Abstract
Ribonuclease H2 (RNase H2) is the major nuclear enzyme involved in the degradation of RNA/DNA hybrids and removal of ribonucleotides misincorporated in genomic DNA. Mutations in each of the three RNase H2 subunits have been implicated in a human auto-inflammatory disorder, Aicardi-Goutières Syndrome (AGS). To understand how mutations impact on RNase H2 function we determined the crystal structure of the human heterotrimer. In doing so, we correct several key regions of the previously reported murine RNase H2 atomic model and provide biochemical validation for our structural model. Our results provide new insights into how the subunits are arranged to form an enzymatically active complex. In particular, we establish that the RNASEH2A C terminus is a eukaryotic adaptation for binding the two accessory subunits, with residues within it required for enzymatic activity. This C-terminal extension interacts with the RNASEH2C C terminus and both are necessary to form a stable, enzymatically active heterotrimer. Disease mutations cluster at this interface between all three subunits, destabilizing the complex and/or impairing enzyme activity. Altogether, we locate 25 out of 29 residues mutated in AGS patients, establishing a firm basis for future investigations into disease pathogenesis and function of the RNase H2 enzyme.
Keywords: Crystal Structure, DNA Repair, DNA Replication, RNA-binding Protein, X-ray Crystallography, Aicardi-Goutières Syndrome, RNase H2
Introduction
RNA/DNA hybrids occur during fundamental cellular processes, such as DNA replication (1), transcription (2), and telomere elongation (3). Ribonuclease H2 (RNase H2)5 is the predominant nuclear enzyme to hydrolyze the RNA strand of such RNA/DNA hybrids. In addition, RNase H2 is able to cleave the 5′-phosphodiester bond of one or more ribonucleotides embedded in a DNA duplex (4, 5). As misincorporated ribonucleotides may be the most abundant non-canonical nucleotides present in genomic DNA (6), their removal by factors including RNase H2 is likely to be critical to maintain genomic integrity.
Eukaryotic RNase H2 has evolved markedly from its prokaryotic ancestry. In both archaea and bacteria, the RNase H2 enzyme consists of a single protein, while eukaryotic RNase H2 is a complex containing three protein subunits, all of which are required for activity (5). The RNASEH2A subunit exhibits significant sequence homology, including the catalytic site residues, with the prokaryotic enzyme, whereas the two auxiliary subunits (RNASEH2B and RNASEH2C) have no known prokaryotic equivalents. The precise role of the auxiliary subunits is not fully defined; however they are thought to serve as a platform for assembly of the RNase H2 complex and to modulate its interaction with other proteins. Indeed, RNASEH2B binds PCNA (7), which is a scaffold and catalyst for DNA-editing enzymes during DNA replication and repair (8).
Mutations in all three subunits of human RNase H2 cause the autoinflammatory disorder Aicardi-Goutières Syndrome (AGS) (9, 10). AGS is an early-onset progressive encephalopathy with similarities to congenital viral infections and shows immunological features similar to those seen in the autoimmune disease systemic lupus erythematosis (11). Inflammation may be triggered by accumulation of nucleic acid byproducts from recently replicated DNA (12), or by increased cellular levels of endogenous retroelements (13). Elevated levels of these cytosolic nucleic acids are thought to be recognized by pattern recognition receptors, which normally act to detect viral infections (13).
Recently, the structure of the murine RNase H2 trimer was solved (14). Despite a high degree of sequence identity between the mouse and human subunits, the authors only located three patient mutations in their structure, stating that the remaining mutations were in disordered loops. We therefore determined the structure of the human RNase H2 trimer to enhance our understanding of how RNase H2 mutations cause disease. We report the crystal structure of human RNase H2 at 4.1 Å resolution. We find that the core of the catalytic domain adopts a canonical RNase H2 fold (15) similar to murine RNASEH2A (14), however we report a different arrangement of the RNASEH2B and RNASEH2C subunits. In addition, we define a more extensive interaction interface for the C terminus of RNASEH2A that involves both auxiliary subunits. Using biochemical assays, we demonstrate that the C termini of RNASEH2A and RNASEH2C form an integral component of the heterotrimer architecture and are important for RNase H2 enzymatic function. Our human structure now allows the mapping of 25 out of 29 AGS mutations, and shows that 7 of these cluster at the interface of the RNASEH2A and RNASEH2C C-terminal regions.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
A polycistronic construct based on pGEX6P1 was made to express the human RNase H2 trimer, allowing simultaneous expression of GST-tagged RNASEH2B, and untagged RNASEH2A and RNASEH2C (supplemental Fig. S1A). Shine-Dalgarno (SD) sequences were introduced by PCR before RNASEH2A and RNASEH2C coding sequences. Similar polycistronic constructs were used for all co-expression, with RNASEH2A, RNASEH2B, or RNASEH2C replaced by alternative coding sequences. Point mutations and internal deletions were introduced using the QuikChange method (Stratagene); C-terminal deletions were created with alternative reverse primers; chimeric Archaeoglobus fulgidus (Afu)/human RNASEH2A was generated by overlap PCR. For a list of constructs generated, see supplemental Table S1.
Recombinant RNase H2 complexes were expressed in Rosetta-2 cells (Novagen). Cells were grown to an optical density of 0.6 at 37 °C, cooled to room temperature, and induced with 0.1 mm IPTG at 20 °C for 14–16 h. Cells were pelleted and lysed by sonication in a PBS buffer containing 0.2% Tween, 10 mm MgCl2, Protease Inhibitor Mixture (Sigma, 50 μl per gram of cell pellet), 100 μg of DNase I and 100 μg of RNase A (Sigma). Cleared lysates were incubated with glutathione-Sepharose 4B beads (GE Healthcare Life Science) for 2 h at 16 °C. GST-tagged and associated proteins were released from the beads by cleavage with PreScission protease (GE Healthcare Life Sciences) for 14–16 h at 4 °C and further purified by size-exclusion chromatography in 150 mm NaCl, 20 mm Tris-HCl pH 7.
Crystallization, Data Collection, and Refinement
As human RNaseH2 containing full-length subunits did not yield crystals of sufficient quality, we improved x-ray diffraction by truncating regions of predicted disorder and introducing point mutations. Our best diffraction data were obtained from crystals of a complex containing RNASEH2B residues 2–226, full-length RNASEH2C, and RNASEH2A with catalytic site mutations D34A and D169A. Importantly, this complex (without the point mutations) retains the same activity as wild-type RNase H2 (supplemental Fig. S1, B and C).
