Abstract
Pancreatic β-cell-restricted expression of insulin is established through several critical cis-regulatory elements located in the insulin gene promoter region. The principal cis elements are A-boxes, E1, and C1/RIPE3b. The β-cell-enriched transcription factors Pdx1 and Beta2 bind to the A-boxes and E1 element, respectively. A β-cell-specific trans-acting factor binding to C1/RIPE3b (termed RIPE3b1 activator) was detected by electrophoretic mobility shift assay and has been identified as MafA, a member of the Maf family of basic leucine zipper (bZip) proteins. Here, ATF2, a member of the ATF/CREB family of basic leucine zipper proteins, was identified as a component of the RIPE3b1 activator. ATF2 alone was unable to bind to the C1/RIPE3b element but acquired binding capacity upon complex formation with MafA. ATF2 also interacted with Pdx1 and Beta2, and co-expression of ATF2, MafA, Pdx1, and Beta2 resulted in a synergistic activation of the insulin promoter. Immunohistochemical analysis of mouse pancreas tissue sections showed that ATF2 is enriched in islet endocrine cells, including β-cells. RNAi-mediated knockdown of MafA or ATF2 in the MIN6 β-cell line resulted in a significant decrease in endogenous levels of insulin mRNA. These data indicate that ATF2 is an essential component of the positive regulators of the insulin gene expression.
Keywords: Gene Expression, Insulin, Pancreatic Islet, Transcription Factors, Transcription Regulation
Introduction
Islet β-cell-restricted expression of the insulin gene is established through several critical cis-regulatory elements located within a 300-bp region of the insulin gene promoter. Numerous studies have identified trans-acting factors that bind to these elements and revealed that β-cell-enriched and ubiquitously expressed factors control β-cell-restricted expression of insulin.
The principal cis-elements involved in regulating β-cell-restricted expression are A-boxes (also known as A1, A3, and GG2 elements in humans) and the E1 and C1/RIPE3b elements (1). A-boxes are AT-rich nucleotide sequence motifs that serve as binding sites for the β-cell-enriched homeodomain transcription factor Pdx1/IPF1/STF1 (2–4). The E1 element is regulated by the β-cell-enriched basic helix-loop-helix factors Beta2/NeuroD, which form a heterodimeric complex with ubiquitously expressed E47 (5). The C1-binding activity in β-cell nuclear extracts, termed RIPE3b1 activator, was identified by EMSA (6, 7), and a β-cell-specific transcription factor, MafA, was subsequently identified as one of the components (8–10). MafA is a member of the Maf family of basic leucine zipper (bZip)3 transcription factors and is expressed exclusively in β-cells in the pancreas (10, 11).
The functions of these β-cell-enriched factors have been studied by detailed expression analysis during development and by use of gene knockouts in mice. For example, MafA expression begins at around embryonic day 13.5 in mice, coinciding with the process of β-cell maturation, and persists until adulthood (12). In mafA knockout mice, islets are normal at birth (12), most likely because of the compensatory action of MafB, which is also expressed in β-cells but at an earlier stage (13). However, upon aging, mafA knockout mice exhibit a deficiency in glucose-stimulated insulin secretion from β-cells and progressive β-cell degeneration by 8–12 weeks of age (12). These observations indicate that MafA regulates maturation, functional maintenance, and survival of β-cells. Biochemical studies have demonstrated that Pdx1, Beta2, and MafA activate the insulin promoter in a synergistic manner by binding to their respective binding sites (14–17). It was also shown that MafA physically interacts with Pdx1 and Beta2 (14).
In this study, we demonstrated that ATF2 (activating transcription factor 2), a member of the ATF/CREB family of bZip proteins, is a component of the RIPE3b1 activator. ATF2 is known to bind to the cAMP-responsive element (CRE) as a homodimer or heterodimer with other bZip proteins, of which c-Jun is the best characterized (for review see Ref. 18). ATF2 is ubiquitously expressed and is involved in a wide variety of cellular processes, including cell cycle progression, apoptosis, cytokine signaling, and genotoxic stress response. Previous studies have shown that ATF2 is involved in the regulation of insulin expression through binding to CRE-like elements in the promoter region (19, 20). Here, we show that, in contrast to binding to CRE, ATF2 alone is incapable of binding to the C1/RIPE3b element, but it acquires binding capacity through complex formation with MafA. ATF2 also interacted with Pdx1 and Beta2, and co-expression of ATF2, MafA, Pdx1, and Beta2 resulted in a synergistic activation of the insulin promoter. RNAi-mediated knockdown of ATF2 in the β-cell-derived cell line MIN6 resulted in a significant decrease in endogenous levels of insulin mRNA. Our data indicate an essential role of ATF2 in the regulation of the insulin gene expression.
