Abstract
Rhodopsin, the visual pigment mediating vision under dim light, is composed of the apoprotein opsin and the chromophore ligand 11-cis-retinal. A P23H mutation in the opsin gene is one of the most prevalent causes of the human blinding disease, autosomal dominant retinitis pigmentosa. Although P23H cultured cell and transgenic animal models have been developed, there remains controversy over whether they fully mimic the human phenotype; and the exact mechanism by which this mutation leads to photoreceptor cell degeneration remains unknown. By generating P23H opsin knock-in mice, we found that the P23H protein was inadequately glycosylated with levels 1–10% that of wild type opsin. Moreover, the P23H protein failed to accumulate in rod photoreceptor cell endoplasmic reticulum but instead disrupted rod photoreceptor disks. Genetically engineered P23H mice lacking the chromophore showed accelerated photoreceptor cell degeneration. These results indicate that most synthesized P23H protein is degraded, and its retinal cytotoxicity is enhanced by lack of the 11-cis-retinal chromophore during rod outer segment development.
Keywords: Receptor Modification, Retina, Retinal Metabolism, Retinoid, Retinoid-binding Protein, Rhodopsin, Vision, Vitamin A
Introduction
Greater understanding of a genetically heterogeneous group of retinal disorders is now possible due to the results of studies that have revealed their causative genes. Many genetic loci can cause such retinopathies (RetNet) (1). Mutations in phototransduction genes, including those in opsin genes (2), constitute one of the major known causes of inherited blinding diseases (3). Among them, retinitis pigmentosa (RP)2 refers to a group that displays genetic heterogeneity and a range of clinical phenotypes (4). RP manifested predominantly by death of rod photoreceptor cells is a progressive disease characterized by night blindness that progresses to loss of peripheral vision and eventually all useful vision over decades (5). Of more than 100 mutant opsins associated with autosomal dominant RP (adRP), the most frequent mutation is P23H (6), accounting for ∼10% of human cases (7, 8).
In vitro studies have shown that the P23H opsin associated with adRP is misfolded and retained in the ER (9–12). Consequently, this protein is not transported to the cell membrane (12) but instead was degraded by the ubiquitin-proteosome system (13). Co-expression of adRP-linked opsin folding-deficient mutants and wild type (WT) opsin resulted in enhanced proteosome-mediated degradation and steady-state ubiquitination of both mutant and WT opsin in an experimental cell line (14). These results imply that in vivo, a misfolded monomer of P23H opsin can also induce co-aggregation with WT rhodopsin preventing rod outer segment (ROS) formation. This dominant negative effect on ROS formation has been considered as the underlying reason for the adRP inheritance of P23H in humans.
The retinal structure in heterozygous transgenic mice and rats expressing the P23H opsin partially mimics that of adRP in humans carrying this mutation (15–19). Mislocalization of the P23H opsin in the retina also has been reported in transgenic animal models (20), and this causes an abnormally reduced response to light (21). Most importantly, photoreceptor cells undergo rapid degeneration even when ROS are formed. Currently available adRP animal models can have differing expression levels of the mutant opsin transgene and WT opsin gene, mixed human and mouse gene products, or an additional mutation in the sensitive N terminus. So, we sought to improve the model by introducing the mouse P23H-opsin mutation in mice by knock-in techniques.
Rod photoreceptors in our P23H knock-in mice displayed regional degeneration and decreased lengths of their ROS, but most survived for several months, even though their OS structures were compromised. No abnormal aggregates were detected in their inner segments. This phenotype mimics changes in the retina of patients with adRP caused by the P23H opsin mutation. Genetic ablation of chromophore production accelerated rod cell degeneration in these mice, suggesting that an interaction between opsin and the mutant opsin proteins prevents normal ROS formation.
MATERIALS AND METHODS
Human Subjects, Psychophysical Testing, Electroretinography, and Retinal Cross-sectional Imaging
Patients with adRP and the P23H mutation in the opsin gene (n = 19) were included in this study (Fig. 1). Informed consent was obtained. Procedures followed the Declaration of Helsinki guidelines and were approved by the institutional review board.
FIGURE 1.
