Abstract
P-glycoprotein (Pgp), a member of the ABC transporter family, functions as an ATP hydrolysis-driven efflux pump to rid the cell of toxic organic compounds, including a variety of drugs used in anti-cancer chemotherapy. We have recently obtained EM projection images of lipid-bound Pgp without nucleotide and transport substrate that showed the two halves of the transporter separated by a central cavity (Lee, J. Y., Urbatsch, I. L., Senior, A. E., and Wilkens, S. (2002) J. Biol. Chem. 277, 40125–40131). Addition of nucleotide and/or substrate lead to a close association of the two halves of the transporter, thereby closing the central cavity (Lee, J. Y., Urbatsch, I. L., Senior, A. E., and Wilkens, S. (2008) J. Biol. Chem. 283, 5769–5779). Here, we used cysteine-mediated disulfide cross-linking to further delineate the structural rearrangements of the two nucleotide binding domains (NBD1 and NBD2) that take place during catalysis. Cysteines introduced at or near the C-terminal ends of NBD1 and NBD2 allowed for spontaneous disulfide cross-linking under nonreducing conditions. For mutant A627C/S1276C, disulfide formation was with high efficiency and cross-linked Pgp retained 30–68% drug-stimulated ATPase activity compared with reduced or cysteine-less Pgp. Two other cysteine pairs (K615C/S1276C and A627C/K1260C) also formed a disulfide but to a lesser extent, and the cross-linked form of these two mutants had lower drug-stimulated ATPase activity. The data suggest that the C-terminal ends of the two NBDs of Pgp are not required to undergo significant motion with respect to one another during the catalytic cycle.
Keywords: ABC Transporter, ATPases, Drug Transport, Membrane Proteins, Multidrug Transporters, Disulfide Cross-linking, P-glycoprotein
Introduction
ATP binding cassette (ABC)2 transporters constitute a superfamily of membrane proteins that couple the hydrolysis of MgATP to substrate translocation across a lipid bilayer. In bacteria, ABC transporters can function in the import of nutrients or export of toxic molecules, whereas in eukaryotes, transport occurs exclusively in the direction of export (1, 2). The 48 ABC transporters encoded by the human genome are responsible for the transport of structurally diverse compounds such as bile acids, carbohydrates, nucleosides, sterols, peptides, inorganic ions, and environmental toxins (3). An important member of the mammalian ABC transporter family is P-glycoprotein, a glycosylated 170-kDa plasma membrane protein that is involved in the export of a large variety of structurally unrelated organic molecules (4). P-glycoprotein (Pgp; ABCB1, and MDR1) is expressed in tissues that function in detoxification such as the liver, placenta, and blood-brain barrier. In certain cancers, a high level expression of Pgp (along with other ABC transporters such as ABCG2 and MRP1) in the plasma membrane can result in the failure of chemotherapy by preventing the mostly hydrophobic anti-cancer drugs from accumulating in the cell thus lowering drug efficacy (5, 6).
Most eukaryotic ABC transporters, including Pgp, are expressed as single polypeptides organized in four domains, two transmembrane domains (TMDs) composed of six α-helices each and two cytoplasmic nucleotide binding domains (NBDs). In Pgp, the four domains are arranged in the order N-TMD1-NBD1-TMD2-NBD2-C with a ∼60-amino acid-long linker connecting the C-terminal end of NBD1 with the N terminus of TMD2 (7). To carry out productive MgATP hydrolysis coupled to drug transport, the two NBDs have to interact in a head-to-tail fashion, thereby sequestering the nucleotide(s) at the NBD1-NBD2 interface. In this so-called “sandwich” configuration, the nucleotide is interacting with the phosphate binding loop (P-loop) of one NBD and the ABC signature motif (LSGGQ) of the other NBD and vice versa (8). High affinity binding of transport substrate occurs in an amphipathic cavity that is formed by the two TMDs toward the cytoplasmic side of the transporter (7). The drug-binding cavity can accommodate a large variety of structurally unrelated molecules and exhibits distinct (but overlapping) binding regions for different subsets of compounds.
Despite ongoing effort, the mechanism by which drug transport from the inner to the outer leaflet of the membrane is coupled to the hydrolysis of one or two molecules of ATP is still not fully understood. Several models have been proposed based on a combination of site-directed mutagenesis and biochemical and biophysical experiments involving transport assays, trapping of catalytic intermediates using inorganic vanadate (Vi) or fluoroaluminate with spin-labeled ATP, and photo affinity labeling with 8-azido nucleotide analogues (9–14). These studies revealed that the two NBDs act in a cooperative manner in that by mutating of a catalytically essential residue in only one of the two NBDs resulted in nearly complete loss of drug stimulated ATPase activity. However, other aspects of the mechanism such as the number of ATP molecules hydrolyzed for each transport event and whether the NBDs must completely dissociate to enable nucleotide and/or drug binding remain controversial.
