Abstract
DNA damage response (DDR) activates a complex signaling network that triggers DNA repair, cell cycle arrest, and/or cell death. Depending on the type and severity of DNA lesion, DDR is controlled by “master” regulators including ATM and ATR protein kinases. Cisplatin, a major chemotherapy drug that cross-links DNA, induces ATR-dependent DDR, resulting in apoptosis. However, it is unclear how ATR is activated. To identify the key regulators of ATR, we analyzed the proteins that associate with ATR after cisplatin treatment by blue native-PAGE and co-immunoprecipitation. The mismatch repair protein hMSH2 was found to be a major ATR-binding protein. Functionally, ATR activation and its recruitment to nuclear foci during cisplatin treatment were attenuated, and DNA damage signaling, involving Chk2, p53, and PUMA-α, was suppressed in hMSH2-deficient cells. ATR activation induced by the DNA methylating agent N-methyl-N-nitrosourea was also shown to be hMSH2-dependent. Intriguingly, hMSH2-mediated ATR recruitment and activation appeared independent of replication protein A, Rad17, and the Rad9-Hus1-Rad1 protein complex. Together the results support a hMSH2-dependent pathway of ATR activation and downstream Chk2/p53 signaling.
Keywords: Apoptosis, DNA Damage, DNA Repair, Kidney, p53, ATR, Cisplatin, Mismatch Repair
Introduction
DNA damage response (DDR)3 is essential for the maintenance of the integrity of the genome (1–5). As a complex multi-step process, DDR involves the recognition of DNA damage, activation of DNA damage-responsive protein kinases, signal amplification by downstream protein kinases, and activation of the effector proteins that trigger various cellular processes. At low levels of DNA damage, activation of DDR results in cell cycle arrest and DNA repair. However, at higher doses, DDR signaling frequently results in cell death by apoptosis (1–5). The phosphoinositide 3-kinase-related protein kinases, ATM (ataxia-telangiectasia mutated) and ATR (ATM- and Rad3-related), are the key regulators of DDR (1–8). Once activated, ATM and ATR regulate an array of substrates including Chk1 and Chk2, which culminate in DNA repair, cell cycle arrest, and/or apoptosis. Although ATM is generally activated by double-stranded DNA breaks, ATR activation can result from different types of DNA lesion including single-stranded breaks, replication stress, base adducts, and DNA cross-links (8, 9). In the canonical model, ATR activation involves the recruitment of the ATR-ATRIP (ATR-interacting protein) and the Rad9-Hus1-Rad1 (9-1-1) protein complexes to the DNA damage site via replication protein A (RPA). As a result, the 9-1-1 complex brings topoisomerase-binding protein-1 (TopBP1, an ATR activator) close to ATR for ATR activation (1, 5, 8). However, alternative pathways of ATR activation may exist. For example, mismatch repair (MMR) proteins have been implicated in ATR activation under certain experimental conditions (10–12).
The MMR system is important for DNA replication and repair (13–18). In mammalian cells, MMR is composed of five MutS homologues (MSH) and four MutL homologue proteins. The function of MMR is to first recognize DNA mismatches and then help recruit DNA repair proteins to the mismatched site (13–18). Although the role of MMR proteins in DNA replication and repair is well recognized, evidence has been reported that these proteins may also function in DNA damage signaling and consequent apoptosis (19–21). Loss of one or more MMR proteins renders the cells resistant to DNA-damaging chemotherapy drugs (22, 23). It has been proposed that MMR proteins may detect DNA lesions and act as signaling mediators for activation of cell death, a function that is independent of DNA repair (19). This possibility is supported by the role of hMSH2 in cisplatin-induced genotoxic stress, where hMSH2 preferentially binds to damaged DNA and the loss of hMSH2 results in resistance to cisplatin-induced cell death (24–26). Remarkably, scattered evidence suggests that MMR proteins may directly mediate ATR activation (10–12). Despite these findings, it remains unclear how MMR proteins participate in ATR activation and the consequent signaling.
Cisplatin is one of the most widely used and most potent chemotherapeutic drugs (26–29). Cisplatin-induced DNA damage is not only responsible for its anti-cancer effects but also contributes to its side effects in normal tissues including nephrotoxicity in kidneys (30–32). Cisplatin treatment results in ATR activation, which in turn phosphorylates and activates Chk2, followed by p53 activation leading to pro-apoptotic gene expression and cell death (30, 33–36). However, it is unclear how ATR is activated during cisplatin treatment. Here we have identified hMSH2 as a key sensor and mediator of ATR activation. Notably, hMSH2 recruits and activates ATR via a novel pathway that appears independent of RPA, Rad17, and the 9-1-1 complex proteins. Importantly, hMSH2-mediated ATR activation seems to specifically account for Chk2/p53 activation and apoptosis.
EXPERIMENTAL PROCEDURES
Materials
Cells
Immortalized wild-type and MSH2-deficient mouse embryonic fibroblasts (MEF) were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 2 mm sodium pyruvate, 2 mm glutamine, 100 IU penicillin, and 100 mg/ml streptomycin as described (37). HEK and HeLa cell lines were purchased from ATCC (Manassas, VA) and maintained in minimal essential medium with 10% fetal bovine serum, 1% glutamine, 1% non-essential amino acids, and 1% antibiotics.