EXPERIMENTAL PROCEDURES
EMSA
The preparation of nuclear extracts of MIN6 (a generous gift from Dr. J.-i. Miyazaki, Osaka University), In1024, and NIH3T3 cells and the EMSA have been described previously (9). The human insulin C1/RIPE3b probe was described previously (9). The probe containing the CRE of the human corticotropin-releasing hormone promoter was generated by annealing two separate oligonucleotides: 5′-TCGTTGACGTCACCAA-3′ and 5′-TTGGTGACGTCAACGA-3′. Antisera used in the EMSA were as follows: anti-CREB/ATF-1 (CREB-1, C-21), anti-ATF2 (N-96), anti-ATF3 (C-19), anti-ATF4 (CREB-2, C-20), anti-MafB (P-20) (Santa Cruz Biotechnology, Inc.), anti-HA (MBL), and anti-FLAG (M2, Sigma). Anti-MafB (P-20) cross-reacts with MafA and was used to detect MafA in MIN6 nuclear extracts, as MafA is the major Maf family member expressed in MIN6 cells.
Immunohistochemistry
Sections of 5-week-old mouse pancreas tissue were incubated with the indicated primary antibodies: anti-ATF2 (N-96), anti-insulin A (C-12), anti-glucagon (N-17), anti-somatostatin (D-20) (Santa Cruz Biotechnology, Inc.), anti-pancreatic polypeptide (Linco Research, Inc.), and anti-MafA (9). Immunoreactive complexes were visualized with Alexa Fluor 488- or Alexa Fluor 594-conjugated secondary antibodies (Invitrogen). Experiments were performed according to the Guidelines for the Care and Use of Laboratory Animals of the Nara Institute of Science and Technology.
Immunoprecipitation
pHygEF2/HA-m-mafA and pHygEF2/HA-h-mafB were described previously (21, 22). To construct pHygEF2/FLAG-ATF2, a cDNA fragment containing the entire open reading frame of mouse ATF2 was amplified from total RNA isolated from MIN6 cells by RT-PCR using the following primers: 5′-AAAGGATCCATGGACTACAAGGATGACGATGACAAGGATATCATGAGTGATGACAAACCCTTTC-3′ and 5′-AAAGCGGCCGCTCAACTTCCTGAGGGCTGTG-3′. The amplified fragment was inserted into the BamHI-NotI sites of the pHygEF2 plasmid. To construct the pHygEF2/ATF2-HA plasmid, the FLAG tag was deleted from pHygEF2/FLAG-ATF2 by BamHI-EcoRV digestion and blunt-end self-ligation, and a HA-encoding double-stranded oligonucleotide, 5′-TCAGGAAGTTACCCATACGATGTTCCAGATTACGCATAGC-3′ and 5′-GGCCGCTATGCGTAATCTGGAACATCGTATGGGTAACTTCC-3′), was inserted into the Bsu36I-NotI sites.
An expression vector for enhanced green fluorescent protein (EGFP) fused to the bZip region of MafA (pHygEF2/EGFP-mafA) was constructed by inserting a BamHI-BsrGI fragment of pEGFP-1 (Clontech Lab) containing entire open reading frame of EGFP and a PmlI-NotI fragment of pHygEF2/HA-m-mafA encoding amino acids 227–359 of MafA into the BamHI-NotI sites of pHygEF2. The amino acid substitution mutants ATF2-L23P and EGFP-MafA-L23P were constructed by site-directed overhang extension PCR mutagenesis (23).
HeLa cells grown in 6-well plates were transfected with a total of 3.2 μg of plasmid using 8 μl of Lipofectamine 2000 reagent (Invitrogen). Whole cell extracts were prepared by cell lysis in 800 μl of NETN buffer (150 mm NaCl, 1 mm EDTA, 10 mm Tris-HCl (pH 7.5), 0.1% Nonidet P-40) containing protease inhibitor mixture (Nacalai Tesque, Inc.). Aliquots of the extracts were subjected to immunoprecipitation using anti-HA-agarose (Roche) or anti-FLAG-agarose (Sigma), followed by immunoblot analysis using anti-HA, anti-FLAG, or anti-GFP (Clontech Lab) antibodies as described previously (9).
GST Pull-down Assay
To generate an expression vector for recombinant MafA fused to GST (GST-MafA), a DNA fragment encoding amino acids 227–359 of mouse MafA was excised from pGEM-T-easy/m-mafA (9) by digestion with PmlI and ApaI and then inserted into the BamHI site of pGEX-5X-3 (GE Healthcare) by blunt-end ligation to create pGEX-5X-3/mafA. GST alone and GST-MafA were expressed in Escherichia coli and purified using glutathione-Sepharose 4B (GE Healthcare).
For in vitro transcription, a plasmid (pBS-II-SK(+)/FLAG-ATF2) containing the cDNA for FLAG-ATF2 was constructed by inserting a BamHI-NotI DNA fragment excised from pHygEF2/FLAG-ATF2 into the BamHI-NotI sites of pBS-II-SK(+). DNA fragments containing the entire open reading frames for ATF7 or CRE-BPa were excised from pact/FLAG-ATF7 or pact/CRE-BPa (generous gifts from Dr. S. Ishii, RIKEN Tsukuba Institute) and then inserted into pBS-II-SK(+). The plasmids were linearized by NotI digestion and subjected to an in vitro transcription reaction using the MEGAscript kit (Ambion). The synthesized RNAs were purified and subjected to in vitro translation using wheat germ extract (Promega) in the presence of [35S]methionine. Programmed extracts (8 μl) were mixed into 800 μl of PBS containing 1% BSA and 0.25% Tween 20 and then subjected to precipitation with GST- or GST-MafA-immobilized glutathione-Sepharose 4B.
