Abstract
Analysis of the mRNA capping apparatus of the malaria parasite Plasmodium falciparum illuminates an evolutionary connection to fungi rather than metazoans. We show that P. falciparum encodes separate RNA guanylyltransferase (Pgt1) and RNA triphosphatase (Prt1) enzymes and that the triphosphatase component is a member of the fungal/viral family of metal-dependent phosphohydrolases, which are structurally and mechanistically unrelated to the cysteine-phosphatase-type RNA triphosphatases found in metazoans and plants. These results highlight the potential for discovery of mechanism-based antimalarial drugs designed to specifically block the capping of Plasmodium mRNAs. A simple heuristic scheme of eukaryotic phylogeny is suggested based on the structure and physical linkage of the triphosphatase and guanylyltransferase enzymes that catalyze cap formation.
Malaria extracts a prodigious toll each year in human morbidity (400 million new cases) and mortality (1 million deaths). Malaria treatment and prevention strategies have been undermined by the spreading resistance of the Plasmodium pathogen to erstwhile effective drugs and of the mosquito vector to insecticides (1, 2). It is anticipated that the Plasmodium falciparum genome project will uncover novel targets for therapy and immunization (3). The most promising drug targets will be those gene products or metabolic pathways that are essential for all stages of the parasite life cycle but either absent from or fundamentally different in the human host and the arthropod vector. Such targets can be identified either by whole-genome comparisons or by directed analyses of specific cellular transactions. In those instances where Plasmodium differs from metazoans, comparisons with other unicellular organisms may provide insights into eukaryotic phylogeny.
Here we conduct a “postgenomics” inquiry into the mRNA capping apparatus of P. falciparum. The m7GpppN cap is formed by three enzymatic reactions: the 5′-triphosphate end of the nascent pre-mRNA is hydrolyzed to a diphosphate by RNA 5′ triphosphatase, the diphosphate end is capped with GMP by GTP:RNA guanylyltransferase, and the GpppN cap is methylated by AdoMet:RNA (guanine-N7) methyltransferase (4). Each of the mRNA capping enzymes is essential for cell growth in budding yeast. The capping apparatus differs in significant respects in metazoans, fungi, and eukaryotic viruses (4). Mammals and other metazoa encode a two-component capping system consisting of a bifunctional triphosphatase-guanylyltransferase polypeptide and a separate methyltransferase polypeptide. Fungi encode a three-component system consisting of separate triphosphatase, guanylyltransferase, and methyltransferase gene products. Viral capping systems are quite variable in their organization; poxviruses encode a single polypeptide containing all three active sites, whereas phycodnaviruses encode a yeast-like capping apparatus in which the triphosphatase and guanylyltransferase enzymes are encoded separately (5). The biochemical mechanisms of the guanylyltransferase and methyltransferase components of the capping apparatus are conserved between fungi, DNA viruses, and mammals. In contrast, the atomic structures and catalytic mechanisms of the fungal (6) and mammalian (A. Changela, C.K.H., A. Martins, S.S., and A. Mondragon, unpublished observations) RNA triphosphatases are completely different. The triphosphatase components of many viral capping enzymes are mechanistically and structurally related to the fungal proteins and not to the host cell triphosphatase (5, 7). Thus it has been suggested that cap formation and RNA triphosphatase in particular are promising targets for antifungal and antiviral drug discovery (6, 8, 9).
Little is known about the organization of the mRNA capping apparatus in the many other branches of the eukaryotic phylogenetic tree. RNA guanylyltransferase has been studied in the kinetoplastids Trypanosoma and Crithidia (10), but the triphosphatase and methyltransferase components have not been identified. Here we report the identification and biochemical characterization of the separately encoded RNA guanylyltransferase and RNA triphosphatase of the malaria parasite P. falciparum.
Materials and Methods
Expression and Purification of P. falciparum RNA Guanylyltransferase.