Antibodies
Goat polyclonal anti-ATR were from Santa Cruz Biotechnology (Santa Cruz, CA); anti-9-1-1 proteins, mouse monoclonal anti-MSH2, anti-p53, rabbit monoclonal anti-MSH2, rabbit polyclonal anti-ATR, anti-Chk2, anti-PUMA (p53 up-regulated modulator of apoptosis), anti-phospho-p53, anti-phospho-Chk1, anti-phospho-Chk2, and anti-phospo-H2AX antibodies were from Cell Signaling Technology (Beverly, MA); mouse monoclonal anti-β-actin was from Sigma; rabbit polyclonal anti-Chk1 was from Epitomics (Burlingame, CA). All of the primary antibodies were used at 1:1000 dilution for Western blot analysis; all of the secondary antibodies were from Jackson ImmunoResearch (West Grove, PA).
Special Reagents
MSH2 and control shRNA plasmids were purchased from SABiosciences Corporation (Frederick, MD). Carbobenzoxy-DEVD-7-amino-4-trifluoromethyl coumarin and 7-amino-4-trifluoromethyl coumarin for caspase assay were purchased from Enzyme Systems Products (Dublin, CA). Unless indicated, all other reagents including cisplatin and N-methyl-N-nitrosourea (MNU) were from Sigma.
Cisplatin and MNU Treatment of Cells
Cisplatin treatment was conducted as described in our recent work (33, 38–40). Cisplatin was used at 25 μm for HeLa cells, 20 μm for MEF cells, and 50 μm for HEK cells. These doses were selected because dose-response experiments showed that at these doses the different cell lines showed 40–50% apoptosis at ∼24 h. MNU was used at 2.5 mm for HeLa cells and MEF cells. After treatment, the cells were morphologically analyzed or harvested to collect cell lysates for various biochemical analyses.
BN-PAGE
BN-PAGE was performed to reveal protein complexes according to standard protocols (41, 42). Briefly, control or cisplatin-treated cells were lysed with a lysis buffer containing 0.2% Triton X-100, 0.2% dodecylmaltoside, 137 mm NaCl, 2 mm EDTA, 10% glycerol, 20 mm Tris-HCl, and protease and phosphatase inhibitors. The cell lysates were then dialyzed and loaded on a 4–15% gradient BN-PAGE. The samples in the wells were further overlaid with Coomassie Blue solution. After electrophoresis, the gel was subjected to Coomassie Blue staining or immunoblot analysis. To identify the ATR-interacting proteins, duplicate samples were run on a single BN-PAGE gel, half of which was used for immunoblot analysis of ATR, whereas the other half was stained with Coomassie Blue to excise the corresponding ATR complex band for mass spectrometry at the Genomics and Proteomics Core Facility of Medical College of Georgia.
shRNA Transfection
The cells were plated to reach 50–60% confluence for transfection with 1 μg of control shRNA or hMSH2 shRNA plasmid using Lipofectamine 2000TM reagent (Invitrogen). G418 was added 16 h later at a final concentration of 800 μg/ml for selection of stably transfected cells. The gene sequences used to design the shRNA are: GAGTTCCTGTCCAAGGTGAA for human hMSH2 and GGAATCTCATTCGATGCATAC for control.
Analysis of Apoptosis and Caspase Activation
Apoptosis was examined by morphological methods as described previously (33, 39, 40). Briefly, the cells were stained with Hoechst 33342 to record cellular morphology by phase contrast microscopy, and nuclear morphology was observed by fluorescence microscopy. The cells with typical apoptotic morphology (cellular shrinkage, nuclear condensation and fragmentation, and formation of apoptotic bodies) were counted to determine the percentage of apoptosis. To measure caspase activity, the cells were extracted with 1% Triton X-100 to collect lysate for an enzymatic reaction containing 50 μm carbobenzoxy-DEVD-7-amino-4-trifluoromethyl coumarin. The fluorescence reading from the enzymatic reaction was used to determine the caspase activity as described previously (33, 39, 40).
Co-immunoprecipitation
The cells were lysed with IP lysis buffer in the presence of protease and phosphatase inhibitors at 4 °C and then subjected to immunoprecipitation as described previously (33, 39). The immunoprecipitates were resuspended in SDS buffer for gel electrophoresis, followed by immunoblot analysis using specific antibodies against ATR and hMSH2.
Biochemical Analysis of Chromatin-associated Proteins
Small scale chromatin isolation was performed as described by others (43). Briefly, the cells were resuspended in buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, protease, and phosphatase inhibitors). Subsequently, 0.1% Triton X-100 was added to incubate the cells at 4 °C for 5–10 min followed by low speed centrifugation to collect nuclei. The nuclei were then washed with buffer A followed by lysis in buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, protease, and phosphatase inhibitors). Chromatin was collected by centrifugation, washed once with buffer B, and resuspended in SDS buffer for electrophoresis and immunoblot analysis.
Immunoblot Analysis
A standard protocol of immunoblot analysis was followed. Briefly, protein concentration was determined by using the bicinchoninic acid reagent (Pierce). Equal amounts (usually 20 μg) of protein were loaded in each lane for electrophoresis, followed by transferring onto polyvinylidene difluoride membranes. The blots were then incubated sequentially in blocking buffer, primary antibody, and the horseradish peroxidase-conjugated secondary antibody. Antigen signal was revealed by using the enhanced chemiluminescence kit from Pierce.
Dual Immunofluorescence Staining
The cells were grown on glass coverslips for indirect immunofluorescence as described previously (33, 39). Briefly, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.4% Triton X-100 in a blocking buffer. The cells were then exposed to two primary antibodies, followed by incubation with a mixture of FITC-labeled goat anti-rabbit and Cy3-labeled donkey anti-goat secondary antibodies. After washes, the coverslips were mounted on slides with Antifade for examination by confocal microscopy using Cy3 and FITC channels.