Luciferase Assay
The luciferase reporter plasmid pGL4/h-ins-p (WT or mut C1), was constructed by inserting a KpnI-HindIII fragment of the pGL2-based h-ins-p-luc plasmid (WT or mut C1) (9) into pGL4.10 (Promega). The core-enh/TATA-luc plasmid was constructed by inserting a double-stranded oligonucleotide (5′-gatcCACCAGGGAAATGGTCCGGAAATTGCAGCCTCAGCCCCCAGCCATCTGCCGA-3′ and 5′-gatcTCGGCAGATGGCTGGGGGCTGAGGCTGCAATTTCCGGACCATTTCCCTGGTG-3′) spanning the human insulin core enhancer elements (-151 to −100 bp) into pRBGP-luc (24). pHygEF2/FLAG-pdx1, pHygEF2/FLAG-beta2, and pEF-Rluc were described previously (9, 25).
HeLa cells grown in 24-well plates were transfected with a total of 800 ng of plasmid DNA using 2 μl of Lipofectamine 2000 reagent (Invitrogen). Where indicated, 10 pmol of a small siRNA targeting human atf2 (Invitrogen) was also included in the transfection mixture. Data represent the averages ± S.E. of two or three independent experiments.
RNAi and Real-time PCR Analysis
MIN6 cells grown in 24-well plates were transfected with a total of 20 pmol synthetic RNA duplex using Lipofectamine 2000 reagent (Invitrogen). The sequences of the oligonucleotides used for targeting mouse mafA were as follows: mafA-#1, 5′-r(CCAUCGAGUACGUCAACGA)dTdT-3′ and 5′-r(UCGUUGACGUACUCGAUGG)dTdT-3′; and mafA#2, 5′-r(GAGGAUCUGUACUGGAUGA)dTdT-3′ and 5′-r(UCAUCCAGUACAGAUCCUC)dTdT-3′. Synthetic RNAs targeting mouse atf2 were purchased from Invitrogen (Atf2-MSS202229 and Atf2-MSS202231). As for a negative control, RNA duplexes targeting GFP were used: 5′-r(GCAAGCUGACCCUGAAGUUC)dTdT-3′; and 5′-r(GAACUUCAGGGUCAGCUUGC)dTdT-3′. Twenty-four hours after transfection, total RNA was prepared using TRIzol reagent (Invitrogen) and then subjected to reverse transcription using oligo-dT primer. The resultant cDNAs were analyzed using a LightCycler 480 system and SYBR Green master mix reagent (Roche Applied Science). The following primer sets were used: insulin-1, 5′-GTGACCAGCTATAATCAGAGGACCA-3′ and 5′-GAGGTGGGCCTTAGTTGCAGTAGTT-3′; insulin-2, 5′-CAAGCAGGAAGCCTATCTTCCAGGT-3′ and 5′-GTTTTATTCATTGCAGAGGGGTAGG-3′; and GAPDH, 5′-GGTGAAGGTCGGTGTGAACGGATTT-3′ and 5′-TCCCGTTGATGACAAGCTTCCCATT-3′. Data represent the averages ± S.E. of three independent experiments.
RESULTS
ATF2 Is a Component of the RIPE3b1 Complex
In1024 is an insulinoma-derived cell line but lacks detectable level of the RIPE3b/C1-binding activity (Fig. 1A, lane 1). In this cell line, the RIPE3b/C1-binding complex can be reconstituted by transfection with an expression vector for HA-tagged full-length MafA (HA-MafA) (Fig. 1A, lane 2). This reconstituted RIPE3b1 complex was reactive toward an anti-HA antibody (Fig. 1A, lane 3). To determine whether the RIPE3b1 complex also contained other, as yet unidentified, protein components, we tested several different antibodies for their ability to react with the reconstituted RIPE3b1 complex in the EMSA. Because MafA possesses a leucine zipper domain that may mediate dimerization with other bZip proteins, we tested antibodies raised against several bZip proteins, including members of the CREB/ATF family (CREB/ATF1, ATF2, ATF3, and ATF4), Fos family (c-Fos, FosB, Fra1, and Fra2), Jun family (c-Jun, JunB, and JunD) and C/EBP family (C/EBPα, C/EBPβ, C/EBPδ, and GADD153) (Fig. 1A, lanes 4–7, and data not shown). Among them, an antibody raised against ATF2 reacted with the RIPE3b1 complex, as indicated by the supershift in the DNA-protein complex upon the addition of the anti-ATF2 antibody (Fig. 1A, lane 5).
FIGURE 1.