Crystals were grown by vapor diffusion at room temperature in 96-well plates with 0.2 μl droplets at 4 °C. Equal volumes of pre-formed RNaseH2 complex (11 mg/ml in 150 mm NaCl, 20 mm Tris, pH 7) and precipitant (20% PEG3350, 0.1 m bis-Tris-propane pH 6.5, 0.2 m KNO3) were mixed. Crystals were cryoprotected with reservoir solution plus 20% ethylene glycol before cryocooling in liquid nitrogen.
Diffraction data were collected at Diamond Light Source (Harwell Science and Innovation Campus, UK) at beamline I04. The data set was integrated and scaled using HKL2000 (16). Pseudo-translational symmetry was detected from a native Patterson map, and as a result MOLREP was used for the molecular replacement strategy (17). Coordinates deposited for the murine RNase H2 (PDB ID: 3KIO) were initially used as a search model for phasing, however limited refinement into the human electron density resulted in poor geometry and statistics (supplemental Table S2). The deposited murine structure was re-refined against deposited structure factors with BUSTER-TNT (18), using non-crystallographic local structure similarity restraints (19) and TLS constraints (where each chain corresponded to a TLS group; supplemental Table S3). As murine RNase H2 (PDB ID: 3KIO) was solved by experimental phasing using selenomethionine derivates (14), we preserved the location but not necessarily the sequence context of these methionines, including those introduced by site-directed mutagenesis (14).
The re-refined murine RNase H2 coordinates were then used as a molecular replacement model for the human structure. To generate a human homology model as a starting point for refinement, point mutations were introduced using COOT into the murine coordinates. Atomic models for the subunits were refined against the 4.1 Å human dataset as individual rigid bodies with PHENIX (20), followed by further cycles of maximum-likelihood positional and B-factor refinement with BUSTER-TNT (18) using TLS and 2-fold NCS restraints. The use of non-crystallographic local structure similarity restraints (19) plus strict stereochemical (18) and B-factor (21) restraints were coupled with highly conservative rebuilding to avoid over-fitting of the experimental data. Because of steric clashes, RNASEH2A:143–166 were excluded from the refinement in one of the two heterotrimers. The analysis and figures presented were generated from the heterotrimer best-resolved in electron density. Final refinement statistics are reported in Table 1.
TABLE 1.
X-ray data collection and refinement statistics for human RNase H2
| Human RNaseH2 | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 212.2, 42.3, 186.9 |
| α, β, γ (°) | 90.0, 98.1, 90.0 |
| Resolution (Å) | 29.8-4.1 (4.4-4.1)a |
| Rmerge | 0.18 (0.69) |
| I/σ(I) | 5.6 (1.7) |
| Completeness (%) | 98.5 (98.7) |
| Redundancy | 3.9 (3.5) |
| Refinement | |
| Number of reflections | 13281 (2371) |
| Rxpctb | 37.4 (28.7) |
| Rfree | 37.9 (28.0) |
| Number of non-H atoms | 7359 |
| Rmsd | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.96 |
| Ramachandran plot | |
| Favored (%) | 94.1 |
| Outliers (%) | 0.5 |
| Average B factors (Å2) | 188.1 |
a Values in parentheses are for highest-resolution shell.
b Rxpct = 100 × (Σhkl ||Fobs| − |Fxpct||/Σhkl |Fobs|), where |Fobs| and |Fxpct| are the observed structure factor amplitude and the expectation of the model structure factor amplitude, respectively (18).
Protein Co-precipitation
Recombinant complexes with one subunit GST-tagged were expressed using polycistronic constructs and purified by affinity chromatography (see above). Affinity-matrix-bound proteins before and after PreScision Protease cleavage were separated by SDS-PAGE (12%) and visualized by Coomassie Brilliant Blue staining.
RNase H2 Activity Assay
Enzyme activity assays were performed using a fluorometric assay as previously described (9). Briefly, 10 μm of 3′ fluorescein-labeled oligonucleotides (GATCTGAGCCTGGGaGCT or gatctgagcctgggagct; uppercase DNA, lowercase RNA) were annealed to a complementary 5′ DABCYL-labeled DNA oligonucleotide (Eurogentec) in 60 mm KCl, 50 mm Tris-HCl, pH 8, by heating for 5 min at 95 °C followed by slow cooling to room temperature. Reactions were performed in 100 μl of buffer (60 mm KCl, 50 mm Tris-HCl, pH 8, 10 mm MgCl2, 0.01% BSA, 0.01% Triton X-100) with 250 nm substrate in 96-well flat-bottomed plates at 24 ± 2 °C for 90 min. Fluorescence was read for 100 ms using a VICTOR2 1420 multilabel counter (Perkin Elmer), with a 480 nm excitation filter and a 535 nm emission filter. Reactions were carried out in triplicate with a range of different protein concentrations. A standard curve (relating fluorescence units to moles of converted substrate) was generated using fluorescein-labeled oligonucleotide annealed to DNA without DABCYL.
Steady State Enzyme Kinetics
Experiments were performed using the fluorometric assay and conditions described above, with 0.1 nm RNase H2 and substrate concentrations ranging from 0 to 1 μm for RNA18:DNA18 substrate or from 0 to 0.5 μm for D14R1D3:D18 substrate. Initial velocities of substrate cleavage were determined and expressed as the rate of the converted substrate concentration (nm) over time (min). Substrate hydrolysis by RNase H2 follows Michaelis-Menten kinetics. Kinetic parameters Km and Vmax were determined using non-linear regression in SigmaPlot 11.0 (Systat Software Inc.).
ThermoFluor Assay
Thermal stability assays were performed using the fluorescence-based thermal shift (ThermoFluor) method as previously described (22). Proteins were diluted in 50 μl volumes to 2 μm in 10 mm HEPES-KOH pH 7.2, 100 mm potassium acetate, 2 mm magnesium acetate, and 5× SYPRO Orange (Invitrogen). Assays were carried out in a Bio-Rad iQ5 rtPCR Thermocycler, heating the samples from 20 °C to 80 °C at 0.5 °C increments, holding each temperature for 30 s. Fluorescence intensity was measured in relative fluorescence units (RFU) using excitation/emission wavelengths of 485 nm/575 nm. Thermal denaturation graphs were plotted as a function of the gradient of protein unfolding [d(RFU)/dT] against the temperature gradient, with the melting temperature (Tm) defined as the temperature at the maximum of d(RFU)/dT.