Statistics
Qualitative data and cell images are representatives of at least three experiments. Quantitative data were expressed as the means ± S.D. Statistical analysis was conducted using GraphPad Prism software. Statistical differences were calculated using Student's t test. p < 0.05 was considered significantly different.
RESULTS
hMSH2 Associates with ATR during Cisplatin Treatment
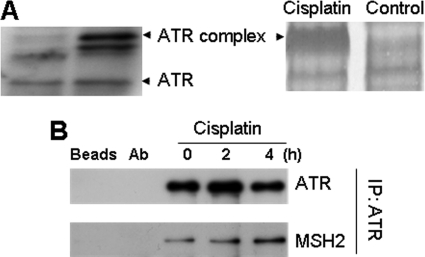
Cisplatin treatment activates ATR to trigger a signaling cascade, leading to apoptosis (33, 34); however, it is unclear how ATR is activated and regulated. To identify the regulators of ATR, we analyzed the proteins that associate with ATR during cisplatin treatment by BN-PAGE. Cell lysates were collected from control and cisplatin-treated cells for BN-PAGE. Immunoblot analysis of ATR following BN-PAGE showed free ATR and ATR complexes at higher molecular weight positions (Fig. 1A, left panel). Compared with control cell lysate, a major ATR complex was notably increased in the lysate of cisplatin-treated cells. This complex was excised from the BN-PAGE gel and subjected to mass spectrometry, which identified several proteins including ATR, ATRIP, and hMSH2, an MMR protein that has been implicated in ATR regulation (10–12). To confirm hMSH2-ATR interaction, we conducted co-immunoprecipitation assays. ATR was immunoprecipitated from untreated or cisplatin-treated cells, and the resultant immunoprecipitates were analyzed for the presence of hMSH2. As shown in Fig. 1B, cisplatin treatment induced hMSH2-ATR association in a time-dependent manner.
FIGURE 1.
hMSH2 associates with ATR during cisplatin treatment of HEK and HeLa cells. A, BN-PAGE analysis. HEK cells were treated with 50 μm cisplatin for 4 h to collect lysate for BN-PAGE followed by Coomassie Blue staining (right panel) or immunoblot analysis of ATR (left panel). The ATR-containing protein complex band that was induced during cisplatin treatment was exercised from the BN-PAGE gel for mass spectrometry. A major protein in this complex was identified to be hMSH2. B, co-IP analysis. HeLa cells were treated with 25 μm cisplatin for 0, 2, and 4 h to collect lysate for IP using a specific anti-ATR antibody. The precipitates were then analyzed for ATR and hMSH2 by immunoblot analysis using specific antibodies. Two controls were included in the co-IP analysis: Beads, protein A/G-agarose beads were incubated with cell lysates in the absence of anti-ATR antibody; Ab, protein A/G-agarose beads were incubated with anti-ATR antibody in the absence of cell lysates.
hMSH2-deficient Cells Are Resistant to Cisplatin-induced Apoptosis and DNA Damage Signaling
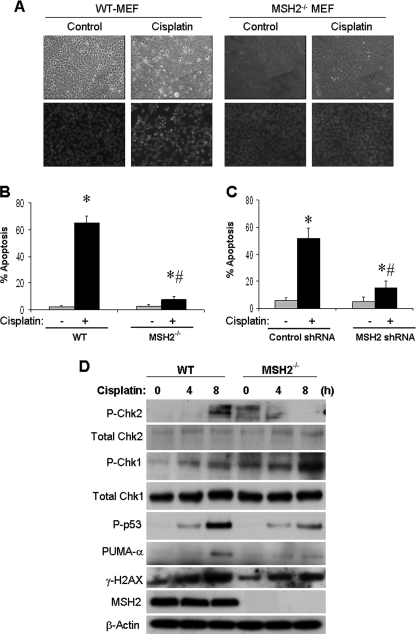
The presence of hMSH2 in ATR complex (Fig. 1) suggests that hMSH2 may regulate ATR and ATR-associated cellular response during cisplatin treatment. A notable cellular response following ATR activation during cisplatin treatment is apoptosis (30, 33–36). To determine the role of hMSH2 in cisplatin-induced apoptosis, wild-type and hMSH2-deficient (hMSH2−/−) MEF cells were incubated with 20 μm cisplatin treatment for 16 h (Fig. 2A). Many wild-type cells showed the typical morphology of apoptosis, including cellular shrinkage, formation of apoptotic bodies, and nuclear condensation and fragmentation. Significantly less apoptosis was induced by cisplatin in hMSH2−/− cells (Fig. 2B). Moreover, cisplatin induced a marked increase in caspase activity in wild-type cells, which was attenuated in hMSH2−/− cells (supplemental Fig. S1). In addition, cisplatin-induced apoptosis in HEK cells was suppressed by hMSH2 knockdown via shRNA (Fig. 2C and supplemental Fig. S2; partial hMSH2 knockdown via shRNA in HEK cells shown in supplemental Fig. S3). Taken together, these results suggest that hMSH2 plays an important role in apoptotic signaling during cisplatin treatment.