ATF2 is a component of the RIPE3b1 activator DNA-binding complex. A, nuclear extracts of untransfected In1024 cells (lane 1) or cells transfected with an expression plasmid for HA-tagged MafA (lanes 2–7) were analyzed by EMSA using a C1/RIPE3b probe in the absence or presence of the indicated antibodies (top). The arrow indicates the reconstituted RIPE3b1 complex; the asterisk (ss) indicates a supershifted complex. B, EMSA of MIN6 nuclear extracts using a labeled RIPE3b/C1 probe in the absence or presence of the indicated antibodies. The arrow indicates the endogenous RIPE3b1 complex; the asterisks (ss1, ss2, and ss3) indicate supershifted complexes. C, nuclear extracts prepared from NIH3T3 cells transfected with an expression plasmid for FLAG-tagged ATF2 were analyzed by EMSA using a RIPE3b/C1 (left panel) or CRE (right panel) probe. The arrows indicate ATF2-CRE complexes; the asterisk (ss) indicates a supershifted complex.
In nuclear extracts of MIN6 insulinoma cells, the endogenous RIPE3b1 complex was detectable by EMSA using the C1/RIPE3b probe (Fig. 1B, lane 1), and the addition of an antibody that recognizes MafA resulted in a supershift of the DNA-protein complex (Fig. 1B, lane 2, ss1) as demonstrated previously (9). The addition of anti-ATF2 antibody also resulted in a supershift of this complex (Fig. 1B, lanes 3–4, ss2), indicating that the native RIPE3b1 complex also contains ATF2. To determine whether ATF2 and MafA were components of the same DNA-binding complex, we added a MafA-reactive antibody and an anti-ATF2 antibody to the EMSA reaction. Simultaneous addition of the two antibodies resulted in decreased mobility of the RIPE3b1 complex (Fig. 1B, lane 5, ss3) relative to the addition of either antibody alone, which indicated that RIPE3b1 is a heteromeric complex containing both MafA and ATF2. We obtained essentially the same result using a nuclear extract prepared from another insulinoma cell line, βTC6 (data not shown).
The C1/RIPE3b element has limited similarity to CRE, the target DNA sequence of members of the ATF/CREB family of transcription factors. We next examined whether ATF2 interacted with the C1/RIPE3b element in the absence of MafA. Nuclear extracts were prepared from NIH3T3 fibroblast cells that were transfected with an expression vector for FLAG-tagged, full-length ATF2 (FLAG-ATF2). When the nuclear extracts were subjected to EMSA, specific C1/RIPE3b binding activity was undetectable (Fig. 1C, left panel), whereas CRE binding activity was readily detectable (Fig. 1C, right panel). These results indicated that ATF2 alone does not bind to C1/RIPE3b but requires the presence of MafA.
ATF2 Interacts with MafA
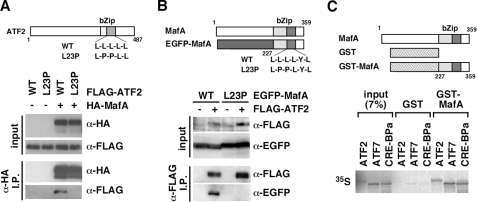
To examine whether ATF2 and MafA interact with each other, FLAG-ATF2 and HA-MafA were expressed in HeLa cells, and then cell extracts were subjected to immunoprecipitation using an anti-HA antibody followed by immunoblot analysis using anti-HA and anti-FLAG antibodies. FLAG-ATF2 was detected in anti-HA immunoprecipitates only when HA-MafA was co-expressed in cells (Fig. 2A), indicating that FLAG-ATF2 forms a complex with HA-MafA. A variant of ATF2 in which the leucine zipper structure was disrupted by amino acid substitutions of two leucine residues (L23P) failed to co-precipitate with HA-MafA.
FIGURE 2.
MafA interacts with ATF2. A, HeLa cells were transfected with expression plasmids for FLAG-tagged wild-type (WT) ATF2 or an L23P mutant of ATF2 (schematically indicated at the top) with or without an expression plasmid for HA-MafA. Cell extracts were subjected to immunoprecipitation with an anti-HA antibody, and the immune complexes (α-HA I.P.) were analyzed by immunoblot using an anti-HA or anti-FLAG antibody. As a control, total cell extract (input) was also analyzed. B, reciprocal immunoprecipitation experiment using EGFP-tagged MafA or an L23P variant of MafA (indicated schematically at the top) and FLAG-ATF2. C, ATF2, ATF7, and CRE-BPa were synthesized in vitro in the presence of [35S]methionine and used in a GST-pull down assay with glutathione-Sepharose-immobilized GST or GST-MafA (indicated schematically at the top). Precipitates were analyzed by SDS-PAGE followed by autoradiography.
In the reciprocal immunoprecipitation analysis, GFP-tagged MafA was co-immunoprecipitated with FLAG-ATF2 (Fig. 2B), and the L23P variant of MafA, in which the leucine zipper was disrupted, failed to bind to FLAG-ATF2. These results indicated that ATF2 and MafA form a complex and that intact leucine zipper domains in both proteins are required for the interaction.
We also examined the interaction between ATF2 and MafA using full-length ATF2 synthesized by in vitro translation using wheat germ extract in the presence of [35S]methionine. In a GST-pull down assay, labeled in vitro-synthesized ATF2 protein was precipitated by a GST-MafA fusion protein containing only the bZip domain of MafA, but not GST alone (Fig. 2C). These results suggested that MafA directly interacts with ATF2, and that the bZip domain of MafA is sufficient for the interaction. In a similar assay, ATF7 and CRE-BPa, which are other members of the ATF2 subfamily, also interacted with GST-MafA (Fig. 2C).