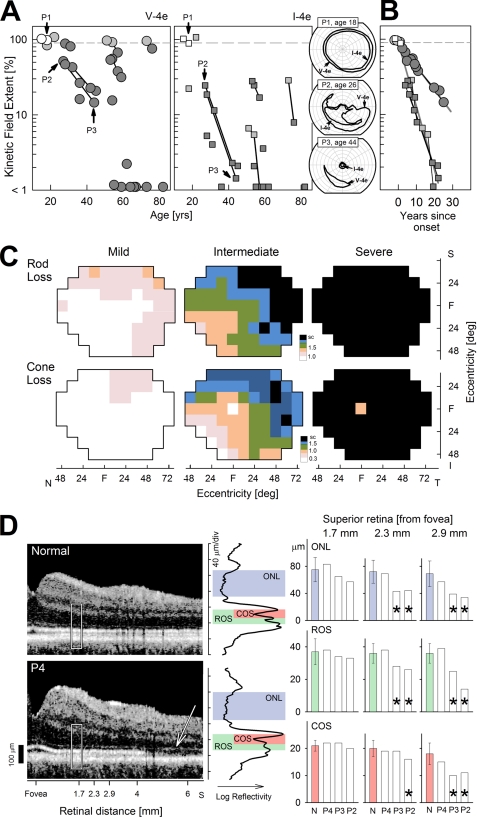
Retinal phenotype of humans with adRP caused by the P23H opsin gene mutation. A, extent of the kinetic visual field (for the larger V-4e stimulus, left, and smaller I-4e, right) is shown as a function of age for 19 patients. Longitudinal data from a subset of the patients are shown as symbols connected by lines. Fields that are normal (>90%) are shown with unfilled symbols, and fields that are abnormal with dark gray symbols; light gray symbols refer to those patients with normal V-4e but abnormally reduced I-4e fields. Inset (right), three representative patients showing a full visual field (P1), altitudinal (superior field) loss (P2), and a small central island with a residual island of nasal field function (P3). B, extent of visual field constriction for the I-4e stimulus shows a faster progression rate compared with that measured with the V-4e stimulus in the subset of patients with longitudinal data. Presumably, visual field constriction in all individuals progresses with an invariant exponential rate (7%/year for V-4e and 18%/year for I-4e) after a variable age of disease onset, which is estimated from the average of the intercepts fit to log-linear data for each target. C, retinal topography of rod sensitivity loss (upper row) and cone sensitivity loss (lower row) demonstrating mild (n = 2; ages 15 and 22), intermediate (n = 5; ages 18–54), and severe (n = 6; ages 30–82) stages of this disease. Rod loss was measured with a 500-nm stimulus (dark-adapted) and cone loss with a 600-nm stimulus (light-adapted). Color changes delineate the 50th percentile contour for a given level of sensitivity loss (specified in log units on the color scale; sc, scotoma). Maps are shown as visual fields of the right eye. S, I, N, and T refer to superior, inferior, nasal, and temporal visual field, respectively. D, retinal laminar architecture of a normal control subject (age 31) and P4 (age 18) imaged with optical coherence tomography along the vertical meridian from the fovea extending 6.5 mm into the superior retina. White rectangle represents location of the LRP shown on the right (black trace). Correspondence between local reflectivity changes on the LRPs and anatomical laminae of ONL, rod, and cone outer segments are shown. The thickness of the laminae was quantified at three neighboring locations (centered at 1.7, 2.3, and 2.9 mm from the fovea in the superior retina) in three patients (P4, P3, and P2) and compared with results in normals (error bars ±2 S.D.). Significant differences from normal are marked with an asterisk. Note the superior retinal region in P4 with remaining ONL showing loss of the signal originating from the connecting cilium between inner and outer segments (arrow).
Psychophysical Testing
Kinetic visual fields were performed with a Goldmann perimeter with large (V-4e, 1.72°) and small (I-4e, 0.11°) targets, and results were quantified (22). Static threshold perimetry in both the dark-adapted (500- and 650-nm stimuli) and light-adapted (600-nm stimulus on 10-cd·m−2 white background) states was performed using an automated perimeter (Humphrey Field Analyzer, San Leandro, CA) and analyzed for photoreceptor mediation and sensitivity losses, as described previously (22–24). Topography of rod and long/middle wavelength cone sensitivity losses was summarized by mapping the frequency of occurrence of a given loss (25, 26). A 3 × 3 moving average filter was applied (excluding the physiological blind spot) to all maps. In addition, foveal value was excluded for rod sensitivity loss maps but used (without applying the moving average) for cone sensitivity loss maps.