FIGURE 2.
hMSH2-deficient cells are resistant to cisplatin-induced apoptosis and DNA damage signaling. A, wild-type and hMSH2−/− MEF cells were left untreated (control) or treated with 20 μm cisplatin for 16 h. The cells were fixed and stained with Hoechst 33342 to record cell and nuclear morphologies. B, wild-type and hMSH2−/− MEF cells were treated as in A. The cells with apoptotic morphology were counted to determine the percentage of apoptosis. C, HEK cells were transfected with control or hMSH2 shRNA. The cells were then left untreated (−) or treated with 50 μm cisplatin (+) for 16 h. The cells were finally fixed and stained with Hoechst 33342 to count the cells with apoptotic morphology to determine the percentage of apoptosis. D, wild-type and hMSH2−/− MEF cells were treated with 20 μm cisplatin for 0, 4, or 8 h to collect whole cell lysate for immunoblot analysis of phosphorylated (threonine 68) Chk2, total Chk2, phosphorylated (serine 345) Chk1, total Chk1, phosphorylated p53, PUMA-α, phosphorylated H2AX (γ-H2AX), hMSH2, and β-actin (sample loading control). The data in B and C are expressed as the means ± S.D. (n = 4). *, p < 0.001 versus control; #, p < 0.001 versus cisplatin-treated wild-type cells.
A major signaling pathway leading to apoptosis during cisplatin treatment involves the sequential activation of ATR, Chk2, p53, and the consequent transcriptional up-regulation of pro-apoptotic genes such as PUMA-α (33). Based on the above observations of hMSH2/ATR association and its role in cisplatin-induced apoptosis, we hypothesized that hMSH2 contributes to the ATR-initiated DNA damage response during cisplatin treatment. To test this possibility, we initially compared cisplatin-induced Chk2 activation in wild-type and hMSH2−/− MEF cells. As shown in Fig. 2D, Chk2 was activated (phosphorylation at threonine 68) after 8 h of cisplatin treatment in wild-type cells, but not in hMSH2−/− cells, suggesting that hMSH2 is required for ATR-mediated Chk2 activation under these conditions. In contrast, cisplatin induced Chk1 activation (phosphorylation at Serine-345) in both wild-type and hMSH2−/− cells (Fig. 2D), suggesting that Chk1 activation during cisplatin treatment is hMSH2-independent. We further showed that cisplatin-induced p53 activation (phosphorylation at serine 15) was abrogated in hMSH2−/− cells, and downstream of p53, PUMA-α induction by cisplatin was also ameliorated in hMSH2−/− cells (Fig. 2D). Furthermore, cisplatin-induced Chk2 and p53 activation and PUMA-α expression were suppressed by partial hMSH2 knockdown in HEK cells (supplemental Fig. S3). Together the results suggest that hMSH2 may interact with ATR and participate in ATR activation during cisplatin treatment, leading to a rapid DNA damage response that results in apoptosis.
hMSH2 Is Required for ATR Accumulation to DNA Damage Sites during Cisplatin Treatment
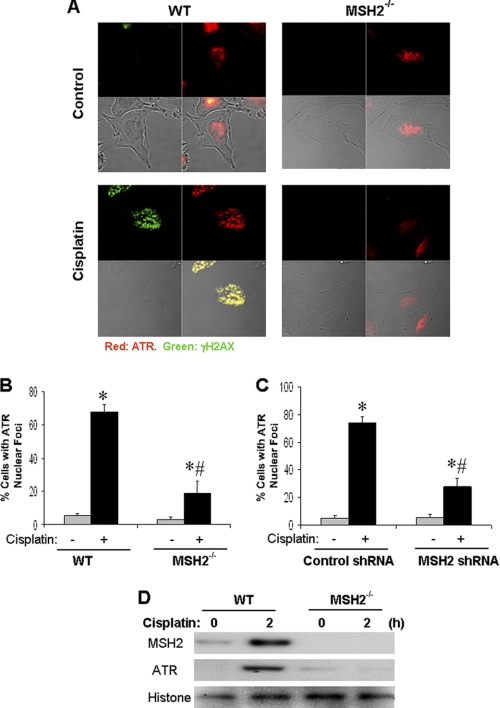
To understand how hMSH2 regulates ATR, we focused on ATR accumulation to nuclear foci or DNA damage sites, a key regulatory step in ATR activation (1, 8). In control cells, ATR staining was diffuse in the nucleus (Fig. 3A). After cisplatin treatment, ATR formed nuclear foci in wild-type cells and co-localized with γH2AX, a DNA damage response protein. However, the accumulation of ATR and γH2AX to nuclear foci was significantly diminished in hMSH2−/− cells (Fig. 3A). To quantify these results, we counted the cells with ATR nuclear foci. As shown in Fig. 3B, 68% of wild-type cells showed ATR nuclear foci, whereas only 22% of hMSH2−/− cells showed ATR nuclear foci after cisplatin treatment.
FIGURE 3.
hMSH2 is required for ATR recruitment to nuclear foci during cisplatin treatment. A, dual immunofluorescence of ATR and γ-H2AX. Wild-type and hMSH2−/− MEF cells were either left untreated or treated with 20 μm cisplatin for 6 h. The cells were then fixed for dual immunofluorescence using antibodies to ATR (red) and γ-H2AX (green). B, cells were treated as in A. The ATR staining was evaluated to determine the percentage of cells with ATR nuclear foci. C, HEK cells were transfected with control or hMSH2 shRNA. The cells were then left untreated (−) or treated with 50 μm cisplatin (+) for 6 h. The cells were fixed for ATR and γ-H2AX dual immunofluorescence to determine the percentage of cells with ATR nuclear foci. D, wild-type and hMSH2−/− cells were untreated or treated with cisplatin for 2 h to isolate the chromatin fraction for immunoblot analysis. The results are representative of at least three separate experiments.