A DNA fragment containing the PGT1 ORF on chromosome 14 was amplified by PCR from total P. falciparum genomic DNA (a gift of Derek deBruin and Jeffrey Ravetch, Rockefeller University) with the use of oligonucleotide primers designed to introduce an NdeI restriction site at the predicted translation start codon and a XhoI site 3′ of the predicted stop codon. Preliminary sequence data for P. falciparum chromosome 14 was obtained from the Institute for Genomic Research website (www.tigr.org). The 1.6-kbp PCR product was digested with NdeI and XhoI and inserted into the T7 RNA polymerase-based expression vector pET16b to generate the plasmid pET-His-Pgt1. The nucleotide sequence of the Plasmodium DNA insert was determined. The predicted amino acid sequence of the 520-aa Pgt1 protein encoded by this plasmid is shown in Fig. 1.
Figure 1.
Amino acid sequences of P. falciparum RNA guanylyltransferase Pgt1 and RNA triphosphatase Prt1. The six nucleotidyltransferase motifs in Pgt1 and phosphohydrolase motifs A, B, and C of Prt1 are highlighted in shaded boxes.
pET-His-Pgt1 was transformed into Escherichia coli BL21-CodonPlus(DE3). A 500-ml culture amplified from a single transformant was grown at 37°C in LB medium containing 0.1 mg/ml ampicillin and 50 μg/ml chloramphenicol until the OD600 reached 0.5. The culture was adjusted to 2% ethanol and then incubated at 17°C for 24 h. Cells were harvested by centrifugation, and the pellet was stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 50 ml of buffer A [50 mM Tris⋅HCl (pH 7.5)/0.25 M NaCl/10% sucrose). Cell lysis was achieved by the addition of lysozyme and Triton X-100 to final concentrations of 100 μg/ml and 0.1%, respectively. The lysate was sonicated to reduce viscosity, and insoluble material was removed by centrifugation. The soluble extract was applied to a 5-ml column of Ni-NTA-agarose resin (Qiagen) that had been equilibrated with buffer A containing 0.1% Triton X-100. The column was washed with 25 ml of the same buffer and then eluted stepwise with 12.5-ml aliquots of buffer B [50 mM Tris⋅HCl (pH 8.0)/0.25 M NaCl/10% glycerol/0.05% Triton X-100) containing 0.05, 0.1, 0.2, 0.5, and 1 M imidazole. The polypeptide compositions of the column fractions were monitored by SDS-PAGE. The ≈70-kDa recombinant Pgt1 polypeptide was recovered in the 0.1 M imidazole fraction, which contained 5 mg of protein. The enzyme preparation was stored at −80°C.
Guanylyltransferase Assay.
Reaction mixtures (20 μl) containing 50 mM Tris⋅HCl (pH 8.0), 5 mM DTT, divalent cation, [α-32P]GTP, and Pgt1 as specified were incubated at 30°C for 10 min. The reactions were quenched with SDS, and the products were resolved by SDS-PAGE. The Pgt1-[32P]GMP adduct was visualized by autoradiography of the dried gel and quantitated by scanning the gel with a FUJIX (Tokyo) PhosphorImager.
Sedimentation Analysis of Pgt1.
An aliquot of the Ni-agarose fraction of Pgt1 (50 μg of protein) was mixed with marker proteins catalase (50 μg), BSA (50 μg), and cytochrome c (50 μg), and the mixture was applied to a 4.8-ml 15–30% glycerol gradient containing 0.5 M NaCl, 50 mM Tris⋅HCl (pH 8.0), 5 mM DTT, and 0.05% Triton X-100. The gradient was centrifuged at 50,000 rpm for 18 h at 4°C in a Beckman SW50 rotor. Fractions (≈0.23 ml) were collected from the bottom of the tube. The polypeptide compositions of the fractions were analyzed by SDS-PAGE. Aliquots (2 μl) of each fraction were assayed for enzyme-GMP formation in a reaction mixture containing 5 mM MnCl2 and 5 μM [α-32P]GTP.
RNA Capping by the Isolated Pgt1-GMP Intermediate.
A reaction mixture (100 μl) containing 50 mM Tris⋅HCl (pH 8.0), 5 mM DTT, 2.5 mM MgCl2, 5 μM [α-32P]GTP, and 10 μg of Pgt1 was incubated for 30 min at 30°C. The mixture was adjusted to 25 mM EDTA and 10% glycerol. The native Pgt1-[32P]GMP complex was resolved from free [α-32P]GTP by gel filtration through a 1-ml column of Sephadex G-50 that had been equilibrated with buffer C [50 mM Tris⋅HCl (pH 8.0)/50 mM NaCl/5 mM DTT/10% glycerol/0.05% Triton X-100). Gel filtration was performed at 4°C. Five-drop fractions were collected serially; the 32P elution profile was determined by Cerenkov counting of each fraction.