Electroretinography
Full field electroretinographies (ERGs) were performed according to published protocols (22).
Retinal Cross-sectional Imaging
Fourier domain (RTVue-100; Optovue Inc., Fremont, CA) and time domain (OCT3; Carl Zeiss Meditec, Inc., Dublin, CA) optical coherence tomography units were used. Our recording and analysis techniques for both systems were published previously (27–29). Scans along the vertical meridian were analyzed with custom programs (MatLab 6.5; MathWorks, Natick, MA). Longitudinal reflectivity profiles (LRPs) making up the optical coherence tomography scans were straightened and laterally averaged to increase the signal to noise ratio. Peaks and transitions between hypo- and hyper-reflective laminae were segmented directly using individual LRPs by the minima and maxima of the signal and its slope. For the outer retinal sublaminae, signal peak assignments were based on previously published work (30).
Animals
Mice were maintained under a 12-h light/12-h dark cycle, the light ranging from 1.2 to 32 lux inside their cages. Prior to ERG analysis, mice were kept under a dim red light (0.03–1.3 lux inside their cages) for at least 2 days. All animal procedures and experiments were approved by the Case Western Reserve University Animal Care Committees and conformed to both the recommendations of the American Veterinary Medical Association Panel on Euthanasia and the Association of Research for Vision and Ophthalmology. Lrat−/− mice (31, 32), hrhoG/hrhoG mice (33), and Rho−/− mice (34) were used for cross-breeding. Because of genetic background differences in each strain, histological data from cross-bred mice were compared with those from their littermates.
P23H Knock-in Mice
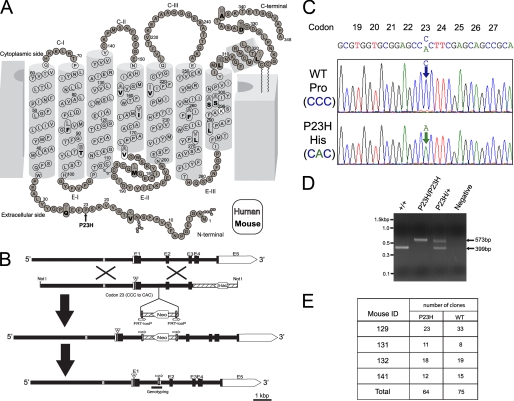
A mouse opsin gene clone from the 129 Sv/Ev Lambda genomic library was used to generate the P23H knock-in targeting vector, which then was validated by full-length sequencing. This vector was linearized by NotI (Fig. 2B, 2nd row) and transfected into iTL1 129 Sv/Ev embryonic stem cells by electroporation. After selection with G418, one recombinant clone was microinjected into blastocysts from which seven P23H knock-in heterozygous mice (P23Hneo/+) (F1) were generated (Fig. 2B, middle row). These were used to generate homozygous mice (P23Hneo/P23Hneo) (F3), which were crossed with B6FVB-Tg (EIIa-cre) C5379Lmgd/J mice (The Jackson Laboratory, Bar Harbor, ME) to generate heterozygous mice without the Neo cassette (P23H/+Cre+/−) (F4). All P23H mice used for the described experiments were offspring of homozygous mice lacking both the Neo cassette and the cre transgene (P23H/P23H Cre−/−) (F5, mouse ID 53 and 62) (Fig. 2B, bottom row).
FIGURE 2.