We further examined ATR nuclear foci formation in hMSH2 knockdown HEK cells. Cisplatin treatment for 6 h induced ATR nuclear foci in 74% of control shRNA-transfected cells but in only 28% hMSH2 shRNA-transfected cells (Fig. 3C and supplemental Fig. S4). To validate the immunostaining results, chromatin was isolated to analyze chromatin-bound ATR by immunoblot analysis. After 2 h of cisplatin treatment, a marked association of both ATR and hMSH2 with the chromatin was observed in wild-type cells, which was blocked in hMSH2−/− cells (Fig. 3D). The results suggest that hMSH2 is required for the recruitment of ATR to nuclear foci during cisplatin treatment to trigger DNA damage signaling.
hMSH2 Recruits ATR to Nuclear Foci Independently of 9-1-1, Rad17, and RPA
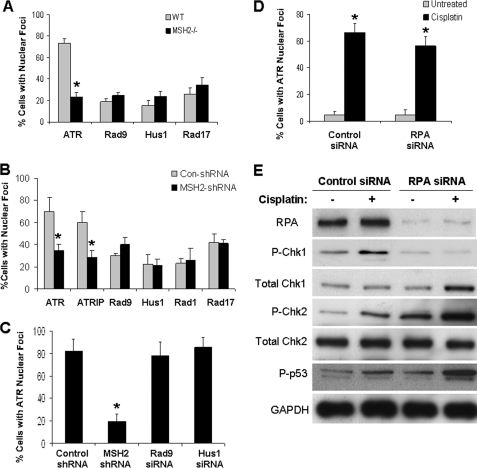
In the canonical pathway of ATR activation, the single-stranded DNA-coating protein RPA binds and recruits ATR/ATRIP to processed DNA damage sites, whereas Rad17 loads the 9-1-1 complex. 9-1-1 recruits TopBP1, which then interacts and activates ATR (1, 5, 8). A role for hMSH2 in this pathway is unclear. To address this, we first determined whether hMSH2 mediates Rad17 or 9-1-1 loading to nuclear foci during cisplatin treatment. In wild-type cells, over 70% of the cells formed ATR nuclear foci during cisplatin treatment, but only 20–30% displayed Rad9/Hus1/Rad17 nuclear foci (Fig. 4A and supplemental Fig. S5). Importantly, the formation of ATR nuclear foci, but not Rad9/Hus1/Rad17 nuclear foci, was suppressed in hMSH2−/− cells (Fig. 4A and supplemental Fig. S5). Moreover, knockdown of hMSH2 in HEK cells did not block the formation of 9-1-1 and Rad17 nuclear foci during cisplatin treatment, although the formation of ATR/ATRIP nuclear foci was attenuated (Fig. 4B). These results suggest that hMSH2, although essential for ATR recruitment, does not mediate Rad17 or 9-1-1 loading to the DNA damage sites under the experimental condition. We then determined the role of 9-1-1 proteins in cisplatin-induced ATR nuclear foci formation. Knockdown of Rad9 and Hus1 (confirmed in supplemental Fig. S6) had little effect on ATR nuclear foci formation, whereas a marked inhibition was demonstrated for hMSH2 knockdown (Fig. 4C). These data suggest that hMSH2 might regulate ATR activation in a 9-1-1 complex-independent manner.
FIGURE 4.
MSH recruits ATR independently of RPA, Rad17, and 9-1-1. A, wild-type and hMSH2−/− cells were treated with 20 μm cisplatin for 4 h and processed for immunofluorescence staining of ATR, Rad9, Hus1, and Rad17. B, HEK cells transfected with control (Con-shRNA) or hMSH2 shRNA were treated with 50 μm cisplatin for 4 h and processed for immunofluorescence of ATR, ATRIP, Rad9, Hus1, Rad1, and Rad17. C, HEK cells were transfected with control shRNA, hMSH2 shRNA, Rad9 siRNA, or Hus1 siRNA. The cells were then treated with 50 μm cisplatin for 4 h and processed for immunofluorescence of indicated proteins. D, HEK cells were transfected with control siRNA or RPA siRNA. The cells were then left untreated or treated with 50 μm cisplatin for 4 h and processed for immunofluorescence of ATR. E, HEK cells were transfected with control siRNA or RPA siRNA. The cells were then left untreated or treated with 50 μm cisplatin for 4 h. Whole cell lysate was collected for immunoblot analysis of indicated proteins. The data in A–C are expressed as the means ± S.D. (n = 3, >200 cells evaluated for each condition). *, p < 0.001 versus wild-type cells in A and B, p < 0.001 versus control shRNA transfected cells in C, and p < 0.001 versus untreated cells in D.
In the canonical pathway of ATR activation, RPA is the direct ATR/ATRIP recruiter (1, 5, 8). We therefore determined whether hMSH2-mediated ATR activation involves RPA. Despite some marginal effects, RPA knockdown did not abrogate cisplatin-induced ATR foci formation in HEK cells (Fig. 4D). We then collected cell lysates to analyze DNA damage signaling (Fig. 4E). RPA knockdown (>90%) by siRNA in this experiment was first confirmed (Fig. 4E). Notably, cisplatin-induced Chk1 activation was attenuated in RPA knockdown cells. However, in these cells Chk2 and p53 activation was not blocked; rather it appeared enhanced (Fig. 4E). Because both Chk1 and Chk2 depend on ATR for activation in this experimental model (33), the results suggest the existence of two ATR activation pathways, one of which is mediated by RPA and relays a signal to Chk1, whereas the other depends on hMSH2 and is responsible for Chk2 activation (see “Discussion”).