Here, we have addressed the question as to whether the two NBDs of Pgp have to come apart for binding of bulky drug molecules as postulated based on the recent x-ray crystal structure of Pgp (7). The introduction of two cysteine residues near the C-terminal ends of the N- and C-terminal NBDs of mouse Pgp lead to spontaneous formation of a disulfide bond. The cross-linked Pgp displayed limited impairment of drug-stimulated ATP hydrolysis activity, indicating that the C-terminal ends of the NBDs can remain in close proximity to one another during the catalytic cycle.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis, Protein Expression, and Purification
Cysteine-less (CL) mouse Mdr3 in the Pichia pastoris expression vector pHIL-D2 (15) was a kind gift from Gregory Tombline and Alan Senior (University of Rochester, Rochester, NY). Cysteine residues were introduced into CL Mdr3 by site-directed mutagenesis using the QuikChange protocol (Stratagene) at locations K615C/S1276C, A627C/K1260C, and A627C/S1276C (hereafter denoted as KS, AK, and AS, respectively). The primers were: A627C (fwd) 5′-gagagaaaagggcatttacttcaaacttgtcatgacacagacaTGTggaaatgaaattg-3′ and (rev) 5′-catccttagatttagcagcttcatttcctaattcaatttcatttccACAtgtctgtgtcatg-3′; K615C (fwd) 5′-cattgtggagcaaggaaatcatgatgagctcatgagagaaTGTggcatttacttc-3′ and (rev) 5′-cctgctgtctgtgtcatgacaagtttgaagtaaatgccACAttctctcatgagc-3′; S1276C (fwd) 5′-gtgcaggctggagcaaagcgcTGTtacgtacatcacc-3′ and (rev) 5′-gagtgatggtgatggtgatgtacgtaACAgcgctttgctcc-3′; K1260C (fwd) 5′-cacccaccagcagctgctggcgcagTGTggcatctac-3′ and (rev) 5′-ctgcacactgaccattgagaagtagatgccACActgcgccagc-3′. Mutagenesis was confirmed by DNA sequencing in the State of New York Upstate Medical University DNA sequencing core facility. Transformation of pHIL-D2-mdr3 into P. pastoris strain GS115 and screening for successful chromosomal integration resulting in Muts variants was done as described previously (16). Fermentor growth of P. pastoris and Pgp purification was carried out following the protocol of Lerner-Marmarosh et al. (17). Briefly, a 1-liter inoculum was grown in minimal glycerol medium to A600 ≥ 2.0 and added to 6 liters of fermenter medium. One liter of glycerol supplemented with Pichia trace metals was gradually added using a peristaltic pump. The rate of feed was increased along with biomass of Pichia trace metals. Upon consumption of the glycerol feed, 1 liter of methanol supplemented with Pichia trace metals was gradually added to the culture. Dissolved oxygen was monitored and maintained at ≥35% (airflow supplemented with pure oxygen as needed), and the pH was maintained between 4.5 and 5.0 with 10% ammonium hydroxide. Cells were harvested by centrifugation at 3000 × g, and cell pellets were resuspended 1:1 (w/w) in 0.3 m Tris, 0.33 m sucrose, 0.1 m 6-aminocaproic acid, 1 mm EDTA, 1 mm EGTA, 2 mm DTT, pH 7.4 (resuspension buffer), and stored at −80 °C. Cell suspensions from harvest were diluted into resuspension buffer lacking sucrose, and ∼90 g of cells were then lysed with 0.5 mm zirconia beads (Biospec) in the presence of protease inhibitors (2 μg/ml leupeptin, 2 μg/ml pepstatin A, 0.5 μg/ml chymostatin, 1 mm PMSF) using a homemade bead beater (12 × 1 min cycles) followed by sonication at 30 watts for 6 × 1 min cycles. Cell debris was removed by centrifugation at 3000 × g and 14,000 × g. Membranes were pelleted by centrifugation at 200,000 × g for 90 min, resuspended in a Dounce homogenizer and passaged through an 18-gauge needle in 10% glycerol, 50 mm Tris, pH 7.4, in the presence of protease inhibitors, and washed membranes (microsomes) were collected by centrifugation at 200,000 × g for 60 min. Microsomes were resuspended in solubilization buffer (50 mm Tris, 50 mm NaCl, 30% glycerol, 10 mm imidazole, pH 7.4, with protease inhibitors) and stored at −80 °C. Microsomes were solubilized at 4 mg protein/ml with 1.2% dodecyl maltoside (DM, Inalco). Membranes were incubated for 15 min on ice and then centrifuged at 64,000 × g for 30 min. Solubilized membranes were incubated with 5 ml of CoNTA (prepared by charging NTA matrix with CoCl2). The Pgp-bound resin was then packed into a column and washed with 20 column volumes of membrane resuspension buffer with 0.1% DM on an ÄKTA FPLC. Pgp was eluted with 300 mm imidazole, 50 mm Tris, 50 mm NaCl, 20% glycerol, 0.1% DM, pH 7.4. The Pgp-containing fractions were collected, diluted 1:3 with 10 mm Tris, 20% glycerol, 0.1% DM, pH 7.8, and passed over a 5-ml DE52 column (Whatman). Pgp was collected in the flow-through, concentrated by ultrafiltration, and further purified by size exclusion chromatography (Superdex 200 16/50) in 20 mm Mops, 10% glycerol, 0.1 mm EDTA, 50 mm NaCl, and 0.1% DM, pH 7.5. Fractions were pooled and concentrated using Viva Spin concentrators with a 50-kDa molecular mass cut-off. Reducing agent was omitted in all purification steps subsequent to cell harvest to obtain disulfide cross-linked Pgp.