Peptide Arrays
Peptides were generated by automatic synthesis (23, 24) on chemically modified cellulose membrane disks derivatized with Fmoc-β-alanine using standard Fmoc (fluoren-9-ylmethoxycarbonyl) chemistry with the MultiPep (Intavis Bioanalytical Instruments AG) (23, 25). After synthesis, side chains were deprotected by incubating the disks in 80% trifluoroacetic acid (TFA), 3% triisopropylsilane (TIPS), 5% H2O, 12% dichloromethane for 1 h at room temperature. This solution was removed and the membrane disks were solubilized in 88.5% TFA, 4% trifluoromethanesulfonic acid, 2.5% TIPS, 5% H2O for 16 h at room temperature. Peptide-cellulose conjugates were precipitated with ice-cold tert-butylmethylether, dried, and dissolved in DMSO. Peptide arrays were created by spotting the peptide-cellulose conjugates on coated microscope slides (CelluSpotsTM, Intavis Bioanalytical Instruments AG) in duplicate.
Slides were blocked in TBST (50 mm Tris pH 7.5, 150 mm NaCl, 0.05% Tween-20) containing 5% BSA and then overlaid with TBST/0.5% BSA containing 0.4 μm recombinant His-RNASEH2BC. Control slides were overlaid with TBST/0.5% BSA only. Bound protein was detected with anti-His-HRP (Qiagen) and visualized by ECL (GE Healthcare Life Sciences).
Fluorescence Anisotropy
Peptide binding experiments were performed at 20 °C in a Perkin Elmer LS55 Luminescence Spectrometer with excitation/emission wavelengths of 480 nm/530 nm and slit widths of 15 nm/20 nm, respectively. The photomultiplier voltage used was 650 V with an integration time of 5 s per measurement. A peptide corresponding to the C terminus of RNASEH2A with an N-terminal fluorescein plus a single glycine linker ([Fluorescein] GSSHRYFLERGLESATSL; Peptide Protein Research) was rehydrated in 150 mm NaCl, 20 mm Tris, pH 8 and diluted in a 900 μl volume to a final concentration of 20 nm. The RNASEH2BC heterodimer (54.4 μm) was titrated into a constantly stirred cuvette containing the fluorescein-labeled peptide. Anisotropy was measured continuously, and the values recorded once measurements were unchanged for five consecutive readings. Dissociation constants (Kd) were calculated by fitting the titration data (corrected for dilution) to a one-site binding equilibrium model in Prism5 (GraphPad Software).
RESULTS
Crystal Structure of the Human RNase H2 Complex
To gain insight into how mutations in RNase H2 cause human disease, we set out to solve the structure of the human RNase H2 heterotrimer. Given significant sequence conservation between human and murine RNase H2 (86, 80, and 65% sequence identity for RNASEH2A, RNASEH2B, and RNASEH2C, respectively) the murine RNase H2 crystal structure (PDB ID: 3KIO) (14) was used as a model for molecular replacement to phase human RNase H2. However, limited refinement of this solution into the human RNase H2 electron density failed to give adequate statistics or geometry (supplemental Table S2). Re-analysis of the coordinates and structure factors deposited in the Protein Data Bank (PDB) for murine RNase H2 established that the published mouse model was not in agreement with the experimental electron density in a number of areas (mRNASEH2C residues 87–116 and 136–144; mRNASEH2B residues 92–102, 146–159, and 267–282; supplemental Fig. S2 and S3) (26). We therefore used BUSTER-TNT to re-refine the deposited murine coordinates against the deposited structure factors. Our new interpretation of the murine model improved the refinement statistics (supplemental Table S3) and produced a better visual fit to the density (supplemental Fig. S3). The two murine models are similar in their secondary structural elements but differ significantly in their connectivity and sequence assignment for all three RNase H2 subunits (see supplemental Fig. S3 and S4, and supplemental text for a detailed description of the differences observed).
We then solved the structure of human RNase H2 to 4.1 Å resolution by molecular replacement using coordinates for this re-refined murine model. To avoid overfitting of the data, the human model was refined using tight geometric and B-factor restraints (21) resulting in refinement residuals Rexpt and Rfree of 37.4 and 37.9%, respectively. The geometry of the final refinement was assessed using MOLPROBITY (27). 94.1% of the residues were in the favored region of the Ramachandran plot; 0.5% were outliers.
As expected from the sequence conservation between murine and human RNase H2, we find their structures to be similar. Superposition of atomic models based on secondary structure matching (28) resulted in a root mean square deviation of 1.01 Å for equivalent Cα atom pairs. The largest difference was found at (human) RNASEH2A residues 195–204. However, due to the low resolution of our human structure, this new conformation was not modeled and the α-helix was truncated at residue 194.
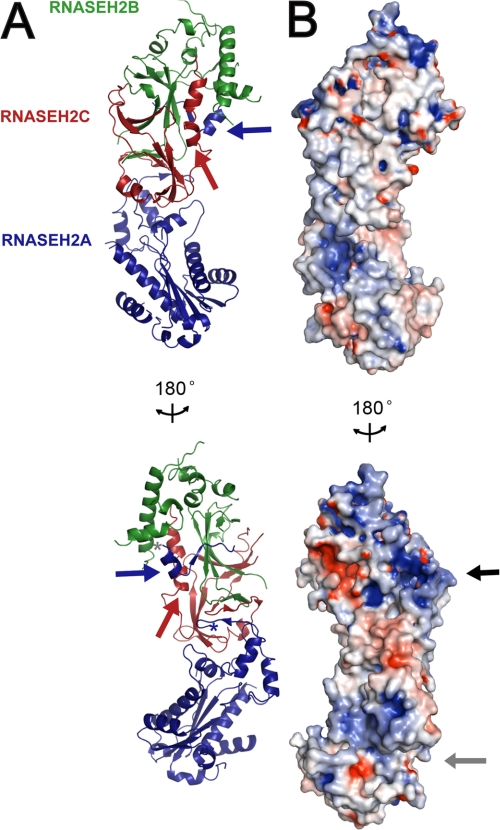
Human RNase H2 is a heterotrimeric complex in which the core domains of the three subunits are linearly stacked (Fig. 1A). The structure of the RNase H2 catalytic subunit is highly conserved between prokaryotes and eukaryotes (14, 15). Indeed, we find that the core domain of human RNASEH2A adopts a canonical RNase H2 fold. In eukaryotes, the sequence of RNASEH2A extends beyond the catalytic core domain to include nearly 50 additional residues. Whereas residues 258–283 are not resolved in our human RNase H2 structure, the extreme C terminus of RNASEH2A (residues 284–299) forms an α/β motif that binds the RNASEH2BC heterodimer, and RNASEH2A residues 250–257 contribute to a β-sheet with RNASEH2C. The auxiliary RNASEH2B and RNASH2C subunits together with the C-terminal extension of RNASEH2A adopt an interwoven triple β-barrel fold centered on the C-terminal half of RNASEH2C. RNASEH2C residues 118–126 and 128–134 form β-strands central to all three β-barrels, whereas residues 143–160 form a kinked α-helix that interacts with secondary structure elements of RNASEH2B and the C terminus of RNASEH2A.