Role of hMSH2 in ATR Recruitment, Activation, and Signaling during MNU Treatment
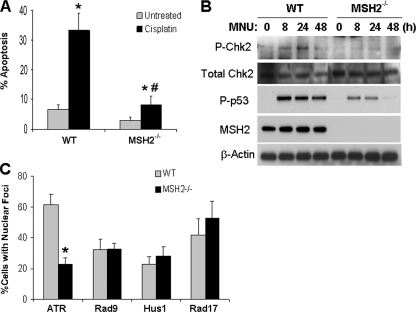
To determine whether hMSH2 regulates ATR in other DNA damage models, we examined the effects of MNU, a DNA methylating agent (44). We first monitored MNU-induced apoptosis in wild-type and hMSH2−/− MEF cells. MNU treatment induced over 30% apoptosis in wild-type cells within 72 h, but only 6% apoptosis in hMSH2−/− cells, supporting a role for hMSH2 in the DNA damage signaling of MNU (Fig. 5A). Moreover, MNU-induced Chk2 and p53 phosphorylation or activation was suppressed in hMSH2−/− cells (Fig. 5B). We further examined ATR accumulation to DNA damage sites. MNU treatment induced ATR nuclear foci in 61% of wild-type cells, which was suppressed to 22% in hMSH2−/− cells (Fig. 5C). In sharp contrast, the formation of Rad9, Hus1, and Rad17 nuclear foci was not affected by hMSH2 deficiency (Fig. 5C).
FIGURE 5.
hMSH2 recruits ATR for activation during MNU treatment. A, wild-type and hMSH2−/− cells were left untreated or treated with 2.5 mm MNU for 72 h. The cells with typical apoptotic morphology were counted to determine the percentage of apoptosis. B, Wild-type and hMSH2−/− cells were treated with 2.5 mm MNU for 0–48 h to collect whole cell lysate for immunoblot analysis of phosphorylated (threonine 68) Chk2, total Chk2, phosphorylated (serine 15) p53, hMSH2, and β-actin. C, wild-type and hMSH2−/− cells were left untreated or treated with 2.5 mm MNU for 12 h. The cells were then processed for immunofluorescence analysis of ATR, Rad9, Hus1, and Rad17. The staining of these proteins was evaluated to determine the percentage of cells with nuclear foci formation for each of the proteins. The data in A and C are expressed as the means ± S.D. (n = 3). In A, *, p < 0.001 versus untreated cells; #, p < 0.001 versus cisplatin-treated wild-type cells. In C, *, p < 0.001 versus cisplatin-treated wild-type cells.
DISCUSSION
Cisplatin and related platinum compounds are widely used chemotherapy drugs for the treatment of solid tumors (26–29). A major mechanism of their therapeutic effects is mediation by cross-linking DNA, leading to stalled or collapsed replication forks, resulting in a rapid DDR and apoptosis (26–29). In the DDR, ATR acts as an upstream signaling trigger, which phosphorylates and activates Chk1 and Chk2 to further activate p53, resulting in the expression of apoptotic genes (e.g. PUMA-α) and apoptosis (26–29, 33). The present study has identified hMSH2, a MMR protein, as a key regulator of ATR during cisplatin-induced DDR and apoptosis. We show that during cisplatin treatment, hMSH2 binds ATR and recruits it to the DNA damage site for activation. This finding provides new insights into the molecular basis of the chemotherapeutic effects of cisplatin and related platinum compounds.
MMR proteins, including hMSH2, have been implicated in DDR-associated apoptosis (19–21). Our present study, using both hMSH2 knock-out and knockdown cells, provides further support for this role of MMR proteins as DNA damage sensors. Moreover, we show that hMSH2 contributes to apoptosis by regulating ATR/Chk2/p53 signaling. As a result, in the cells with complete loss or reduced hMSH2 protein, cisplatin- and MNU-induced ATR/Chk2/p53 activation was ameliorated, which was accompanied by the suppression of pro-apoptotic gene expression and lower apoptosis (Figs. 2 and 5). Thus, although both p53-dependent and -independent mechanisms may underlie the apoptotic resistance of MMR-deficient cells (20, 45, 46), in our study the effect appears largely p53-dependent.
Two models have been proposed to account for the MMR involvement in DNA damage signaling and apoptosis (13, 17, 19, 47). The futile cycle model emphasizes DNA repair as the single function of MMR. According to this model, a futile attempt of the MMR system to repair damaged DNA creates strand breaks, leading to DDR and apoptosis. The direct signaling model, however, proposes two distinct functions for MMR: DNA repair and DNA damage signaling. In this model, MMR proteins may directly mediate DNA damage signaling to result in cell death. Apparently, these two models are not mutually exclusive and are both supported by good experimental evidence (13, 17, 20, 21, 47). Our results suggest that during cisplatin and MNU treatment, hMSH2 directly participates in ATR recruitment and activation, leading to DNA damage signaling followed by apoptosis. These observations not only support a direct role for MMR proteins in DNA damage signaling but have also pinpointed ATR as a key regulatory target.