Protein Concentration Determination
Protein concentration was determined by gel densitometry (ImageJ) using cysteine-less Pgp as a standard. The cysteine-less Pgp concentration was determined by UV absorption at A280 using the calculated extinction coefficient of 109,000 m−1 cm−1.
Fluorescein Maleimide Labeling
Cross-linked mutants AS, KS, and AK were reduced with 2.5 mm tris(2-carboxyethyl)phosphine, pH 7, for 30 min followed by reaction with 10 μm fluorescein maleimide in 50 mm Mops buffer, pH 7, 1 mm EDTA, 10% glycerol. Excess label was quenched with 1 mm cysteine followed by 0.5 mm N-ethyl maleimide. For labeling of oxidized samples, tris(2-carboxyethyl)phosphine and cysteine were omitted and the reaction was stopped with N-ethyl maleimide only. For SDS-PAGE of labeled protein, DTT was omitted from the gel loading buffer. Fluorescein fluorescence was imaged with a DocIt system using 366 nm UV light for excitation.
ATPase Activity Assay
ATPase activity assays were carried out using 20 μg Pgp premixed with 1:1 (w/w) Escherichia coli lipids (Avanti) in an ATP-regenerating system (13). Pgp and lipids with or without 20 mm DTT were incubated at 20 °C for 5 min and added to the assay at 37 °C, and the absorbance was measured until a basal activity slope was obtained. For ATPase stimulation, drugs were added from stock solutions in dimethyl sulfoxide or methanol. For reduced samples, 10 mm DTT was included in the assay mix. Each condition was measured at least in triplicate (from two or more independent preparations for mutant AS and for one preparation for mutants KS and AK). For vanadate trapping, 10 μm vanadate was included in the ATPase assay. Both DTT and vanadate at the concentrations used had no measurable effect on the ATP-regenerating system.
RESULTS
Placement of Cysteine Mutants
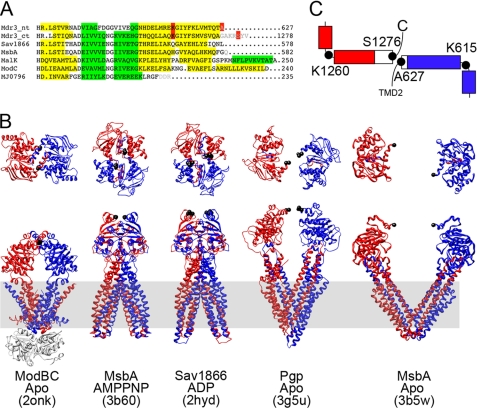
To address the question whether the NBD separation seen in the crystal structure of Pgp is necessary for drug binding-induced ATP hydrolysis, we introduced cysteines at the C-terminal regions of NBD1 and NBD2 to facilitate a reversible covalent link between the two NBDs without modifying the catalytic sites as in the earlier experiments involving NBD disulfide links (18–20). In studying the amino acid sequences and secondary structures of several NBDs for which crystal structures are available: MsbA (Protein Data Bank codes 3B60 and 3B5W; 21), Pgp (Protein Data Bank code 3G5U; 7), MalK (Protein Data Bank code 2R6G; 22), ModC (Protein Data Bank code 2ONK; 23), Sav1866 (Protein Data Bank code 2HYD; 24), and MJ0796 (Protein Data Bank code 1L2T; 8), we observed that there is a degree of variability in the length of the C-terminal α helical region of the NBDs (Fig. 1A). Although the NBDs with longer C-terminal regions (from the bacterial importers ModBC and MalFGK2) are in close association, the NBDs with shorter C-terminal α helices (exporters Sav1866, MsbA, and Pgp) show varying degrees of association depending upon presence of nucleotide and/or substrate (Fig. 1B).
FIGURE 1.
Secondary and tertiary structure analysis of Pgp and comparison to other ABC transporters. A, sequence alignment between NBDs using ClustalW (version 2.0; 34). Secondary structure (α helices in yellow; β sheets in green) was assigned from the available crystal structures or by using the PsiPred server (residues not resolved in the structures as shown in gray letters). Highlighted in red are residues Ala627, Ser1276, Lys615, and Lys1260. B, Top and side views of several ABC transporters: ModAB2C2 (Protein Data Bank code 2ONK), MsbA (Protein Data Bank code 3B60), Sav1866 (Protein Data Bank code 2HYD), and Pgp (Protein Data Bank code 3G5U). Colored in blue is TMD1-NBD1, and red is TMD2-NBD2. The residues homologous to AS in Pgp are in black spheres. The lipid bilayer is indicated in gray. C, schematic of the NBD1 and NBD2 C-terminal α helices. Black circles indicate the positions of the cysteine mutants, and red/blue boxes represent α helices that are resolved in the crystal structure. Note that NBD1 and NBD2 are only resolved up to Thr626 and Ala1271, respectively, and the α helical parts represented by white boxes are based on secondary structure prediction.