FIGURE 1.
Crystal structure of the heterotrimeric RNase H2 complex. A, ribbon diagram representation of human RNase H2 reveals a linear arrangement of subunits where the core of the catalytic domain (RNASEH2A; blue) is stacked on the interwoven auxiliary RNASEH2B (green) and RNASEH2C (red) subunits. The C terminus of RNASEH2A, residues 284–299 (blue arrow), extends from the core domain and bridges the other two subunits. The first residue of the extension, RNASEH2A:284, and the last residue of the core domain, RNASEH2A:257, are indicated by gray and blue asterisks, respectively. The C terminus of RNASEH2C (red arrow) forms a kinked α-helix at the intersubunit interface. B, electrostatic surface of human RNase H2 indicates two positively charged patches: one in the catalytic active site groove (gray arrow) and the second on the interwoven RNASEH2BC heterodimer (black arrow). Electrostatic surface potential was calculated using PDB2PQR (34) and colored by potential (+9kT/e to −9 kT/e; electropositive in blue and electronegative in red) on the solvent accessible surface using the APBS tool (35). These and all other molecular graphics were generated using PyMOL (DeLano Scientific LLC).
Analysis of the electrostatic surface potential of RNase H2 reveals two positively charged patches on the same face of the molecule (Fig. 1B). The first region corresponds to the active site binding groove of the catalytic RNASEH2A subunit and is similar to that observed for Archaeoglobus fulgidus (Afu) RNase HII (15). The second basic patch occurs in the vicinity of the central β-barrel formed by the RNASEH2B and RNASEH2C subunits, and may indicate an additional substrate binding site.
The RNASEH2A C Terminus Is Necessary for an Enzymatically Active Complex
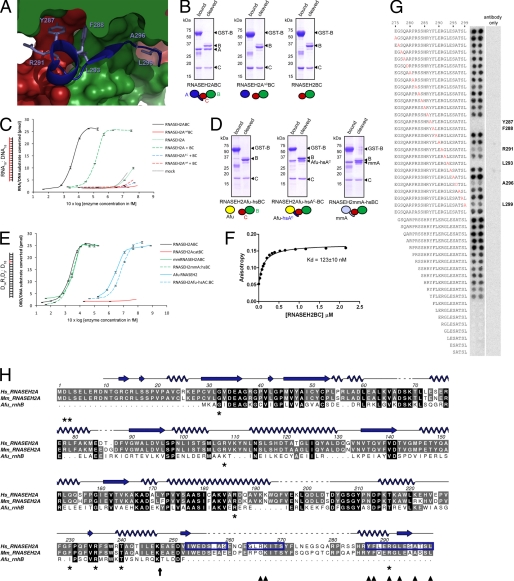
In the crystal structure of human RNase H2 the C terminus of RNASEH2A extends from the core catalytic domain and bridges the two auxiliary subunits (Fig. 2A). To assess the functional relevance of this interaction, we examined the effect of mutant and chimeric RNASEH2A on complex assembly and enzyme activity. GST-tagged RNASEH2B was used to co-purify complexes by affinity chromatography. When co-expressed, wild-type RNASEH2A, RNASEH2B, and RNASEH2C subunits assemble to form an enzymatically active complex (Fig. 2). In contrast, deletion of the RNASEH2A C-terminal extension (residues 248–299; RNASEH2AΔC) entirely abolished heterotrimer formation, resulting in isolation of the RNASEH2BC heterodimer only (RNASEH2AΔCBC; Fig. 2B). RNASEH2AΔC could be purified (supplemental Fig. S5), indicating that lack of assembly was not due to misfolding of the truncated protein. Proteins purified after RNASEH2AΔCBC expression did not exhibit enzyme activity (data not shown). This is in agreement with the requirement for all three subunits for substrate cleavage by eukaryotic RNase H2 (5, 7). However, in vivo assembly is not a pre-requisite for an enzymatically active complex, as we could reconstitute RNase H2 activity by mixing recombinant RNASEH2A with separately purified RNASEH2BC in vitro (Fig. 2C). In contrast, mixing of RNASEH2AΔC with RNASEH2BC did not show activity above background (Fig. 2C). We therefore conclude that the RNASEH2A C terminus is essential for the formation of an enzymatically active RNase H2 heterotrimer.
FIGURE 2.
The C terminus of RNASEH2A is essential and sufficient for its interaction with RNASEH2B and -C. A, surface representation of the interface between human RNASEH2B (green), RNASEH2C (red) and the C terminus of RNASEHA:284–299 (blue ribbon). Amino acids found to be essential for binding in the peptide array (Tyr-287, Phe-288, Arg-291, Leu-293, Ala-296) are shown as sticks. B, Coomassie Blue stained SDS-PAGE gels show purification of GST-tagged RNASEH2B (GST-B) and associated proteins. Schematics illustrate subunit compositions of expressed complexes. Complexes composed of full-length subunits (RNASEH2ABC or RNASEH2BC) or with RNASEH2A truncated after residue 247 (RNASEH2AΔCBC) were bound to glutathione-Sepharose (bound) and released by cleavage with Precision Protease (cleaved). C, reconstitution of enzyme activity after mixing of RNASEH2A and RNASEH2BC in vitro. RNA18:DNA18 substrate was cleaved with increasing concentrations of purified proteins. + BC (dashed lines) indicates addition of RNASEH2BC heterodimer; RNASEH2Acat and RNASEH2AcatBC indicate D34A and D169A point mutations in RNASEH2A (catalytically inactive); RNASEH2AΔC indicates truncated RNASEH2A:1–247; mock indicates background activity from the cleaved fraction of a GST-only purification. D, affinity purification after co-expression of human GST-RNASEH2B and RNASEH2C with either murine RNASEH2A (RNASEH2mmA-hsBC), A. fulgidus RNaseHII (RNASEH2Afu-hsBC) or Afu RNaseHII fused to residues 248–299 of human RNASEH2A (RNASEH2Afu-hsAC-BC). E, cleavage of D14R1D3:D18 substrate by chimeric RNase H2 complexes described under D, as well as wild-type human RNase H2 (RNASEH2ABC), murine RNase H2 (mmRNASEH2ABC) and A. fulgidus RNaseHII (Afu RNASEH2). F, fluorescence anisotropy of a fluorescein-labeled peptide (RNASEH2A:283–299) as a function of increasing concentrations of RNASEH2BC. G, peptide array corresponding to RNASEH2A residues 275–299 probed with His-tagged RNASEH2BC and anti-His-HRP. Alanine point mutations are printed in red; RNASEH2A residues essential for binding are indicated in bold; “antibody only” shows background binding of anti-His-HRP to the array. H, sequence alignment for human (Hs) and Mus musculus (Mm) RNASEH2A with RNase HII from Archeoglobus fulgidus (Afu_rnhB). Secondary structure elements for human RNASEH2A are shown in blue (arrows for β-strands; loops for α-helices). The vertical black arrow marks where RNASEH2AΔC was truncated; dark blue boxes highlight peptides sufficient for binding RNASEH2BC; dark blue arrowheads indicate residues essential for binding in the peptide arrays described above; * mark residues mutated in AGS. This and all other sequence alignments were generated using ALINE (36).