In 2003, Wang and Qin (10) demonstrated hMSH2/ATR interaction during DNA damage induced by the methylating agent N-methyl-N′-nitro-N-nitrosoguanidine. Interestingly, in this model hMSH2/ATR was suggested to initiate two different signaling pathways for Chk1 and SMC1 (structure maintenance of chromosome) phosphorylation (10). Using an in vitro assay, Yoshioka et al. (11) further showed that the binding of ATR to mismatched DNA sequences requires MMR proteins. The latest study by Liu et al. (12) used nuclear extracts and recombinant proteins to systematically analyze the molecular interaction between MMR proteins and checkpoint proteins. MMR proteins were shown to directly interact with ATR, TopBP1, and Chk1 (12). Our present study demonstrates ATR/hMSH2 interaction during cisplatin-induced DNA damage by both BN-PAGE and co-IP analysis (Fig. 1). Importantly, we show that hMSH2 is critical for ATR recruitment to nuclear foci or DNA damage sites, a key event for ATR activation (1, 5, 8).
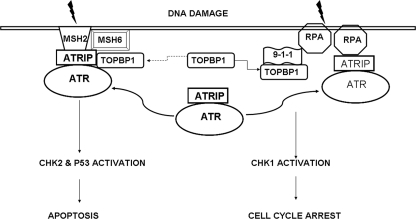
ATR activation by DNA damage depends on its recruitment to DNA damage sites and subsequent interaction with proteins at the nuclear foci (1, 5, 8). In the canonical pathway, ATR activation involves two protein complexes: the ATR/ATRIP complex and the 9-1-1 complex. During DNA damage, ATR/ATRIP is recruited to the DNA damage site via RPA, which also directs the loading of 9-1-1 via Rad17-RFC. The loading of the 9-1-1 complex brings TopBP1 (an ATR activator) close to ATR for its activation (8). Although this pathway is widely appreciated, alternative pathways of ATR recruitment and activation may exist (8, 12). Our results show that hMSH2 recruits ATR to nuclear foci for activation, and this might be independent of 9-1-1 complex and RPA proteins. Interestingly, although knockdown of hMSH2 in HEK cells diminished cisplatin-induced ATR accumulation and activation in nuclear foci, knockdown of RAD9 and HUS1 was without effect (Fig. 4C). Moreover, hMSH2 deficiency in MEF and HEK293 cells resulted in decreased ATR nuclear foci formation, without affecting the translocation of Rad17 and 9-1-1 proteins (Fig. 4, A and B). Finally, knockdown of RPA only had marginal inhibitory effects on ATR nuclear foci formation, further suggesting ATR activation by a RPA/9-1-1-independent mechanism (Fig. 4D). Interestingly, in the RPA knockdown cells, the activation of Chk2 and p53 was largely preserved, but the activation of Chk1 was almost completely attenuated (Fig. 4E). Based on these observations, we speculate that there are two pathways of ATR activation (Fig. 6). One is the canonical pathway, involving RPA, Rad17, and 9-1-1, which leads to Chk1 activation. The other pathway is initiated by hMSH2, which recruits and activates ATR, resulting in Chk2 and p53 activation. Of note, these two pathways may lead to different cellular responses. For example, in our study, hMSH2 deficiency made the cells more resistant to apoptosis (Fig. 2), whereas knockdown of RPA and Rad9 sensitized the cells to apoptosis during cisplatin treatment (supplemental Fig. S7). Whether one or both pathways are activated may depend on the type and extent of DNA damage. Further dissection of the ATR activation pathways and delineation of their potential cross-talk may shed significant new light on the cellular response of DNA damage and lead to refined therapeutic protocols.
FIGURE 6.
RPA/9-1-1- and hMSH2-mediated ATR activation in DNA damage signaling. DNA damage signaling involving ATR activation may take two pathways, which are respectively initiated by RPA/9-1-1 and hMSH2. RPA/9-1-1 and hMSH2 separately recruit ATR/ATRIP to DNA damage sites for ATR activation, leading to the activation of Chk1 and Chk2, culminating in cell cycle arrest and apoptosis. TopBP1 is required for ATR activation by the RPA/9-1-1 pathway; however, its role in hMSH2-mediated ATR activation is unclear (dashed line).
This work was supported, in whole or in part, by National Institutes of Health Grants DK087843 and DK058831 (to Z. D.) and CA067007 and GM080176 (to R. F.). This work was also supported by funds from the Department of Veterans Affairs (to Z. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- DDR
- DNA damage response
- 9-1-1
- Rad9-Hus1-Rad1
- RPA
- replication protein A
- MMR
- mismatch repair
- MSH
- MutS homologues
- Chk2
- Checkpoint kinase 2
- BN
- blue native
- MNU
- N-methyl-N-nitrosourea
- MEF
- mouse embryonic fibroblasts
- TopBP1
- topoisomerase-binding protein-1
- IP
- immunoprecipitation.