ATF2 Is Enriched in Endocrine Cells of Pancreatic Islets
ATF2 has been shown to be expressed ubiquitously and not in a tissue-specific manner. A previous report indicates, however, that ATF2 is highly expressed in islet β-cells (19). To analyze the expression of ATF2 in the pancreas in more detail, we examined pancreas tissue sections from 5-week-old mice by fluorescence immunohistochemistry using anti-ATF2 antibody. To identify cell types within the islets, the sections were co-stained using antibodies for several endocrine cell markers, i.e. insulin for β-cells, glucagon for α-cells, somatostatin for δ-cells, and pancreatic polypeptide for γ-cells. Anti-ATF2 immunoreactivity was observed in the nuclei of cells within islets (Fig. 3A). Nuclear staining with the anti-ATF2 antibody was also observed in some non-islet cells, but the signals were weaker than in islet cells. ATF2-expressing cells within the islets were also positive for insulin, glucagon, somatostatin, or pancreatic polypeptide. These results indicated that ATF2 is abundantly expressed in endocrine β-, α-, δ-, and γ-cells in the pancreas. As previously demonstrated (11), MafA was exclusively expressed in the nuclei of β-cells (Fig. 3B). Thus, ATF2 and MafA are co-expressed in the nuclei of β-cells of the pancreas.
FIGURE 3.
ATF2 is abundantly expressed in endocrine cells in the pancreas. A, sections of adult mouse pancreas tissue were analyzed by immunohistochemistry using an anti-ATF2 antibody (red) in combination with an anti-insulin (Ins), anti-glucagon (Glu), anti-somatostatin (Som), or anti-pancreatic polyleltide (Ppy) antibody (green). DNA was visualized by staining with DAPI (blue, lower panels). B, double immunostaining of mouse pancreas sections with an anti-MafA antibody (red) in combination with an anti-insulin or anti-glucagon antibody (green).
The Core Enhancer Elements of the Insulin Promoter Are Activated by ATF2
To investigate the role of ATF2 in the RIPE3b1 activator complex, we carried out a luciferase reporter gene assay. HeLa cells were transfected with a luciferase reporter gene driven by the human insulin promoter together with various combinations of expression plasmids for ATF2 and MafA, and Pdx1 and Beta2, two other important insulin promoter activators. Pdx1 binds A1, A3, and GG2 elements, and Beta2 binds E1. The recipient HeLa cells expressed endogenous ATF2, but not the islet-enriched transcription factors MafA, Pdx1, and Beta2.
MafA or Pdx1 plus Beta2 alone weakly activated the insulin promoter, whereas the combination of all three factors resulted in a synergistic increase in promoter activity (Fig. 4A, lanes 1, 3, 5, and 7), as demonstratedpreviously (11). ATF2 alone had only a marginal effect on insulin promoter activity, and co-expression with MafA resulted in weak activation (Fig. 4A, lanes 2 and 4). More strikingly, co-expression of all four factors, ATF2, MafA, Pdx1, and Beta2, resulted in strong transactivation (Fig. 4A, lane 8), and a mutation in the C1 element significantly reduced this level of transactivation activity (lanes 9–11). These results suggested that the binding of ATF2 and MafA to the C1 element is crucial for the synergistic action of all four factors to achieve maximum activation of the insulin promoter. We also observed that ATF2 enhanced the insulin promoter activity with Pdx1/Beta2 even in the absence of MafA (Fig. 4A, lane 6). Given that ATF2 alone does not bind to the C1 element, this synergistic effect may be mediated through other previously reported ATF2-target cis elements such as CRE1, CRE2, and CRE3 (20).
FIGURE 4.
ATF2 activates the insulin promoter. A, HeLa cells were transfected with 100 ng of a reporter gene (pGL4/h-ins-p, top) driven by the wild-type (WT) or mutant (mut C1) human insulin promoter together with pEF-Rluc (25 ng) and various combinations of expression plasmids for HA-MafA (25 ng), FLAG-Pdx1 (25 ng), FLAG-Beta2 (25 ng), and FLAG-ATF2 (600 ng). Data represent the averages ± S.E. of three independent experiments. Statistical significance was calculated by analysis of variance (ANOVA). *, p < 0.02; **, p < 0.01). B, HeLa cells were transfected with 100 ng of a reporter plasmid (core-enh/TATA-luc, top) containing the core enhancer region of the insulin promoter and the TATA-box of the rabbit β-globin gene promoter along with pEF-Rluc (25 ng) and various combinations of the indicated expression plasmids. Data represent the averages ± S.E. of two independent experiments. Statistical significance was calculated by ANOVA. *, p < 0.05; **, p < 0.01). C, HeLa cells were transfected with 100 ng of the core-enh/TATA-luc reporter plasmid, pEF-Rluc (25 ng), and expression plasmids for HA-MafA, FLAG-Pdx1, and FLAG-Beta2 (25 ng each) together with the indicated siRNAs (10 pmol). Aliquots of the cell extracts were analyzed by immunoblot using anti-ATF2 antibody (upper panel) to confirm knockdown of endogenous ATF2. TATA-binding protein (TBP) was analyzed as a loading control. Data represent the averages ± S.E. of three independent experiments. Statistical significance was calculated by ANOVA. *, p < 0.01).