An aliquot (25 μl) of the gel-filtered Pgt1-[32P]GMP complex (recovered in the void volume of the G-50 column) was incubated for 30 min at 30°C in a reaction mixture (100 μl) containing 50 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 5 mM DTT, and 75 pmol of 5′ diphosphate-terminated poly(A). The reaction products were then extracted once with phenol and once with chloroform-isoamyl alcohol (24:1). RNA was recovered from the aqueous phase by ethanol precipitation and resuspended in 20 μl of 10 mM Tris⋅HCl (pH 8.0), 1 mM EDTA. Aliquots (4 μl) were digested with 5 μg of nuclease P1 for 60 min at 37°C followed by digestion with 1 unit of calf intestine alkaline phosphatase for 60 min at 37°C. The digests were analyzed by TLC on polyethyleneimine-cellulose plates developed with 0.45 M ammonium sulfate. The radiolabeled material was visualized by autoradiography.
Expression and Purification of P. falciparum RNA Triphosphatase.
A DNA fragment containing the PRT1 ORF was amplified by PCR from total P. falciparum genomic DNA with the use of oligonucleotide primers designed to introduce an NcoI restriction site at the predicted translation start codon and a BamHI site 3′ of the predicted stop codon. The PCR product was digested with NcoI and BamHI and cloned into plasmid pYX132. The nucleotide sequence of the Plasmodium DNA insert was determined. The predicted amino acid sequence of the 596-aa Prt1 protein is shown in Fig. 1. A deletion mutant PRT1-CΔ140 lacking the C-terminal 140 aa was generated by PCR amplification with a primer designed to introduce a new stop codon and a BamHI site immediately downstream. The C terminus of the Prt1-CΔ140 polypeptide is indicated by the dot above the sequence in Fig. 1. The PCR product was digested with NcoI and BamHI, the 5′ overhangs were filled in with DNA polymerase, and the DNA was inserted into the filled-in BamHI site of pET28-His/Smt3 (a gift of Chris Lima, Cornell Medical College) so as to fuse the ORF in-frame to N-terminal His6/Smt3.
pET-His/Smt3-Prt1(CΔ140) was transformed into E. coli BL21-CodonPlus(DE3). A 200-ml culture amplified from a single transformant was grown at 37°C in LB medium containing 60 μg/ml kanamycin and 100 μg/ml chloramphenicol until the A600 reached 0.5. The culture was adjusted to 2% ethanol and 0.4 mM isopropyl β-d-thiogalactoside and then incubated at 17°C for 16 h. Recombinant Prt1 was isolated from the soluble bacterial extract by Ni-agarose chromatography as described above for Pgt1. The recombinant His/Smt3-Prt1 polypeptide was recovered in the 0.1 M imidazole eluate fraction. The enzyme preparation was stored at −80°C.
Triphosphatase Assay.
Phosphohydrolase reaction mixtures (10 μl) containing 50 mM Tris⋅HCl (pH 7.5), 5 mM DTT, 1 mM [γ-32P]ATP or 2 μM γ-32P-labeled poly(A), 2 mM MnCl2 or MgCl2, and recombinant Prt1 (0.1 M imidazole eluate) as specified were incubated for 15 min at 30°C. An aliquot (2.5 μl) of the mixture was applied to a polyethyleneimine-cellulose TLC plate, which was developed 0.5 M LiCl, 1 M formic acid. The radiolabeled material was visualized by autoradiography, and 32Pi formation was quantitated by scanning the TLC plate with a PhosphorImager.