REFERENCES
- 1. Harper J. W., Elledge S. J. (2007) Mol. Cell. 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 2. Bartek J., Bartkova J., Lukas J. (2007) Oncogene 26, 7773–7779 [DOI] [PubMed] [Google Scholar]
- 3. Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 4. Jackson S. P., Bartek J. (2009) Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 6. Abraham R. T. (2004) DNA Repair 3, 883–887 [DOI] [PubMed] [Google Scholar]
- 7. Hurley P. J., Bunz F. (2007) Cell Cycle 6, 414–417 [DOI] [PubMed] [Google Scholar]
- 8. Cimprich K. A., Cortez D. (2008) Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shiloh Y. (2003) Nat. Rev. Cancer 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y., Qin J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15387–15392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshioka K., Yoshioka Y., Hsieh P. (2006) Mol. Cell 22, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Y., Fang Y., Shao H., Lindsey-Boltz L., Sancar A., Modrich P. (2010) J. Biol. Chem. 285, 5974–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiricny J. (2006) Nat. Rev. Mol. Cell Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]
- 14. Li G. M. (2008) Cell Res. 18, 85–98 [DOI] [PubMed] [Google Scholar]
- 15. Modrich P. (2006) J. Biol. Chem. 281, 30305–30309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jun S. H., Kim T. G., Ban C. (2006) FEBS J. 273, 1609–1619 [DOI] [PubMed] [Google Scholar]
- 17. O'Brien V., Brown R. (2006) Carcinogenesis 27, 682–692 [DOI] [PubMed] [Google Scholar]
- 18. Kolodner R. D., Marsischky G. T. (1999) Curr. Opin. Genet. Dev. 9, 89–96 [DOI] [PubMed] [Google Scholar]
- 19. Fishel R. (1999) Nat. Med. 5, 1239–1241 [DOI] [PubMed] [Google Scholar]
- 20. Gong J. G., Costanzo A., Yang H. Q., Melino G., Kaelin W. G., Jr., Levrero M., Wang J. Y. (1999) Nature 399, 806–809 [DOI] [PubMed] [Google Scholar]
- 21. Zhang H., Richards B., Wilson T., Lloyd M., Cranston A., Thorburn A., Fishel R., Meuth M. (1999) Cancer Res. 59, 3021–3027 [PubMed] [Google Scholar]
- 22. Aebi S., Kurdi-Haidar B., Gordon R., Cenni B., Zheng H., Fink D., Christen R. D., Boland C. R., Koi M., Fishel R., Howell S. B. (1996) Cancer Res. 56, 3087–3090 [PubMed] [Google Scholar]
- 23. Meyers M., Theodosiou M., Acharya S., Odegaard E., Wilson T., Lewis J. E., Davis T. W., Wilson-Van Patten C., Fishel R., Boothman D. A. (1997) Cancer Res. 57, 206–208 [PubMed] [Google Scholar]
- 24. Mello J. A., Acharya S., Fishel R., Essigmann J. M. (1996) Chem. Biol. 3, 579–589 [DOI] [PubMed] [Google Scholar]
- 25. Guggenheim E. R., Xu D., Zhang C. X., Chang P. V., Lippard S. J. (2009) ChemBioChem 10, 141–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang D., Lippard S. J. (2005) Nat. Rev. Drug Discov. 4, 307–320 [DOI] [PubMed] [Google Scholar]
- 27. Cepeda V., Fuertes M. A., Castilla J., Alonso C., Quevedo C., Pérez J. M. (2007) Anticancer Agents Med. Chem. 7, 3–18 [DOI] [PubMed] [Google Scholar]
- 28. Einhorn L. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4592–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siddik Z. H. (2003) Oncogene 22, 7265–7279 [DOI] [PubMed] [Google Scholar]
- 30. Pabla N., Dong Z. (2008) Kidney Int. 73, 994–1007 [DOI] [PubMed] [Google Scholar]
- 31. Arany I., Safirstein R. L. (2003) Semin. Nephrol. 23, 460–464 [DOI] [PubMed] [Google Scholar]
- 32. Price P. M., Safirstein R. L., Megyesi J. (2009) Kidney Int. 76, 604–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pabla N., Huang S., Mi Q. S., Daniel R., Dong Z. (2008) J. Biol. Chem. 283, 6572–6583 [DOI] [PubMed] [Google Scholar]
- 34. Jiang M., Dong Z. (2008) J. Pharmacol. Exp. Ther. 327, 300–307 [DOI] [PubMed] [Google Scholar]
- 35. Jiang M., Wei Q., Wang J., Du Q., Yu J., Zhang L., Dong Z. (2006) Oncogene 25, 4056–4066 [DOI] [PubMed] [Google Scholar]
- 36. Seth R., Yang C., Kaushal V., Shah S. V., Kaushal G. P. (2005) J. Biol. Chem. 280, 31230–31239 [DOI] [PubMed] [Google Scholar]
- 37. McIlhatton M. A., Tyler J., Burkholder S., Ruschoff J., Rigas B., Kopelovich L., Fishel R. (2007) Cancer Res. 67, 10966–10975 [DOI] [PubMed] [Google Scholar]
- 38. Jiang M., Pabla N., Murphy R. F., Yang T., Yin X. M., Degenhardt K., White E., Dong Z. (2007) J. Biol. Chem. 282, 2636–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brooks C., Wei Q., Feng L., Dong G., Tao Y., Mei L., Xie Z. J., Dong Z. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11649–11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brooks C., Wei Q., Cho S. G., Dong Z. (2009) J. Clin. Invest. 119, 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Swamy M., Siegers G. M., Minguet S., Wollscheid B., Schamel W. W. (2006) Sci. STKE 2006, pl4. [DOI] [PubMed] [Google Scholar]
- 42. Wittig I., Braun H. P., Schägger H. (2006) Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 43. Méndez J., Stillman B. (2000) Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wyatt M. D., Pittman D. L. (2006) Chem. Res. Toxicol. 19, 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Topping R. P., Wilkinson J. C., Scarpinato K. D. (2009) J. Biol. Chem. 284, 14029–14039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Toft N. J., Winton D. J., Kelly J., Howard L. A., Dekker M., te Riele H., Arends M. J., Wyllie A. H., Margison G. P., Clarke A. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3911–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang J. Y., Edelmann W. (2006) Cancer Cell 9, 417–418 [DOI] [PubMed] [Google Scholar]