To examine the role of ATF2 independently of its effects mediated by CRE-like elements, we constructed a reporter plasmid that spanned only the core-enhancer elements (GG2, C1, and E1) of the insulin promoter (core-enh/TATA-luc). MafA, Pdx1, and Beta2 alone had only a marginal effect on the reporter activity (Fig. 4B, lanes 1, 3, 5, and 7), whereas combinations of two or three of these factors resulted in an additive or synergistic increase in the reporter activity (Fig. 4B, lanes 9, 11, 13, and 15). ATF2 alone had no effect on the reporter activity (Fig. 4B, lane 2). We observed strong transactivation when ATF2 was co-expressed with MafA, Pdx1, and Beta2 (Fig. 4B, lane 16). Conversely, siRNA-mediated knockdown of endogenous ATF2 (Fig. 4C, upper panel) resulted in a significant decrease of MafA/Pdx1/Beta2-stimulated luciferase activity (Fig. 4C, lower panel). These results indicated that ATF2 is capable of enhancing the insulin promoter activity independently of CRE-like elements.
To our surprise, ATF2 synergistically activated the core enhancer activity together with Pdx1 and Beta2 alone (Fig. 4B, lanes 6 and 8) or in combination (lane 14) even in the absence of MafA, although the maximum activation requires co-expression of MafA. These results suggested that, in addition to ATF2 binding to the C1/RIPE3b element in complex with MafA, ATF2 is capable of activating the insulin promoter through interaction with Pdx1 and Beta2.
ATF2 Interacts with Pdx1 and Beta2
We next examined whether ATF2 physically interacts with Pdx1 and Beta2. FLAG-tagged Pdx1 or Beta2 was expressed together with ATF2-HA in HeLa cells, and the cell extracts were subjected to immunoprecipitation using an anti-FLAG antibody. Subsequent immunoblot analysis showed that ATF2-HA was co-immunoprecipitated with FLAG-Pdx1 and, to a lesser extent, with FLAG-Beta2 (Fig. 5A), indicating that ATF2 forms a complex with Pdx1 and Beta2, even in the absence of MafA. We confirmed that MafA also forms a complex with Pdx1 and Beta2 (Fig. 5B), as demonstrated previously (14). These results together suggested that ATF2, MafA, Pdx1, and Beta2 form a multi-protein complex to facilitate insulin gene transcription.
FIGURE 5.
ATF2 interacts with Pdx1 and Beta2. HeLa cells were transfected with expression plasmids for HA-tagged ATF2 (ATF2-HA) (A) or HA-MafA (B) with or without an expression plasmid for FLAG-tagged Pdx1 (FLAG-Pdx1) or Beta2 (FLAG-Beta2). Cell extracts were subjected to immunoprecipitation with an anti-FLAG antibody, and the immune complexes (α-FLAG I.P.) were analyzed by immunoblot using an anti-HA or anti-FLAG antibody. As a control, total cell extract (input) was also analyzed.
Knockdown of ATF2 in β-Cells Leads to Decreased Insulin Expression
To investigate the role of ATF2 and MafA in the regulation of insulin gene expression in β-cells, MIN6 cells were treated with siRNAs that targeted ATF2 and MafA, respectively. Immunoblotting analysis confirmed that endogenous ATF2 and MafA protein levels were reduced to 50–60% of the level in control siRNA-treated cells by treatment with their respective siRNAs (Fig. 6A). Insulin promoter activity was also significantly reduced to about 60–70% of control levels in both ATF2- and MafA-knockdown MIN6 cells (Fig. 6B). Quantitative RT-PCR analysis demonstrated that the mRNA levels of insulin-1 and insulin-2 were significantly reduced by knockdown of either MafA or ATF2 (Fig. 6C), indicating that both MafA and ATF2 are required for insulin gene expression.
FIGURE 6.
ATF2 is required for insulin gene expression in β-cells. A, MIN6 cells were transfected with a control siRNA or a mixture of two siRNAs designed to target ATF2 or MafA. Cell extracts were analyzed by immunoblot using an anti-ATF2 or anti-MafA antibody. TATA-binding protein (TBP) was analyzed in parallel as a loading control. Relative band intensities were quantified by ImageJ software. n.s., nonspecific cross-reacting material. B, MIN6 cells were co-transfected with a luciferase reporter plasmid (pGL4/h-ins-p, 30 ng) and pEF-Rluc (15 ng) together with mixtures of ATF2- or MafA-specific siRNAs (20 pmol). Data represent the averages ± S.E. of three independent experiments. Statistical significance was calculated by ANOVA. *, p < 0.001). C, total RNA was isolated from MIN6 cells transfected with the indicated siRNAs and subjected to quantitative RT-PCR analysis using specific primers for insulin-1 or insulin-2. Relative expression levels were calculated by the comparative ΔCt method using GAPDH for normalization. Data represent the averages ± S.E. of three independent experiments. Statistical significance was calculated by ANOVA. *, p < 0.001).