The C Terminus of RNASEH2A Is a Eukaryotic Adaptation Allowing Binding of Accessory Subunits
Prokaryotes which lack the auxiliary RNASEH2B and RNASEH2C subunits also lack the C-terminal extension of RNASEH2A present in eukaryotes. It is therefore not surprising that Afu RNase HII is unable to bind the human RNASEH2BC heterodimer despite sharing 27% sequence identity with human RNASEH2A (Fig. 2D). In contrast, a chimeric Afu RNase HII to which C-terminal residues 248–299 of human RNASEH2A were fused allowed efficient heterotrimer formation (Fig. 2D). This complex was able to cleave RNase H2 substrates (though with reduced activity compared with Afu RNase HII alone; Fig. 2E). Furthermore, murine RNASEH2A could bind human RNASEH2BC (Fig. 2D), showing that residues essential for interaction are conserved between the mouse and human catalytic subunits. This mouse/human chimeric RNase H2 was functional and exhibited enzyme activity similar to both wild-type mouse and human RNase H2 (Fig. 2E).
RNASEH2A Residues Critical for Interaction with the Accessory Subunits
The extended C terminus of RNASH2A is central to the inter-subunit triple β-barrel architecture formed between all three RNase H2 subunits. As the chain tracing of this region (RNASEH2A:284–299) differs from that previously reported for murine RNase H2 (14), we performed biochemical and biophysical assays to corroborate our atomic model. In addition, these assays define specific residues involved in the interaction between the RNASEH2A C terminus and the accessory subunits.
Firstly, to confirm binding of the C-terminal 17 residues of RNASEH2A to the RNASEH2BC heterodimer, we assessed the anisotropy of a fluorescein-labeled peptide (RNASEH2A:283–299) in the presence of increasing concentrations of RNASEH2BC heterodimer (Fig. 2F). The peptide bound the heterodimer both with high affinity and specificity (Kd = 123 ± 10 nm), and the addition of purified full-length RNASEH2A successfully competed for RNASEH2BC binding while the RNASEH2AΔC C-terminal truncation mutant had no effect (data not shown).
Secondly, peptide arrays were used to examine the interaction between the RNASEH2A C terminus and the RNASEH2BC heterodimer in more detail. Use of such peptide arrays has proved an accurate method for the mapping of protein-protein interactions (29, 30). RNASEH2A peptides covering residues Ile-253–Ala-259 and Tyr-287–Leu-299 were sufficient to bind RNASEH2BC (Fig. 2G and supplemental Fig. S6A). Furthermore, systematic mutational analysis of a peptide corresponding to RNASEH2A:275–299 identified residues Tyr-287, Phe-288, Arg-291, Leu-293, and Ala-296 as essential for binding (Fig. 2G). This is in agreement with our structural model, which shows direct involvement for the side chains of these amino acids in inter-subunit interactions. All of these residues are conserved between human and mouse (Fig. 2H), consistent with the formation of a functional complex between mouse RNASEH2A and human RNASEH2BC. Significantly, substitution of one of these residues (Arg-291) has been found to cause AGS (10).
Furthermore, though unresolved in our crystal structure, peptide array analysis indicates that RNASEH2A residues Gly-264—Thr-270 are also involved in RNASEH2BC binding. We show that Arg-266 and Lys-267 are essential for binding, and mutagenesis of these two residues (R266A/K267A) in recombinant RNase H2 leads to reduced enzyme activity, without affecting complex stability (supplemental Fig. S6). In conclusion, our data demonstrate that the RNASEH2A C terminus has a key role in enzyme function, being critical both for complex formation and enzymatic activity.
RNASEH2C as a Platform for Complex Assembly
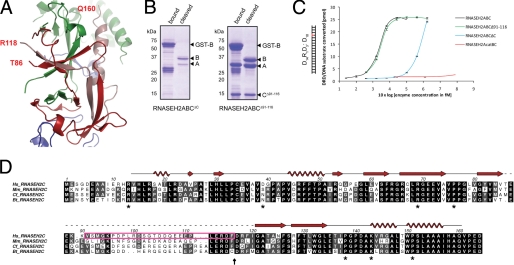
The human RNase H2 structure shows that the C terminus of RNASEH2C (Arg-118—Gln-160) forms an integral component of the heterotrimer architecture. Residues in the C-terminal kinked helix (RNASEH2C:143–160) contact both RNASEH2A and RNASEH2B, while β-strands formed by RNASEH2C residues Arg-118—Phe-126 and Arg-128—Leu-134 are part of two central β-sheets which contribute to the inter-subunit β-barrel architecture (Fig. 3A). Given that the chain tracing of this entire region (RNASEH2C: 87–160) is completely different from that previously reported for murine RNase H2 (14), we proceeded to confirm the functional relevance of the interaction interfaces defined by our structure. We examined complex integrity and enzyme activity of a deletion mutant lacking the C-terminal residues 117–164 of human RNASEH2C (residues that our human structure predicts are integral to heterotrimer formation). Removal of these residues abolished heterotrimer formation (Fig. 3B; RNASEH2ABCΔC) and over 99.5% of enzyme activity was lost (Fig. 3C). In contrast, deletion of RNASEH2C residues Val-91—Phe-116 had no effect on complex stability or enzymatic function: all three subunits were purified in equimolar amounts (Fig. 3B; RNASEH2ABCΔ91–116) and exhibited enzyme activity similar to that of wild-type RNase H2 (Fig. 3C). The previously published murine RNase H2 structure predicted that these residues should be critical for complex formation as they were central to the interwoven triple β-barrel fold (14). However, in our human and re-refined murine structures these residues were not modeled and we predicted them to form a disordered loop. Comparative sequence analysis shows that residues 91–111 are poorly conserved in mammals, in contrast with the rest of the RNASEH2C protein and the extreme C terminus in particular (Fig. 3D). Therefore, our structural, biochemical and primary sequence analyses are all consistent with RNASEH2C residues 87–117 forming a disordered loop, while RNASEH2C residues 118–160 comprise an integral part of the heterotrimer, essential for complex formation and enzymatic activity.