In the Pgp crystal structure, electron density at the C-terminal ends of NBD1 and NBD2 was resolved up to residues Thr626 and Ala1271, respectively (3G5U; 7). These residues are shown as black spheres in the Pgp model in Fig. 1B. However, based on secondary structure prediction, the terminal α-helices in NBD1 and NBD2 are expected to continue for one and seven more residues in NBD1 and NBD2, respectively (Fig. 1A). Based on the crystal structure of Pgp (7), Ala627 and Ser1276 would likely be in close proximity if the α helices were to be extended in a straight fashion beyond the resolved electron density (Fig. 1C). To test this prediction, we replaced these two residues with cysteines to be able to create a reversible covalent link. In addition to the mutations A627C and S1276C, we also placed cysteine residues at the beginning of the C-terminal α helix of NBD1 (Lys615) in combination with S1276C and at the end of the C-terminal α helix of NBD1 (Ala627) in combination with the beginning of the C-terminal α helix of NBD2 (Lys1260) to test whether the C-terminal α helices of NBD1 and NBD2 can adopt a similar parallel arrangement as seen in ModC (Fig. 1B).
Purification of Cross-linked Pgp
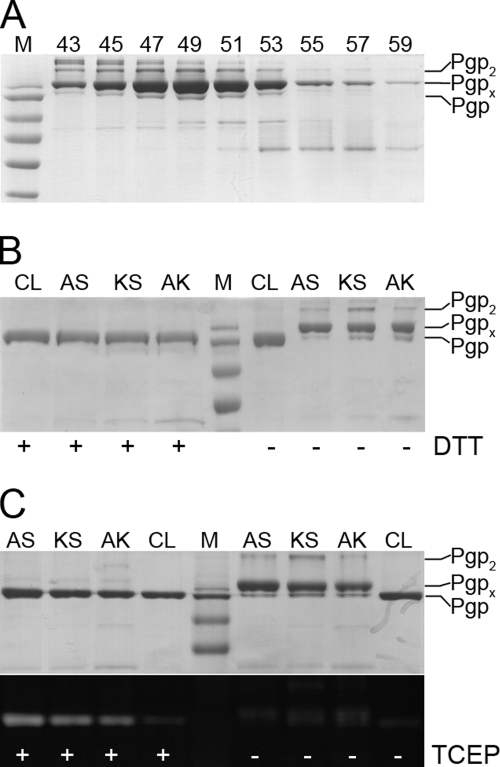
When purifying double cysteine mutant A627/S1276 (AS) in presence of 1 mm β-mercaptoethanol, we noticed spontaneous but incomplete cross-link formation after the CoNTA affinity purification step. Efforts to increase cross-link yield by addition of 5′,5′-dithiobis-(z-nitrobenzoic acid) or CuCl2 and/or drugs and nucleotides were unsuccessful, possibly due to irreversible oxidation/modification of one or both of the cysteine residues during CoNTA column chromatography (data not shown). However, by purifying the double cysteine mutants AS in absence of added reducing agents, cross-link yield could be increased to near completeness (85–90% with 5–8% uncross-linked and/or dimeric Pgp). Fig. 2A shows representative SDS-PAGE of a gel filtration profile of disulfide cross-linked mutant AS. As can be seen, cross-linked (Pgpx) and uncross-linked Pgp co-elute in all fractions, though some separation of dimers (Pgp2) and higher oligomers can be observed. Very similar profiles were obtained for the other two mutant proteins after cross-linking (data not shown). Fig. 2B shows SDS-PAGE of all cross-linked and reduced double cysteine mutants. Cross-linked and reduced Pgp migrate at 170 and 140 kDa, respectively, similar to what has been observed for cross-links involving cysteines in the P-loop or ABC signature motifs (18–20). Unlike mutant AS, mutants KS and AK purified under similar conditions reached a maximum cross-link yield of 60–80% with 15–20% uncross-linked and intermolecular cross-links. Fluorescein labeling of oxidized and reduced mutants AS, KS, and AK confirmed that the disulfide cross-link is present in solution and not an artifact of denaturing polyacrylamide gel electrophoresis (lower panel in Fig. 2C).
FIGURE 2.