DISCUSSION
ATF2, a member of the ATF/CREB family of bZip proteins, is ubiquitously expressed and is a regulator of a wide variety of cellular processes, including cell cycle progression, apoptosis, cytokine signaling, and genotoxic stress response. In the current study, we demonstrated that ATF2 is capable of activating the insulin promoter through interaction with MafA, Pdx1, and Beta2, the principal β-cell-enriched transcriptional activators.
Previous studies have shown that ATF2 is involved in the regulation of insulin expression through binding to CRE-like elements in the promoter region (19, 20). We demonstrated here that ATF2 binds to the C1/RIPE3b element as a heteromeric complex with MafA. MafA has been identified as a component of the RIPE3b1 activator, an important trans-acting regulator of insulin gene expression (8–10). It was shown previously by EMSA that C1/RIPE3b binding can be reconstituted in non-β-cells by ectopic expression of MafA (8, 9). We showed here that both the reconstituted RIPE3b1 complex as well as the endogenous RIPE3b1 complex in β-cells contained ATF2. ATF2 alone was unable to bind to the C1/RIPE3b element but acquired binding capacity in the presence of MafA. As a wide variety of cells express endogenous ATF2, it seems that expression of MafA alone in non-β-cells is sufficient to reconstitute RIPE3b1 activity.
Previous studies have demonstrated that MafA, Pdx1, and Beta2 activate the insulin promoter in a synergistic manner by binding to their respective binding sites (14–17). By a series of luciferase gene reporter assays, we showed here that overexpression of ATF2 in non-β-cells (HeLa) together with MafA, Pdx1, and Beta2 results in synergistic activation of the insulin promoter. In the β-cell line MIN6, ATF2 knockdown resulted in decreased insulin promoter activity and decreased expression of insulin-1 and insulin-2 mRNA. These results indicate that ATF2 is an important transactivator of insulin gene expression. We found that ATF2 interacts with Pdx1 and Beta2 and is capable of activating the insulin promoter even in the absence of MafA, although the maximum activation requires the presence of MafA. We also showed here that MafA physically interacts with Pdx1 and Beta2, as reported previously (14). As protein-protein interaction of transcription factors is one strategy to achieve synergistic transcriptional activation, it appears that ATF2, MafA, Pdx1, and Beta2 form a multi-protein complex and facilitate insulin gene expression through the core enhancer elements.
Gene knockout studies in mice have revealed the detailed functions of the β-cell-enriched factors during embryonic development and in adults. Global deletion of pdx1 causes pancreas agenesis (26, 27), whereas β-cell-specific deletion results in β-cell dysfuncion and diabetes (28). In beta2 knockout mice, the number of β-cells is decreased and islet cells fail to mature (29). mafA knockout mice exhibit progressive β-cell degeneration upon aging (12). In this study, we showed that ATF2 is abundantly expressed in islet endocrine cells (α-, β-, γ-, and δ-cells). Considering that ATF2 is an important regulator of insulin gene expression, this transcription factor may also play an essential role in the development of islet cells. Re-examination of the development and function of islet endocrine cells in atf2 knockout mice is warranted to determine whether ATF2 is involved in islet morphology and function.
Many diverse extracellular stimuli such as cytokines (e.g. interleukin 1β), hormones (e.g. glucagon-like peptide 1), fatty acids, and oxidative stress affect insulin gene expression and β-cell physiology through signaling pathways involving JNK or p38 (30–33). ATF2 activity has been shown to be regulated by these kinases; thus, there are a number of diverse stimuli that may potentially affect insulin gene expression through modulation of ATF2 to regulate. The elucidation of ATF2 function and regulation in the context of the RIPE3b1 activator complex will be important for a better understanding of β-cell physiology and the pathogenesis of diabetes.
Acknowledgments
We thank Dr. Jun-ichi Miyazaki (Osaka University) for the MIN6 cell line and Dr. Shunsuke Ishii (RIKEN Tsukuba Institute) for the pact/FLAG-ATF7 and pact/CRE-BPa plasmids.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (DECODE), for Scientific Research (C), and for the Global COE Program in NAIST from MEXT, Japan, a grant from the Mitsubishi Foundation, and a grant from Takeda Science Foundation (to K. K.). This work was also supported by the Postdoctoral Fellowship for Foreign Researchers from the Japan Society for the Promotion of Science (to S. H.).
- bZip
- basic leucine zipper
- CRE
- cAMP responsive-element(s)
- EGFP
- enhanced green fluorescent protein
- ANOVA
- analysis of variance.