FIGURE 3.
The RNASEH2C C terminus, but not residues 91–116, are essential for complex formation and enzyme activity. A, RNASEH2C (red and pink ribbons), together with RNASEH2A (blue ribbons) and RNASEH2B (green ribbons), form a triple β-barrel. End points of the disordered loop in RNASEH2C (residues Thr-86 and Arg-118) are indicated. The chain is continuous from residue Arg-118 to Gln-160 (pink ribbon). B, Coomassie Blue-stained SDS-PAGE gels show purification of GST-tagged RNASEH2B (GST-B) and associated proteins. Affinity purification after coexpression of GST-RNASEH2B and RNASEH2A with either RNASEH2CΔC (left panel) or RNASEH2CΔ91–116 (right panel). C, RNASEH2ABCΔ91–116 shows the same activity as wild-type RNase H2 (RNASEH2ABC). In contrast, RNASEH2ABCΔC is more than 100-fold less active. Point mutations D34A/D169A (RNASEH2AcatBC) abolish all detectable RNase H2 activity. D, sequence alignment for human (Hs), Mus musculus (Mm), Canis familiaris (Cf), and Bos taurus (Bt) RNASEH2C. Secondary structure elements for human RNASEH2C are shown in red (arrows for β-strands; spirals for α helices). The magenta box marks amino acids deleted in RNASEH2CΔ91–116; the vertical black arrow marks where RNASEH2CΔC was truncated; * mark residues mutated in AGS.
Structural Implications of AGS Mutations
Point mutations in all three subunits of RNase H2 cause the auto-inflammatory disorder AGS, although it remains uncertain how these amino acid substitutions cause disease (7, 31). We are now able to assign 25 of the 29 disease-causing substitutions to the human structure (Fig. 4A), and find that a significant number of affected residues cluster in and around the C termini of the RNASEH2A and RNASEH2C subunits (Fig. 4B), regions we show to be critical for heterotrimer formation.
FIGURE 4.
AGS mutations at the RNASEH2A/RNASEH2C interface lead to reduced protein stability and enzyme activity. A, crystal structure of human RNase H2 is shown as ribbons and colored as in Fig. 1. Residues mutated in AGS are shown as yellow sticks. Nitrogen and oxygen atoms of highlighted residues are blue and red, respectively. B, close-up showing AGS affected mutations in the vicinity of the RNASEH2C C-terminal kinked helix. The left panel highlights residues in our human RNase H2 crystal structure while the right panel shows the corresponding residues for our re-refined murine model. (Arrows indicate the direction of the helix dipole running from positive to negative). C, fluorescence-based thermal stability assay was used to measure the melting temperatures of recombinant AGS mutant complexes. All mutations that cluster at the heterotrimer interface destabilize RNase H2. Error bars indicate the standard deviation of 10 independent measurements. D, most of these AGS mutations also cause reduced enzyme activity. Cleavage of D14R1D3:D18 substrate with increasing amounts of RNase H2 was measured in triplicate.
This region contains seven affected residues: five in RNASEH2C (R69W, P76L, P138L, K143I, and P151S), one in RNASEH2A (R291H) and one in RNASEH2B (A177T). These all influence the geometry of a kinked helix at the RNASEH2C C terminus. Firstly, we predict that the R69W mutation will disrupt the orientation of an RNASEH2C β-strand that packs against the kinked helix, as the Arg-69 side chain forms a salt-bridge with mRNASEH2C:D55 in the murine structure (Fig. 4B, right panel), anchoring it to a core β-sheet. Although the aspartic acid at Asp-55 is not conserved in humans, two other negatively charged amino acids (Glu-57 and Glu-60) are available to bind this arginine residue.
Secondly, three of the mutations (P76L, P138L, and P151S) target proline residues at transition points between secondary structure elements. Prolines have a unique structural role due to their rigid backbone dihedral angle enforced by a cyclic side chain. Together, the three affected proline residues influence the geometry of the kinked helix: Pro-138 commences the loop (138PGP140) that interrupts the β-turn preceding the kinked C-terminal helix; Pro-151 facilitates the bend, while its side chain packs against that of Pro-76 (which initiates a loop, 76PPG78, that changes the direction of a β-strand).
Thirdly, side chains of two mutated residues (RNASEH2C:K143 and RNASEH2A:R291) are involved in the binding of RNASEH2A to the kinked helix. Lys-143 is located at the first turn of the kinked helix and the amino group of its side chain caps a short helix at the RNASEH2A C terminus, strengthening the RNASEH2A helix dipole. This interaction locks the side chain of RNASEH2C:K143 into place, thus anchoring the kinked helix. Notably RNASEH2A:R291 also forms part of this interface, as its guanidinium group forms a salt bridge with the backbone carbonyl oxygen of RNASEH2C:G146 (Arg-148 in mouse).
Finally, we predict the A177T mutation in RNASEH2B to disrupt the interface between a RNASEH2B α-helix and the RNASEH2C kinked helix. The affected alanine residue sits in a hydrophobic pocket and packs against RNASEH2C:A156 of the kinked helix. Notably, the A177T substitution in RNASEH2B is the most common mutation, found in more than a quarter of AGS patients (10).
In summary, we predict all seven residues to influence the conformation of the RNASEH2C kinked helix and to have an important role at an interface bridging all three subunits. We would therefore expect mutations in these residues to reduce stability of the RNase H2 complex; a prediction we assessed using a fluorescence-based thermal shift assay, which measures the melting temperatures of purified recombinant human RNase H2 proteins. All seven AGS mutations caused reduced thermal stability of recombinant RNase H2 complex compared with wild-type RNase H2 (Fig. 4C). Furthermore, with the exception of RNASEH2B:A177T and RNASEH2C:P138L, mutations reduced cleavage of RNA/DNA hybrids (Fig. 4D and data not shown). In contrast to the small effect on the thermal stability of the complex, K143I and R291H had the most pronounced effect on enzyme activity, with an 85 and 94% reduction in specific activity respectively (Fig. 4D). This is particularly striking given their distance from the catalytic center and predicted substrate binding groove. Measurements of enzyme kinetics for RNase H2 with K143I and R291H mutations (Table 2) indicate that their enzyme activities are decreased due to reduced turnover (lower kcat) and not reduced substrate affinity (no change in Km). In conclusion, all these mutations either disrupt complex stability and/or in vitro enzymatic activity. Atomic coordinates have been deposited at the RCSB Protein Data bank under ID codes 3P56 for human RNase H2 and 3P5J for the re-refined murine RNase H2.
TABLE 2.