Results and Discussion
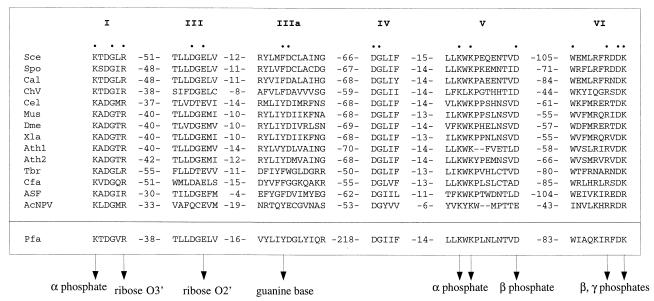
We identified a candidate P. falciparum mRNA guanylyltransferase—a 520-aa polypeptide encoded by a continuous ORF on chromosome 14 (Fig. 1). The signature features of mRNA guanylyltransferases are a ping-pong reaction mechanism of nucleotidyl transfer through a covalent enzyme-(lysyl-N)-GMP intermediate and a set of six conserved peptide motifs (I, III, IIIa, IV, V, and VI) involved in GTP binding and catalysis (4). The Plasmodium guanylyltransferase (henceforth named Pgt1) contains all six catalytic motifs in the standard order and spacing (Fig. 2), except that the 218-aa interval between motifs IIIa and IV of Pgt1 is exceptionally long. This segment in Pgt1 consists of reiterative tracts of hydrophilic amino acids and has no counterpart in other capping enzymes. The hydrophilic segment is predicted, based on the crystal structure of Chlorella virus guanylyltransferase (11), to comprise a large surface loop. All of the amino acids within the six motifs that are essential for the function of Saccharomyces cerevisiae RNA guanylyltransferase Ceg1 (12) are conserved in the Plasmodium protein, as are the residues that make direct contact with the GTP substrate in the Chlorella virus guanylyltransferase–GTP cocrystal (11) (Fig. 2).
Figure 2.
Guanylyltransferase signature motifs present in metazoan, plant, and viral capping enzymes are conserved in P. falciparum Pgt1. The amino acid sequences are aligned for the capping enzymes of S. cerevisiae (Sce), S. pombe (Spo), C. albicans (Cal), Chlorella virus PBCV-1 (ChV), Caenorhabditis elegans (Cel), mouse (Mus), Drosophila melanogaster (Dme), Xenopus laevis (Xle), Arabidopsis thaliana (Ath), Trypanosoma brucei gambiense (Tbr), Crithidia fasciculata (Cfa), African swine fever virus (ASF), and AcNPV baculovirus (AcNPV). The motifs of the P. falciparum (Pfa) guanylyltransferase are highlighted below the other aligned sequences. The numbers of amino acid residues separating the motifs are indicated. The amino acids of the Sce guanylyltransferase that are essential for function in vivo are denoted by dots. Specific contacts between amino acid side chains and the GTP substrate in the ChV capping enzyme–GTP cocrystal are indicated by arrowheads.
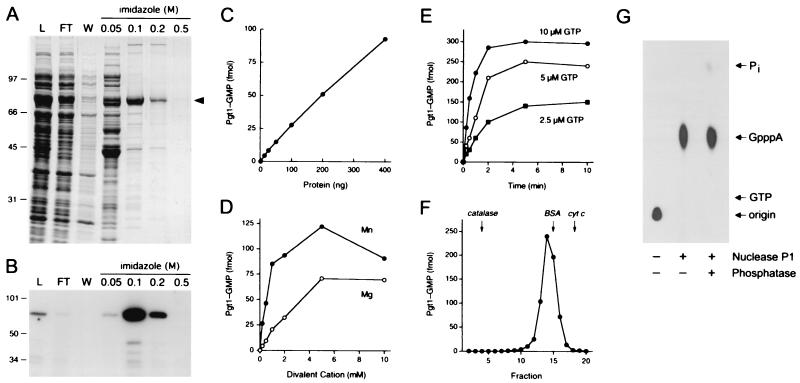
We produced Pgt1 in bacteria as an N-terminal His10-tagged fusion, which allowed for rapid purification of Pgt1 based on the affinity of the tag for immobilized nickel. The ≈70-kDa Pgt1 polypeptide was adsorbed to Ni-agarose and eluted with 0.1 M imidazole (Fig. 3A). Guanylyltransferase activity was measured by reaction of the protein with [α-32P]GTP in the presence of a divalent cation to form the covalent Pgt1–GMP intermediate (Fig. 3B). Enzyme-guanylate formation was linear with respect to Pgt1 concentration (Fig. 3C) and was strictly dependent on a divalent cation cofactor—either manganese or magnesium (Fig. 3D). Other divalent cations—calcium, cobalt, copper and zinc—did not support guanylyltransferase activity (data not shown). Pgt1 formed a covalent intermediate with [α-32P]GTP but was unable to do so with [α-32P]ATP (not shown). The rate and extent of formation of the covalent intermediate were proportional to GTP concentration and leveled off at 10 μM GTP (Fig. 3E and data not shown). We calculated that ≈20% of the input enzyme molecules were labeled with GMP during the reaction with 10 μM GTP