Purification and fluorescein maleimide labeling of cross-linked and reduced double cysteine mutant Pgp. A, SDS-PAGE of size exclusion chromatography fractions of cross-linked mutant AS separated on a Superdex 200 column (16/50, Äkta FPLC) in 20 mm Mops, 10% glycerol, 0.1 mm EDTA, 50 mm NaCl, 0.1% DM, pH 7.5 (0.8 ml/min). Markers (from top to bottom) are 170, 130, 95, 72, 55, and 43 kDa. B, SDS-PAGE of 1.5 μg of CL, cross-linked mutants AS, KS, and AK in absence and presence of DTT. Based on gel densitometry using ImageJ software, the yield of intramolecular cross-link formation was estimated to 85–90% for AS, 60–70% for KS, and 77% for AK. C, fluorescein maleimide labeling of CL and cross-linked AS, KS, and AK in presence (left) and absence (right) of the reducing agent tris(carboxyethyl)phosphine (TCEP). Upper panel, Coomassie stain; lower panel, fluorescence image. As can be seen, significant labeling occurs only in presence of tris(2-carboxyethyl)phosphine, indicating that the disulfide cross-link is present in solution and not a result of SDS-PAGE. The weak fluorescence signal of cross-linked and cysteine-less protein is due to nonspecific labeling of e.g. lysine side chains.
Characterization of Drug-stimulated ATPase Activities
To test the effect of covalently linking NBD1 and NBD2 on catalytic activity, drug-stimulated ATPase activities of cross-linked mutants AS, KS, and AK were measured in the absence or presence of reducing agent. For comparison, CL Pgp from mouse (15) was used as a standard. The data are summarized in Fig. 3A (for mutant AS) and Fig. 3B (for KS and AK). No significant effect on activity was observed for CL upon addition of DTT (data not shown). We tested a variety of drugs with different affinities for Pgp, fold stimulation of ATPase activity, and molecular sizes: colchicine (399 Da; 40 μm), verapamil (455 Da; 200 μm), paclitaxel (854 Da; 50 μm), vinblastine (909 Da; 5 μm), and cyclosporin A (1203 Da; 5 μm) (25, 26).
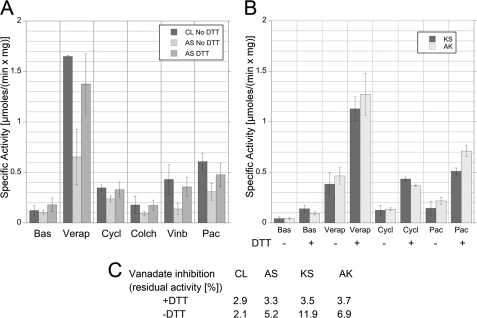
FIGURE 3.
ATPase characterization of Pgp mutants AS, AK, and KS. Using an ATP-regenerating system, specific ATPase activities were determined for CL, cross-linked AS, AK, KS, and DTT-reduced AS, AK, KS. For each activity assay, 20 μg of protein was incubated with 20 μg E. coli lipids in absence or presence of 20 mm DTT. The protein/lipid mixture was then added to an ATPase assay at 37 °C. In the assay, 10 mm DTT was present to maintain the cysteines in the reduced state; DTT was omitted for ATPase characterization of the cross-linked species. The concentrations of drugs used are as follows: verapamil (Verap; 455 Da; 200 μm), cyclosporin A (Cycl; 1203 Da; 5 μm), colchicine (Colch; 399 Da; 40 μm), vinblastine (Vinb; 909 Da; 5 μm), and paclitaxel (Pac; 854 Da; 50 μm). A, drug-stimulated ATPase activities of CL and AS, reduced and cross-linked. B, specific ATPase activities of AK and KS using verapamil, cyclosporin A, and paclitaxel, reduced and cross-linked. C, residual ATPase activities after trapping with orthovanadate (the values are an average of two or more measurements). Vanadate trapping was induced by addition of 10 μm vanadate to the ATPase assay in presence of verapamil.
The level of ATPase activity in presence of different size drugs may reveal the exclusion of large drugs when the motion of the NBDs that is required for drug binding is restricted by the covalent cross-link. For instance, if the cross-linked Pgp has normal drug-stimulated activity for small compounds but a reduced activity for larger compounds, then it would indicate that Pgp must be able to open beyond what is possible due to the cross-link to permit drug binding for larger compounds as suggested based on the crystal structure (7). As can be seen from Fig. 3A, this was not the case for mutant AS. Basal activity for CL and cross-linked AS is about the same with a slight increase of AS activity upon reduction of the disulfide bridge (a slight increase of basal activity is also seen for CL upon addition of DTT (data not shown)). Verapamil leads to about a 13.5- and 7-fold stimulation of ATPase in CL and cross-linked AS Pgp, respectively. Reduction of the disulfide bond in mutant AS results in a verapamil-stimulated ATPase activity that is ∼85% of the activity of CL Pgp, indicating that the two cysteine mutations on their own have little effect on verapamil-stimulated ATPase activity. There is significant ATPase stimulation in cross-linked AS also for the largest of the drugs tested, cyclosporin A, and for the intermediately sized paclitaxel (2.3- and 3-fold compared with 2.9- and 5-fold for CL, respectively). ATPase stimulation of cross-linked AS by these drugs (verapamil, paclitaxel, and cyclosporin A) suggests that the NBDs do not have to come apart for drug stimulated catalysis beyond what is still possible in presence of the covalent disulfide cross-link between the C-terminal ends of the NBDs. Little to no activation of cross-linked AS was observed with the drugs colchicine and vinblastine, indicating that binding of these molecules might occur at a site that is not accessible in the cross-linked enzyme.