Kinetic parameters for human RNase H2 with and without AGS mutations
| Km | kcat | kcat/Km | |
|---|---|---|---|
| nm | min−1 | min−1nm−1 | |
| D14R1D3:DNA18 | |||
| Wild type | 28 ± 2.7a | 24 ± 0.7 | 0.9 |
| A:R291H | 27 ± 1.8 | 9.5 ± 0.2 | 0.3 |
| C:K143I | 30 ± 3.4 | 21 ± 0.8 | 0.7 |
| RNA18:DNA18 | |||
| Wild type | 143 ± 1.7 | 209 ± 0.8 | 1.5 |
| A:R291H | 144 ± 4.5 | 59 ± 0.6 | 0.4 |
| C:K143I | 158 ± 3.4 | 126 ± 0.9 | 0.8 |
a Values indicate standard errors.
DISCUSSION
Our structure of the human RNase H2 complex identifies novel structural elements of the RNASEH2BC heterodimer and catalytic RNASEH2A subunit relevant to enzyme function and disease. We establish that the C-terminal extension of RNASEH2A spans the two auxiliary subunits and is necessary for the formation of an enzymatically active complex. The C terminus of RNASEH2C also contributes to the unique interwoven architecture of the heterotrimer, by forming a helix that bridges the RNASEH2A and RNASEH2B subunits. Analysis of human disease mutations that cluster in this region demonstrates the importance of this interface for complex stability and enzymatic activity.
Role of the RNASEH2A C terminus in Enzymatic Activity
Our data establish that the RNASEH2A C-terminal tail is required for enzymatic activity. A unique adaptation of eukaryotic RNase H2 enzymes, this C-terminal tail permits enzyme activity only when accessory subunits are bound. Strikingly, substitution of specific residues in this C-terminal extension (R291H, K266A/R267A) impair enzyme activity, with minimal or no effect on complex stability (Fig. 4 and supplemental Fig. S6). Furthermore, mutation of a directly interacting residue of the RNASEH2C subunit (K143I) has the same effect. Notably, all these residues are distant from the catalytic center. These residues may therefore influence enzyme activity by forming an additional substrate-binding site distinct from the proposed RNASEH2A binding groove (14, 15). Alternatively, they could influence RNase H2 quaternary structure to alter enzymatic activity.
Spatial and temporal regulation of DNA replication and repair pathways is essential for genome stability in eukaryotic cells. Intriguingly, the RNASEH2BC heterodimer can exist as a stable subcomplex in vitro (Fig. 2) (7). As the RNASEH2A protein is present at much lower levels than the other subunits,6 this catalytic subunit might bind only when and where enzyme activity is required.
AGS Mutations Cause RNase H2 Dysfunction
We are able to map the majority of AGS mutations onto the human RNase H2 structure (86%). This provides a significant advance in understanding the structural context of these mutations in human disease. It is evident that some mutations directly affect enzyme catalysis (7, 9, 31, 32). It has also been established that other AGS mutations do not dramatically alter enzymatic activity in vitro (7, 31) and our findings are consistent with these studies. However, we find that mutations with no or limited effects on in vitro enzyme activity (RNASEH2B:A177T, and RNASEH2C:R69W, P76L, and P138L) significantly destabilize the RNase H2 complex, as measured by a reduction in thermal stability of recombinant RNase H2. Though such instability does not necessarily have a direct in vivo correlation, these mutations may lead to diminished RNase H2 levels and impaired cellular activity. Hence, our data are consistent with the notion that human mutations cause AGS through reduced enzymatic activity, in an analogous manner to disease mutations in TREX1 (33). Future examination of other human mutations may well provide additional insight into the biology of RNase H2. For instance, mutations in surface residues (e.g. RNASEH2A:T240M, RNASEH2B: K162T and V185G, and RNASEH2C:R13H) could impair interactions with nucleic acid substrates or other proteins.
In summary, our findings provide important structural insight into the requirements for RNase H2 complex integrity and regulation of enzyme activity, relevant to the cellular process of RNA/DNA hybrid degradation and the pathogenesis of systemic autoimmune disease. Additionally, the structure of the RNase H2 complex provides a framework for future investigation of RNase H2 function and its regulation in the eukaryotic cell.
Acknowledgments
We thank J. Ren and K. Harlos (Oxford) for assistance in crystallographic data collection and refinement, N. M. Devine (INP, Glasgow) for help with the peptide arrays, E. Blackburn (Edinburgh Biophysical Characterization Facility; use of this facility was supported by The Wellcome Trust, the Scottish University Life Sciences Alliance and the BBSRC), and S. Corless (IGMM) for technical assistance; and T. Walter, E. Seiradake, B. Janssen, N. Hastie, D. Stuart, and members of the Jackson Laboratory for useful discussions. We thank the ESRF and Diamond Light Source for synchrotron access.
This research was funded in part by a Medical Research Council Senior Clinical Fellowship grant (to A. P. J.) and Cancer Research UK grant (to E. Y. J.). The work was also supported by the Wellcome Trust Core Award (Grant 075491/Z04).
The atomic coordinates and structure factors (codes 3P5J and 3P56) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3.
M. A. M. Reijns and A. P. Jackson, unpublished data.
- RNase H2
- ribonuclease H2
- AGS
- Aicardi-Goutières Syndrome.