and 5 mM MnCl2. The reaction with 10 μM GTP displayed pseudo-first-order kinetics with an apparent rate constant of 1.4 min−1. The native size of Pgt1 was analyzed by glycerol gradient sedimentation with internal standards. The guanylyltransferase activity sedimented as a single peak of 4.6 S, which suggested that Pgt1 is a monomer in solution (Fig. 3F). The activity profile coincided with the distribution of the Pgt1 polypeptide (not shown). That Pgt1 is a bona fide capping enzyme was verified by isolating the Pgt1-[32P]GMP intermediate by gel filtration and demonstrating that it catalyzed transfer of the GMP to diphosphate-terminated poly(A) to form a GpppA cap structure that was liberated from the RNA by digestion with nuclease P1 and was resistant to alkaline phosphatase (Fig. 3G).
Figure 3.
Characterization of P. falciparum RNA guanylyltransferase. (A) Pgt1 purification. Aliquots (15 μl) of the soluble bacterial lysate (L), the Ni-agarose flow-through (FT), wash (W), and indicated imidazole eluates were analyzed by SDS-PAGE. The fixed gel was stained with Coomassie brilliant blue dye. The positions and sizes (in kDa) of marker polypeptides are shown on the left. The Pgt1 protein is indicated by the arrowhead on the right. (B) Guanylyltransferase activity. Reaction mixtures contained 5 mM MgCl2, 0.17 μM [α-32P]GTP, and 1 μl of the protein fractions specified above the lanes. (C) Enzyme titration. Reaction mixtures contained 5 mM MgCl2, 0.17 μM [α-32P]GTP, and Pgt1 as specified. (D) Divalent cation requirement. Reaction mixtures contained 0.17 μM [α-32P]GTP, 200 ng of Pgt1, and either MgCl2 or MnCl2 as specified. (E) Kinetics. Reaction mixtures (100 μl) containing 50 mM Tris⋅HCl (pH 8.0), 5 mM DTT, 5 mM MnCl2, 1 μg of Pgt1, and 2.5, 5, or 10 μM [α-32P]GTP were incubated at 30°C. Aliquots (10 μl) were withdrawn at the times indicated and quenched immediately with SDS. (F) Sedimentation of Pgt1 in a glycerol gradient. The guanylyltransferase activity profile is shown. The peaks of the internal marker proteins are indicated by arrows. (G) RNA cap formation. The RNA reaction product was analyzed by TLC before and after digestion with nuclease P1 and alkaline phosphatase. The positions of the chromatographic origin, GpppA, GTP, and Pi are indicated on the right.
Motif I of Pgt1 (62-KxDGxR-67) contains the lysine nucleophile to which GMP becomes covalently attached during the guanylyltransferase reaction. The position of Lys-62 relative to the N terminus of Pgt1 is typical of the monofunctional guanylyltransferases of fungi and Chlorella virus. (The motif I lysine is located at positions 70, 67, 67, and 84 in the S. cerevisiae, Schizosaccharomyces pombe, Candida albicans, and Chlorella virus enzymes, respectively.) The Plasmodium enzyme conspicuously lacks the ≈200-aa N-terminal RNA triphosphatase domain present in metazoan and higher plant capping enzymes (4). Metazoan RNA triphosphatases belong to a distinct branch of the cysteine phosphatase enzyme superfamily, and they are easily identified by their primary structure (4). We did not find an ORF encoding a homolog of the metazoan RNA triphosphatase located upstream of the PGT1 gene on the P. falciparum chromosome 14 contig (nor was such an ORF found elsewhere in the P. falciparum genome database at National Center for Biotechnology Information). Thus we surmise that Plasmodium does not encode a metazoan-type mRNA capping enzyme.