For cross-linked mutants AK and KS, basal activity was significantly lower compared with CL and AS, whereas the reduced forms had comparable values (Fig. 3B). Drug stimulation of ATPase activities for mutants AK and KS were characterized using verapamil, cyclosporin A, and paclitaxel. Overall, significantly lower values for drug-stimulated activities were measured for cross-linked KS and AK (for example 0.38 μmoles·min−1·mg−1 for cross-linked KS compared with 0.65 μmol·min−1·mg−1 for AS in presence of verapamil). The values for cross-linked mutants KS and AK are likely even lower considering that part of the activity in these mutant enzymes is contributed by uncross-linked enzyme. However, upon reduction of the disulfide bond, drug stimulated activity in KS and AK is similar compared with CL and reduced AS, again indicating that the mutations themselves have no effect on activity.
As a further test of functional NBD communication, cross-linked mutants AS, KS, and AK were vanadate-trapped in the presence and absence of DTT, and the residual measured ATPase activity was compared with the activity of vanadate-trapped CL. In the presence of 10 μm orthovanadate, verapamil stimulated ATPase activity in CL and reduced AS, KS, and AK decreased by ∼96–97%. In absence of DTT, mutants AS, KS, and AK maintained 5.2, 11.9, and 6.9% verapamil-stimulated ATPase activity, respectively (Fig. 3C). This shows that cross-linked mutant AS traps vanadate almost as efficiently as CL, again indicating that the cooperativity of the two ATP binding sites is preserved even when the two NBDs are covalently linked at their very C-terminal ends. However, vanadate trapping is less efficient for cross-linked mutants KS and AK, suggesting that the disulfide in these mutants fixes the NBDs in a conformation that is less favorable for NBD communication.
DISCUSSION
Over the past decade, x-ray crystal structures of several full-length ABC transporters that were crystallized under different substrate conditions have been solved. The structures revealed that ABC transporters can adopt so called out- and inward facing conformations in which the NBDs are either found to interact intimately in a head-to-tail fashion (nucleotide-bound MalFGK2 (22) Sav1866 (24) and MsbA (21)) or to be more open (nucleotide-free MsbA (21), MalFGK2 (27), and ModBC (23)). In two of the structures, both from importers, the interaction of the NBDs is maintained by a closely interacting C-terminal α helix, as in case of ModBC, or an extra domain, as in case of MalFGK2. Disulfide cross-linking the extra domains in MalK had no effect on ATPase and maltose transport activity, indicating that at least for this bacterial importer, the NBDs can stay together during the catalytic cycle (28). On the other hand, the bacterial exporters Sav1866 and MsbA as well as mammalian Pgp seem to lack a structural element that could hold the NBDs together near their C-terminal ends in the absence of nucleotide, and it is possible that the lack of such feature may be in part responsible for the wide open conformation observed in the crystal structure of E. coli MsbA (21).
Of course, it should be noted not only that the crystal structures represent static images of highly dynamic transporters but also that the apo forms themselves may not represent physiologically relevant conformations given cytoplasmic MgATP concentrations. The question therefore arises as to how much the two NBDs can or must move in relation to each other during the catalytic cycle.
In the crystal structure of nucleotide free Pgp, a 20 Å separation is seen between the two NBDs for both the “apo” and drug-bound transporter (7). However, because larger drug molecules did not appear to bind to Pgp that was crystallized in the absence of drug, the authors of the study speculated that an even greater separation of the NBDs (as seen in apo-MsbA from E. coli (21)) may be required for binding of bulky transport substrates (7). Biochemical evidence in support of such a mechanism had been reported from cross-linking experiments that involved disulfide bond formation between cysteines in the P-loop and/or ABC signature motifs (18–20). In one study, it was shown that one of the naturally occurring cysteines in the P-loops (of e.g. NBD1) and a cysteine introduced into the adjacent ABC signature motif (NBD2) could form a disulfide bond (19) and that binding of large transport substrates, such as cyclosporin A, slowed down disulfide formation (20). On the other hand, disulfide bond formation was also observed between the cysteines in both P-loops (18) and taken together, these cross-link experiments not only suggested that the NBDs of Pgp have to come apart to bind bulky substrates, but also that the NBDs have to be quite flexible as the two P-loops in crystal structures of the nucleotide bound NBD sandwich are ∼30 Å apart (e.g. ADP-bound Sav1866 or AMPPNP-bound MsbA). However, as the cysteines involved in these cross-link experiments were part of the catalytic sites and therefore likely affected nucleotide binding, the inhibitory effect of the cross-links on ATPase activities were difficult to interpret in terms of the question as to how much NBD motion is required for catalysis.