REFERENCES
- 1. Bernardo A. S., Hay C. W., Docherty K. (2008) Mol. Cell. Endocrinol. 294, 1–9 [DOI] [PubMed] [Google Scholar]
- 2. Ohlsson H., Karlsson K., Edlund T. (1993) EMBO J. 12, 4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Petersen H. V., Serup P., Leonard J., Michelsen B. K., Madsen O. D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10465–10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marshak S., Totary H., Cerasi E., Melloul D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15057–15062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Naya F. J., Stellrecht C. M., Tsai M. J. (1995) Genes Dev. 9, 1009–1019 [DOI] [PubMed] [Google Scholar]
- 6. Shieh S. Y., Tsai M. J. (1991) J. Biol. Chem. 266, 16708–16714 [PubMed] [Google Scholar]
- 7. Sharma A., Fusco-DeMane D., Henderson E., Efrat S., Stein R. (1995) Mol. Endocrinol. 9, 1468–1476 [DOI] [PubMed] [Google Scholar]
- 8. Olbrot M., Rud J., Moss L. G., Sharma A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6737–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kataoka K., Han S. I., Shioda S., Hirai M., Nishizawa M., Handa H. (2002) J. Biol. Chem. 277, 49903–49910 [DOI] [PubMed] [Google Scholar]
- 10. Matsuoka T. A., Zhao L., Artner I., Jarrett H. W., Friedman D., Means A., Stein R. (2003) Mol. Cell. Biol. 23, 6049–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kataoka K., Shioda S., Ando K., Sakagami K., Handa H., Yasuda K. (2004) J. Mol. Endocrinol. 32, 9–20 [DOI] [PubMed] [Google Scholar]
- 12. Zhang C., Moriguchi T., Kajihara M., Esaki R., Harada A., Shimohata H., Oishi H., Hamada M., Morito N., Hasegawa K., Kudo T., Engel J. D., Yamamoto M., Takahashi S. (2005) Mol. Cell. Biol. 25, 4969–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Artner I., Blanchi B., Raum J. C., Guo M., Kaneko T., Cordes S., Sieweke M., Stein R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3853–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao L., Guo M., Matsuoka T. A., Hagman D. K., Parazzoli S. D., Poitout V., Stein R. (2005) J. Biol. Chem. 280, 11887–11894 [DOI] [PubMed] [Google Scholar]
- 15. Aramata S., Han S. I., Yasuda K., Kataoka K. (2005) Biochim. Biophys. Acta 1730, 41–46 [DOI] [PubMed] [Google Scholar]
- 16. Docherty H. M., Hay C. W., Ferguson L. A., Barrow J., Durward E., Docherty K. (2005) Biochem. J. 389, 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaneto H., Matsuoka T. A., Nakatani Y., Miyatsuka T., Matsuhisa M., Hori M., Yamasaki Y. (2005) J. Biol. Chem. 280, 15047–15052 [DOI] [PubMed] [Google Scholar]
- 18. van Dam H., Castellazzi M. (2001) Oncogene 20, 2453–2464 [DOI] [PubMed] [Google Scholar]
- 19. Ban N., Yamada Y., Someya Y., Ihara Y., Adachi T., Kubota A., Watanabe R., Kuroe A., Inada A., Miyawaki K., Sunaga Y., Shen Z. P., Iwakura T., Tsukiyama K., Toyokuni S., Tsuda K., Seino Y. (2000) Diabetes 49, 1142–1148 [DOI] [PubMed] [Google Scholar]
- 20. Hay C. W., Ferguson L. A., Docherty K. (2007) Biochim. Biophys. Acta 1769, 79–91 [DOI] [PubMed] [Google Scholar]
- 21. Han S. I., Aramata S., Yasuda K., Kataoka K. (2007) Mol. Cell. Biol. 27, 6593–6605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanai K., Reza H. M., Kamitani A., Hamazaki Y., Han S. I., Yasuda K., Kataoka K. (2010) Genes Cells 15, 971–982 [DOI] [PubMed] [Google Scholar]
- 23. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 24. Igarashi K., Kataoka K., Itoh K., Hayashi N., Nishizawa M., Yamamoto M. (1994) Nature 367, 568–572 [DOI] [PubMed] [Google Scholar]
- 25. Kataoka K., Yoshitomo-Nakagawa K., Shioda S., Nishizawa M. (2001) J. Biol. Chem. 276, 819–826 [DOI] [PubMed] [Google Scholar]
- 26. Jonsson J., Carlsson L., Edlund T., Edlund H. (1994) Nature 371, 606–609 [DOI] [PubMed] [Google Scholar]
- 27. Offield M. F., Jetton T. L., Labosky P. A., Ray M., Stein R. W., Magnuson M. A., Hogan B. L., Wright C. V. (1996) Development 122, 983–995 [DOI] [PubMed] [Google Scholar]
- 28. Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H. (1998) Genes Dev. 12, 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Naya F. J., Huang H. P., Qiu Y., Mutoh H., DeMayo F. J., Leiter A. B., Tsai M. J. (1997) Genes Dev. 11, 2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abdelli S., Abderrahmani A., Hering B. J., Beckmann J. S., Bonny C. (2007) Diabetologia 50, 1660–1669 [DOI] [PubMed] [Google Scholar]
- 31. Kemp D. M., Habener J. F. (2001) Endocrinology 142, 1179–1187 [DOI] [PubMed] [Google Scholar]
- 32. Solinas G., Naugler W., Galimi F., Lee M. S., Karin M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16454–16459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaneto H., Xu G., Fujii N., Kim S., Bonner-Weir S., Weir G. C. (2002) J. Biol. Chem. 277, 30010–30018 [DOI] [PubMed] [Google Scholar]