The similarities between the Plasmodium and fungal guanylyltransferases prompted us to search for a P. falciparum polypeptide resembling fungal RNA triphosphatases. The S. cerevisiae RNA triphosphatase Cet1 exemplifies a growing family of metal-dependent phosphohydrolases that includes the RNA triphosphatases encoded by other fungi (C. albicans and S. pombe) (7, 8, 13), by algal virus PBCV-1 (5), and by several groups of animal viruses (poxviruses, African swine fever virus, and baculoviruses) (9, 14, 15). The yeast/viral triphosphatase family is defined by two glutamate-rich peptide motifs (A and C) that are essential for catalytic activity and comprise the metal-binding site and by an intervening basic peptide motif (B) that is implicated in binding of the 5′ triphosphate moiety of the substrate (Fig. 4). The crystal structure of S. cerevisiae Cet1 reveals that the active site is located within the hydrophilic core of a topologically closed 8-stranded β-barrel—the so-called triphosphate tunnel (6). The β-strands comprising the tunnel are displayed over the Cet1 amino acid sequence in Fig. 4.
Figure 4.
Structural conservation among fungal and Plasmodium RNA triphosphatases. The amino acid sequence of P. falciparum Prt1 is aligned with the sequences of the catalytic domains of fungal RNA triphosphatases of S. cerevisiae Cet1, C. albicans CaCet1, S. cerevisiae Cth1, and S. pombe Pct1. Gaps in the alignment are indicated by dashes. The β-strands that form the triphosphate tunnel of Cet1 are denoted above the sequence. Peptide segments with the highest degree of conservation in all five proteins are highlighted by the shaded boxes. Hydrophilic amino acids that comprise the active site within the tunnel are denoted by dots. Conserved motifs A (β1), B (β9), and C (β11) that define the metal-dependent RNA triphosphatase family are indicated below the sequence. The polyasparagine insert in Prt1 is omitted from the alignment and denoted by a triangle under the sequence between strands β6 and β7.
A psi-blast search (16) initially identified a short segment of weak similarity between Cet1 and the hypothetical P. falciparum polypeptide PFC0985c encoded on chromosome 3 (blast score 42) (17). The similarity between PFC0985c and the other fungal RNA triphosphatases was statistically significant after the first iteration of the search (blast score 122). We then aligned the Plasmodium and fungal protein sequences manually with the use of the tertiary structure of Cet1 and known structure–activity relationships for fungal RNA triphosphatases as a guide (6–8). We thereby identified in the Plasmodium protein counterparts of all eight β-strands of the Cet1 triphosphate tunnel (Fig. 4). The 596-aa Plasmodium protein contains a 162-aa segment between strands β6 and β7, consisting mainly of polyasparagine and acidic residues, that has no counterpart in other RNA triphosphatases (Fig. 1). Reference to the Cet1 structure (6) suggests that this segment is a surface loop emanating from the roof of the tunnel. The instructive point is that the 12 catalytically important hydrophilic amino acids within the tunnel that comprise the active site of fungal RNA triphosphatases (8, 18) are conserved in the Plasmodium protein (Fig. 4), which strongly suggests that this protein (henceforth named Prt1) is the RNA triphosphatase component of the Plasmodium mRNA capping apparatus.
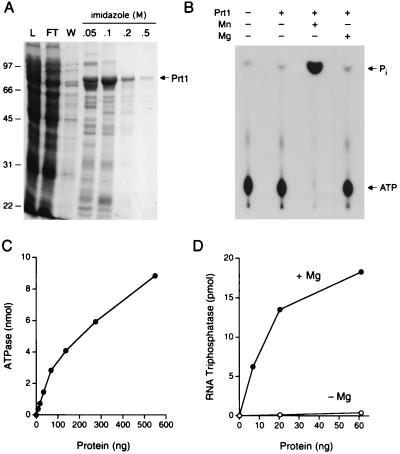
We produced a 456-aa N-terminal domain of Prt1 in bacteria as a His6-tagged fusion and isolated the recombinant Prt1 protein by Ni-agarose chromatography (Fig. 5A). Prt1 displayed the signature biochemical feature of the fungal RNA triphosphatase family (7)—it catalyzed the hydrolysis of the γ phosphate of ATP in the presence of manganese (Fig. 5B). Activity was absolutely dependent on a metal cofactor, and, as with the fungal enzymes (7, 8, 13), magnesium was ineffective in supporting ATP hydrolysis by Prt1 (Fig. 5B). ATPase activity increased with Prt1 concentration (Fig. 5C). Prt1 also catalyzed the magnesium-dependent hydrolysis of the γ phosphate of triphosphate-terminated RNA (Fig. 5D).