The only structural information for nucleotide-bound Pgp comes from EM of two-dimensional crystals. EM projection images of two-dimensional crystals of lipid bilayer reconstituted mouse and human Pgp showed the two halves of the transporter in close proximity with a central stain-filled cavity (29), much like the cavity seen in the crystal structure (7). When adding nucleotides in presence or absence of transport substrate to the lipid bilayer bound two-dimensional crystals of Pgp, the two halves of the transporter appeared to get closer together, closing the central cavity seen in the apo images (29, 30). Substrate-dependent structural changes were also observed in images of two-dimensional crystals of detergent solubilized hamster Pgp (31), and taken together, the EM data suggest that substrate binding to apo-Pgp involves notable motion of the NBDs, consistent with the earlier disulfide cross-linking studies (18–20).
However, contrary to a mechanism involving complete dissociation of the NBDs, there is biochemical evidence that suggests that the NBDs remain closely associated during steady state turnover, with release of ADP and phosphate and subsequent binding of MgATP at one catalytic site while the second site has nucleotide occluded (tightly bound) (11, 12). A more or less tight association of the NBDs during steady state turnover is also more easily rationalized in terms of the observed cooperativity of the two active sites of Pgp (32).
Alternatively, if the NBDs do not remain in close contact as seen in the apo crystal structures of ModBC, MsbA and Pgp, then it is possible that the NBDs rapidly sample different conformations until MgATP binding to both NBDs results in the closed sandwich configuration followed by hydrolysis of one or both ATP molecules, product release and NBD dissociation. Interestingly, Loo et al. (33) have recently shown that cross-linking of the TMDs at the cytoplasmic side results in a 3-fold increase of basal ATPase activity without affecting drug-stimulated ATPase to a significant degree. It is possible that restricting the motion of the NBDs near their N-terminal ends results in a greater probability of generating the ATP-bound sandwich configuration, which in turn may explain the increase in basal activity (33).
Here, we have shown that the two NBDs of mouse Pgp can be covalently linked via cysteine residues near their C-terminal ends and consistent with the crystal structure of Pgp, the cross-link is most efficient for double cysteine mutant AS. Interestingly, cross-linked AS retained significant drug stimulated ATPase activity for most of the drugs tested, including verapamil, paclitaxel, and the largest molecule assayed, cyclosporin A. This suggests that binding of these drugs does not require an opening of the NBDs beyond what is seen in the crystal structure of the apo- and drug-bound transporter (7). However, for two of the drugs tested, vinblastine and colchicine, little to no activation was observed, and it cannot be ruled out that binding and/or ATPase activation of some drug molecules requires a motion (e.g. twisting) of the TMDs and/or NBDs that may be restricted in the cross-linked enzyme.
Cysteine pairs KS and AK, even though at greater distance based on the crystal structure compared with AS, are still able to cross-link with 60–80% efficiency, consistent with highly dynamic NBDs as mentioned above. On the other hand, basal and drug-stimulated ATPase activity as well as the ability to efficiently trap vanadate are significantly impaired in cross-linked mutants KS and AK compared with AS, suggesting that the cross-link in these two mutants locks the two NBDs in a configuration that is unfavorable for NBD cooperativity and maximum turnover.
Taken together, our findings support recent models in which the NBDs remain in close association during steady state turnover due to occluded nucleotide at one catalytic site. In addition, the data suggest that the conformation of Pgp as seen in the crystal structure is competent for productive binding of some drugs, including large molecules such as cyclosporin A. Other transport substrates such as colchicine and vinblastine may require a conformation different from the one seen in the crystal structure but it appears that the size of the transport substrate alone may not be the sole determining factor in this process.
This work was supported, in whole or in part, by National Institutes of Health Grants CA100246 and GM058600 (to S. W.).
- ABC
- ATP-binding cassette
- Pgp
- P-glycoprotein
- DM
- n-dodecyl β-d-maltoside
- fwd
- forward
- rev
- reverse
- KS
- K615C/S1276C
- AK
- A627C/K1260C
- AS
- A627C/S1276C
- TMD
- transmembrane domain
- NBD
- nucleotide-binding domain
- CL
- cysteine-less.