REFERENCES
- 1. Machida Y., Okazaki T., Okazaki R. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 2776–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li X., Manley J. L. (2005) Cell 122, 365–378 [DOI] [PubMed] [Google Scholar]
- 3. Förstemann K., Lingner J. (2005) EMBO Rep. 6, 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eder P. S., Walder J. A. (1991) J. Biol. Chem. 266, 6472–6479 [PubMed] [Google Scholar]
- 5. Jeong H. S., Backlund P. S., Chen H. C., Karavanov A. A., Crouch R. J. (2004) Nucleic Acids Res. 32, 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chon H., Vassilev A., DePamphilis M., Zhao Y., Zhang J., Burgers P. M., Crouch R. J., Cerritelli S. M. (2009) Nucleic Acids Res. 37, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maga G., Hubscher U. (2003) J. Cell Sci. 116, 3051–3060 [DOI] [PubMed] [Google Scholar]
- 9. Crow Y., Leitch A., Hayward B., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., Baumann C., Baxter P., Bertini E., Chandler K., Chitayat D., Cau D., Déry C., Fazzi E., Goizet C., King M. D., Klepper J., Lacombe D., Lanzi G., Lyall H., Martínez-Frías M., Mathieu M., McKeown C., Monier A., Oade Y., Quarrell O., Rittey C., Rogers R., Sanchis A., Stephenson J., Tacke U., Till M., Tolmie J., Tomlin P., Voit T., Weschke B., Woods C. G., Lebon P., Bonthron D. T., Ponting C. P., Jackson A. P. (2006) Nat. Genet. 38, 910–916 [DOI] [PubMed] [Google Scholar]
- 10. Rice G., Patrick T., Parmar R., Taylor C. F., Aeby A., Aicardi J., Artuch R., Montalto S. A., Bacino C. A., Barroso B., Baxter P., Benko W. S., Bergmann C., Bertini E., Biancheri R., Blair E. M., Blau N., Bonthron D. T., Briggs T., Brueton L. A., Brunner H. G., Burke C. J., Carr I. M., Carvalho D. R., Chandler K. E., Christen H. J., Corry P. C., Cowan F. M., Cox H., D'Arrigo S., Dean J., De Laet C., De Praeter C., Dery C., Ferrie C. D., Flintoff K., Frints S. G., Garcia-Cazorla A., Gener B., Goizet C., Goutieres F., Green A. J., Guet A., Hamel B. C., Hayward B. E., Heiberg A., Hennekam R. C., Husson M., Jackson A. P., Jayatunga R., Jiang Y. H., Kant S. G., Kao A., King M. D., Kingston H. M., Klepper J., van der Knaap M. S., Kornberg A. J., Kotzot D., Kratzer W., Lacombe D., Lagae L., Landrieu P. G., Lanzi G., Leitch A., Lim M. J., Livingston J. H., Lourenco C. M., Lyall E. G., Lynch S. A., Lyons M. J., Marom D., McClure J. P., McWilliam R., Melancon S. B., Mewasingh L. D., Moutard M. L., Nischal K. K., Ostergaard J. R., Prendiville J., Rasmussen M., Rogers R. C., Roland D., Rosser E. M., Rostasy K., Roubertie A., Sanchis A., Schiffmann R., Scholl-Burgi S., Seal S., Shalev S. A., Corcoles C. S., Sinha G. P., Soler D., Spiegel R., Stephenson J. B., Tacke U., Tan T. Y., Till M., Tolmie J. L., Tomlin P., Vagnarelli F., Valente E. M., Van Coster R. N., Van der Aa N., Vanderver A., Vles J. S., Voit T., Wassmer E., Weschke B., Whiteford M. L., Willemsen M. A., Zankl A., Zuberi S. M., Orcesi S., Fazzi E., Lebon P., Crow Y. J. (2007) Am. J. Hum. Genet. 81, 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramantani G., Kohlhase J., Hertzberg C., Innes A. M., Engel K., Hunger S., Borozdin W., Mah J. K., Ungerath K., Walkenhorst H., Richardt H. H., Buckard J., Bevot A., Siegel C., von Stulpnagel C., Ikonomidou C., Thomas K., Proud V., Niemann F., Wieczorek D., Hausler M., Niggemann P., Baltaci V., Conrad K., Lebon P., Lee-Kirsch M. A. (2010) Arthritis Rheum. 62, 1469–1477 [DOI] [PubMed] [Google Scholar]
- 12. Yang Y. G., Lindahl T., Barnes D. E. (2007) Cell 131, 873–886 [DOI] [PubMed] [Google Scholar]
- 13. Stetson D. B., Ko J. S., Heidmann T., Medzhitov R. (2008) Cell 134, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shaban N. M., Harvey S., Perrino F. W., Hollis T. (2010) J. Biol. Chem. 285, 3617–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chapados B. R., Chai Q., Hosfield D. J., Qiu J., Shen B., Tainer J. A. (2001) J. Mol. Biol. 307, 541–556 [DOI] [PubMed] [Google Scholar]
- 16. Otwinowski Z., Minor W. (1997) Macromolecular Crystallography, Part A, Academic Press, New York [Google Scholar]
- 17. Vagin A., Teplyakov A. (2010) Acta Crystallogr. D. Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 18. Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S. M., Bricogne G. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 19. Smart O., Brandl M., Flensburg C., Keller P., Paciorek W., Vonrhein C., Womack T., Bricogne G. (2008) Annual Meeting of the ACA, 137, Knoxville, TN [Google Scholar]
- 20. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tronrud D. E. (1996) J. Appl. Crystallogr. 29, 100–104 [Google Scholar]
- 22. Nettleship J. E., Brown J., Groves M. R., Geerlof A. (2008) Methods Mol. Biol. 426, 299–318 [DOI] [PubMed] [Google Scholar]
- 23. Winkler D. F., Hilpert K., Brandt O., Hancock R. E. (2009) Methods Mol. Biol. 570, 157–174 [DOI] [PubMed] [Google Scholar]
- 24. Dikmans A., Beutling U., Schmeisser E., Thiele S., Frank R. (2006) Qsar Combinatorial Science 25, 1069–1080 [Google Scholar]
- 25. Frank R. (2002) J. Immunol. Methods 267, 13–26 [DOI] [PubMed] [Google Scholar]
- 26. Kleywegt G. J., Harris M. R., Zou J. Y., Taylor T. C., Wählby A., Jones T. A. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2240–2249 [DOI] [PubMed] [Google Scholar]
- 27. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krissinel E., Henrick K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 29. Li X., MacLeod R., Dunlop A. J., Edwards H. V., Advant N., Gibson L. C., Devine N. M., Brown K. M., Adams D. R., Houslay M. D., Baillie G. S. (2009) FEBS Lett. 583, 3310–3316 [DOI] [PubMed] [Google Scholar]
- 30. Meng D., Lynch M. J., Huston E., Beyermann M., Eichhorst J., Adams D. R., Klussmann E., Houslay M. D., Baillie G. S. (2009) J. Biol. Chem. 284, 11425–11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perrino F. W., Harvey S., Shaban N. M., Hollis T. (2009) J. Mol. Med. 87, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rohman M. S., Koga Y., Takano K., Chon H., Crouch R. J., Kanaya S. (2008) FEBS J. 275, 4836–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crow Y., Hayward B. E., Parmar R., Robins P., Leitch A., Ali M., Black D. N., van Bokhoven H., Brunner H., Hamel B., Corry P., Cowan F., Frints S., Klepper J., Livingston J., Lynch S., Massey R., Meritet J., Michaud J., Ponsot G., Voit T., Lebon P., Bonthron D., Jackson A., Barnes D., Lindahl T. (2006) Nat. Genet. 38, 917–920 [DOI] [PubMed] [Google Scholar]
- 34. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) Nucleic Acids Res. 32, W665–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bond C. S., Schüttelkopf A. W. (2009) Acta Crystallogr. D. Biol. Crystallogr. 65, 510–512 [DOI] [PubMed] [Google Scholar]