Figure 5.
Characterization of P. falciparum RNA triphosphatase. (A) Prt1 purification. Aliquots (15 μl) of the soluble bacterial lysate (L), the Ni-agarose flow-through (FT), wash (W), and indicated imidazole eluates were analyzed by SDS-PAGE. The fixed gel was stained with Coomassie brilliant blue dye. (B) Manganese-dependent NTP hydrolysis. ATPase reaction mixtures contained 0.6 μg of Prt1 and divalent cations as specified. The reaction products were analyzed by TLC and visualized by autoradiography. The positions of [γ-32P]ATP and 32Pi are indicated. (C) Prt1 titration. ATPase reaction mixtures contained 2 mM MnCl2 and Prt1 as specified. (D) RNA triphosphatase activity. Reaction mixtures contained 2 μM γ-32P-labeled poly(A), either 2 mM MgCl2 or no added divalent cation, and recombinant Prt1 as specified.
As a member of the metal-dependent RNA triphosphatase protein family, Prt1 is an attractive antimalarial drug target because (i) the active site structure and catalytic mechanism of this protein family are completely different from the RNA triphosphatase domain of the metzoan capping enzymes and (ii) metazoans encode no identifiable homologs of the fungal or Plasmodium RNA triphosphatases. Thus a mechanism-based inhibitor of Prt1 should be highly selective for the malaria parasite and have a minimal effect on either the human host or the mosquito vector. Given the central role of the mRNA cap in eukaryotic gene expression, an antimalarial drug that targets Prt1 would presumably be effective at all stages of the parasite's life cycle. Moreover, the structural similarity between Prt1 and the fungal RNA triphosphatases raises the exciting possibility of achieving antifungal and antimalarial activity with a single class of mechanism-based inhibitors.
Capping enzymes are a good focal point for considering eukaryotic evolution because the mRNA cap structure is ubiquitous in eukaryotic organisms but absent from the bacterial and archaeal kingdoms. Thus any differences in the capping apparatus between taxa would reflect events that postdate the emergence of ancestral nucleated cells. The enzymes that catalyze the basic nucleic acid transactions (DNA replication, DNA repair, RNA synthesis, and RNA processing) are generally conserved in lower and higher eukaryotes. Yet, in the case of the capping apparatus, we see a complete divergence of the triphosphatase component and the physical linkage of the triphosphatase and guanylyltransferase in unicellular and multicellular organisms.
This divergence suggests a heuristic scheme of eukaryotic phylogeny based on two features of the mRNA capping apparatus: the structure and mechanism of the triphosphatase component (metal-dependent “fungal” type versus metal-independent cysteine-phosphatase type) and whether the triphosphatase is physically linked in cis to the guanylyltransferase component. By these simple criteria, relying on “black-and-white” differences in the same metabolic pathway, one arrives at relationships among taxa that are different from those suggested by comparisons of sequence variations among proteins that are themselves highly conserved in all eukaryotes (19). For example, capping-based phylogeny would place metazoans in a common lineage with Viridiplantae (exemplified by Arabidopsis) because all of these multicellular organisms have a cysteine-phosphatase type RNA triphosphatase fused in cis to their guanylyltransferase. Fungi and now Plasmodia (which are classified as Apicomplexa along with pathogenic parasites Toxoplasma and Cryptosporidia) fall into a different lineage distinguished by a “Cet1-like” RNA triphosphatase that is physically separate from RNA guanylyltransferase. In contrast, the protein sequence variation-based scheme places fungi in the same supergroup as metazoa and puts the Apicomplexa nearer to plants (19).