REFERENCES
- 1. Holland I. B., Blight M. A. (1999) J. Mol. Biol. 293, 381–399 [DOI] [PubMed] [Google Scholar]
- 2. Rees D. C., Johnson E., Lewinson O. (2009) Nat. Rev. Mol. Cell Biol. 10, 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dean M., Hamon Y., Chimini G. (2001) J. Lipid Res. 42, 1007–1017 [PubMed] [Google Scholar]
- 4. Gottesman M. M., Ling V. (2006) FEBS Lett. 580, 998–1009 [DOI] [PubMed] [Google Scholar]
- 5. Gottesman M. M., Fojo T., Bates S. E. (2002) Nat. Rev. Cancer 2, 48–58 [DOI] [PubMed] [Google Scholar]
- 6. Sharom F. J. (2008) Pharmacogenomics 9, 105–127 [DOI] [PubMed] [Google Scholar]
- 7. Aller S. G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P. M., Trinh Y. T., Zhang Q., Urbatsch I. L., Chang G. (2009) Science 323, 1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., Hunt J. F. (2002) Mol. Cell 10, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Urbatsch I. L., Tyndall G. A., Tombline G., Senior A. E. (2003) J. Biol. Chem. 278, 23171–23179 [DOI] [PubMed] [Google Scholar]
- 10. Al-Shawi M. K., Omote H. (2005) J. Bioenerg. Biomembr. 37, 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sauna Z. E., Kim I. W., Nandigama K., Kopp S., Chiba P., Ambudkar S. V. (2007) Biochemistry 46, 13787–13799 [DOI] [PubMed] [Google Scholar]
- 12. Siarheyeva A., Liu R., Sharom F. J. (2010) J. Biol. Chem. 285, 7575–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Urbatsch I. L., Sankaran B., Weber J., Senior A. E. (1995) J. Biol. Chem. 270, 19383–19390 [DOI] [PubMed] [Google Scholar]
- 14. Delannoy S., Urbatsch I. L., Tombline G., Senior A. E., Vogel P. D. (2005) Biochemistry 44, 14010–14119 [DOI] [PubMed] [Google Scholar]
- 15. Tombline G., Urbatsch I. L., Virk N., Muharemagic A., White L. B., Senior A. E. (2006) Arch. Biochem. Biophys. 445, 124–128 [DOI] [PubMed] [Google Scholar]
- 16. Beaudet L., Urbatsch I. L., Gros P. (1998) Methods Enzymol. 292, 397–413 [DOI] [PubMed] [Google Scholar]
- 17. Lerner-Marmarosh N., Gimi K., Urbatsch I. L., Gros P., Senior A. E. (1999) J. Biol. Chem. 274, 34711–34718 [DOI] [PubMed] [Google Scholar]
- 18. Urbatsch I. L., Gimi K., Wilke-Mounts S., Lerner-Marmarosh N., Rousseau M. E., Gros P., Senior A. E. (2001) J. Biol. Chem. 276, 26980–26987 [DOI] [PubMed] [Google Scholar]
- 19. Loo T. W., Bartlett M. C., Clarke D. M. (2002) J. Biol. Chem. 277, 41303–41306 [DOI] [PubMed] [Google Scholar]
- 20. Loo T. W., Bartlett M. C., Clarke D. M. (2003) J. Biol. Chem. 278, 1575–1578 [DOI] [PubMed] [Google Scholar]
- 21. Ward A., Reyes C. L., Yu J., Roth C. B., Chang G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oldham M. L., Khare D., Quiocho F. A., Davidson A. L., Chen J. (2007) Nature 450, 515–521 [DOI] [PubMed] [Google Scholar]
- 23. Hollenstein K., Frei D. C., Locher K. P. (2007) Nature 446, 213–216 [DOI] [PubMed] [Google Scholar]
- 24. Dawson R. J., Locher K. P. (2006) Nature 443, 180–185 [DOI] [PubMed] [Google Scholar]
- 25. Liu R., Sharom F. J. (1996) Biochemistry 35, 11865–11973 [DOI] [PubMed] [Google Scholar]
- 26. Delannoy S., Urbatsch I. L., Tombline G., Senior A. E., Vogel P. D. (2005) Biochemistry 44, 14010–14019 [DOI] [PubMed] [Google Scholar]
- 27. Khare D., Oldham M. L., Orelle C., Davidson A. L., Chen J. (2009) Molecular Cell 33, 528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Samanta S., Ayvaz T., Reyes M., Shuman H. A., Chen J., Davidson A. L. (2003) J. Biol. Chem. 278, 35265–35271 [DOI] [PubMed] [Google Scholar]
- 29. Lee J. Y., Urbatsch I. L., Senior A. E., Wilkens S. (2002) J. Biol. Chem. 277, 40125–40131 [DOI] [PubMed] [Google Scholar]
- 30. Lee J. Y., Urbatsch I. L., Senior A. E., Wilkens S. (2008) J. Biol. Chem. 283, 5769–5779 [DOI] [PubMed] [Google Scholar]
- 31. Rosenberg M. F., Velarde G., Ford R. C., Martin C., Berridge G., Kerr I. D., Callaghan R., Schmidlin A., Wooding C., Linton K. J., Higgins C. F. (2001) EMBO J. 20, 5615–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Senior A. E., al-Shawi M. K., Urbatsch I. L. (1995) FEBS Lett. 377, 285–289 [DOI] [PubMed] [Google Scholar]
- 33. Loo T. W., Bartlett M. C., Clarke D. M. (2010) Biochem. Biophys. Res. Commun. 395, 436–440 [DOI] [PubMed] [Google Scholar]
- 34. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]