholar]
- 30. Liu H., Gauld J. W. (2008) J. Phys. Chem. B112, 16874–16882 [DOI] [PubMed] [Google Scholar]
- 31. Cleland W. W. (1970) in The Enzymes (Boyer P. ed) Vol. II, pp. 1–65, Academic Press, New York [Google Scholar]
- 32. Cleland W. W., Northrop D. B. (1999) Methods Enzymol. 308, 3–27 [DOI] [PubMed] [Google Scholar]
- 33. Arnez J. G., Augustine J. G., Moras D., Francklyn C. S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7144–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gernert D. (2007) J. Sci. Expl. 21, 135–140 [Google Scholar]
- 35. Sherlin L. D., Perona J. J. (2003) Structure 11, 591–603 [DOI] [PubMed] [Google Scholar]
- 36. Bullock T. L., Uter N., Nissan T. A., Perona J. J. (2003) J. Mol. Biol. 328, 395–408 [DOI] [PubMed] [Google Scholar]
- 37. Bullock T. L., Rodríguez-Hernández A., Corigliano E. M., Perona J. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7428–7433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guth E. C., Francklyn C. S. (2007) Mol. Cell 25, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eriani G., Delarue M., Poch O., Gangloff J., Moras D. (1990) Nature 347, 203–206 [DOI] [PubMed] [Google Scholar]
- 40. Schall O. F., Suzuki I., Murray C. L., Gordon J. I., Gokel G. W. (1998) J. Org. Chem. 63, 8661–8667 [Google Scholar]
- 41. Stockbridge R. B., Wolfenden R. (2009) J. Biol. Chem. 284, 22747–22757 [DOI] [PMC free article] [PubMed] [Google Scholar]