Assuming that multicellular organisms evolved from unicellular ancestors, we envision an early gene rearrangement event that transferred a cysteine-phosphatase domain into the same transcription unit as the guanylyltransferase, leading to creation of the triphosphatase-guanylyltransferase fusion protein that we see today in multicellular eukaryotes. The fusion presumably allowed for the loss of a Cet1-like enzyme from the early metazoan/plant genome or the divergence of such a protein to a point that it is no longer discernible as Cet1-like. The alternative explanation, which adheres to the sequence-based scheme, would be that plants and metazoans independently experienced this gene fusion in distant branches of the phylogenetic tree.
It is conceivable that, as more eukaryotic genomes are sequenced, we will see some species containing a Cet1-like triphosphatase fused to a guanylyltransferase, others with a cysteine-phosphatase-type RNA triphosphatase that participates in cap formation but is physically separate from the guanylyltransferase, and yet others that encode a novel class of RNA triphosphatase. Nonetheless, a survey of current unicellular genome databases suggests that other protozoans such as Dictyostelium and the pathogenic parasite Trypanosoma do indeed have ORFs encoding polypeptides that resemble fungal RNA triphosphatases. Thus mechanism-based antimalarial inhibitors of Plasmodium RNA triphosphatase may be effective against a battery of other unicellular parasites that cause human disease.
Abbreviations
- Pgt
Plasmodium guanylyltransferase
- Prt
Plasmodium RNA triphosphatase
References
- 1.Newton P, White N. Annu Rev Med. 1999;50:179–192. doi: 10.1146/annurev.med.50.1.179. [DOI] [PubMed] [Google Scholar]
- 2.Enserink M. Science. 2000;287:1956–1958. doi: 10.1126/science.287.5460.1956. [DOI] [PubMed] [Google Scholar]
- 3.Gardner M J. Curr Opin Genet Dev. 1999;9:704–708. doi: 10.1016/s0959-437x(99)00032-5. [DOI] [PubMed] [Google Scholar]
- 4.Shuman S. Prog Nucleic Acid Res Mol Biol. 2000;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 5.Ho C K, Gong C, Shuman S. J Virol. 2001;75:1744–1750. doi: 10.1128/JVI.75.4.1744-1750.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lima C D, Wang L K, Shuman S. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- 7.Ho C K, Pei Y, Shuman S. J Biol Chem. 1998;273:34151–34156. doi: 10.1074/jbc.273.51.34151. [DOI] [PubMed] [Google Scholar]
- 8.Pei Y, Lehman K, Tian L, Shuman S. Nucleic Acids Res. 2000;28:1885–1892. doi: 10.1093/nar/28.9.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho C K, Martins A, Shuman S. J Virol. 2000;74:5486–5494. doi: 10.1128/jvi.74.12.5486-5494.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva E, Ullu E, Kobayashi R, Tschudi C. Mol Cell Biol. 1998;18:4612–4619. doi: 10.1128/mcb.18.8.4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Håkansson K, Doherty A J, Shuman S, Wigley D B. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 12.Wang S P, Deng L, Ho C K, Shuman S. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei Y, Schwer B, Hausmann S, Shuman S. Nucleic Acids Res. 2001;29:387–396. doi: 10.1093/nar/29.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin J, Dong W, Guarino L A. J Virol. 1998;72:10011–10019. doi: 10.1128/jvi.72.12.10011-10019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross C H, Shuman S. J Virol. 1998;72:10020–10028. doi: 10.1128/jvi.72.12.10020-10028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman S, Lawson D, Basham D, Brown D, Chillingworth T, Churcher C M, Craig A, Davies R M, Devlin K, Feltwell T, et al. Nature (London) 1999;400:532–538. doi: 10.1038/22964. [DOI] [PubMed] [Google Scholar]
- 18.Pei Y, Ho C K, Schwer B, Shuman S. J Biol Chem. 1999;274:28865–28874. doi: 10.1074/jbc.274.41.28865. [DOI] [PubMed] [Google Scholar]
- 19.Baldauf S L, Roger A J, Wenk-Siefart I, Doolittle W F. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]