Two-dimensional model of rhodopsin, targeting strategy, and molecular characterization of P23H knock-in mice. A, two-dimensional model of human and mouse rhodopsin. The 18 residues that are not conserved between human and mouse are indicated with white and bold black letters, respectively. Residues of transmembrane helices are indicated by open circles. Lys296, indicated by white filled circle, makes a Schiff base linkage with 11-cis-retinal. The RefSeq data base accession numbers for the presented proteins are NP_000530.1 and NP_663358.1 for human and mouse opsin, respectively, and the two-dimensional model was modified from Ref. 92. Amino acid sequence of mouse and human opsin are well conserved including Pro23. The P23H mutation (indicated by an arrow) is located on the extracellular or intradiscal face of the protein near the Asn15 N-linked glycosylation site. B, targeting strategy for P23H opsin knock-in mice. Top, mouse opsin consists of five exons (E1–5). The opsin enhancer region 2 kb upstream of the opsin gene is indicated by gray line. 2nd row, the targeting vector contained 6.4 kb of 5′ and 1.9 kb of 3′ homologous sequence with C to A transversion at codon 23 (A inside arrowhead). Vector backbone sequences are indicated by hatched lines, and β-lac stands for the β-lactamase gene. 3rd row, as a result of homologous recombination, codon 23 in exon 1 was changed to CAC and the flippase recognition target (FRT)-loxP flanked neomycin resistance gene (Neo) was inserted after exon 1. Bottom row, neomycin was removed from targeted locus by cre-loxP recombination. By this knocking-in method, we generated mice with a point mutation at codon 23, which encode the P23H opsin. This mouse lacks the neomycin cassette but has a 174-bp vector backbone insertion in intron between exon 1 and exon 2. C, chromatogram of opsin cDNA from a P23H/+ mouse. The cDNA sequence chromatograms of two clones from one PND 26 P23H/+ mouse retina confirm the presence of both mutant and WT mRNAs. D, genotyping. PCR primers were designed to amplify the intron where the 174 bp of vector backbone sequence was inserted (Fig. 2B, bottom). The WT allele produced 399 bp and the P23H opsin allele produced 573 bp of PCR products. The negative control displayed no template contamination. Identities of PCR products were confirmed by sequencing (supplemental data 2). E, cDNA ratios of the P23H opsin to WT opsin. A total of 139 opsin cDNA clones isolated from four heterozygous mice (mouse ID 129, 131, 132, and 141, PND 26) were sequenced and used to identify ratios of P23H and WT opsin to subtotal clone numbers in each mouse. There was no statistically significant difference between P23H and WT opsin ratios (p = 0.62 between WT and P23H).
Opsin Locus Sequencing
Tails from mouse ID 53 and 62 were incubated with 50 μg of proteinase K (Fisher) and 0.01% SDS in 507 μl of 10 mm Tris, pH 8.0, at 60 °C for 1 h and then at 37 °C for 16 h in individual tubes. Genomic DNA was purified twice with phenol, which then was removed with chloroform. Genomic DNA was precipitated by adding 2 μl of nucleic acid carrier (Pellet Paint Co-Precipitant, Novagen, Gibbstown, NJ), 50 μl of 3 m sodium acetate, pH 5.2, and 550 μl of isopropyl alcohol. Precipitated genomic DNA was rinsed with 80% ethanol and dissolved in Tris-HCl, pH 7.4, and 1 mm EDTA. The opsin locus, including the entire targeted region, was amplified with the following: 0.5 μm 17392, 5′-CGTCTCCTGCCCAGATGAAGTTTGTAT-3′; 0.5 μm 28219, 5′-GCAGAGAAAACCAGGGCAGGTATGTAA-3′; 1 unit of DNA polymerase (Takara LA Taq, Takara, Otsu, Shiga, Japan); 2.5–3.5 mm MgCl2, 0.4 mm dNTP, and 55–136 ng of mouse ID 53 or 62 genome in 20-μl reaction mixtures placed in individual tubes. PCR amplifications were performed with an initial 1-min denaturation at 94 °C, 35 cycles at 94 °C (20 s), and 68 °C (15 min), and the last elongation at 72 °C (10 min). Amplified PCR products (∼11 kb) were gel-purified and cloned into pCR-XL-TOPO vector (Invitrogen) according to manufacturer's instructions except that 0.2 m NaCl and 11 mm MgCl2 were added for ligation. Cloned PCR products were sequenced in one direction with DNA sequencing primers at ∼700-bp intervals and an ABI 3730xl sequencer (Applied Biosystems, Carlsbad, CA). Sequences are shown in supplemental data 1. We also confirmed by the genotyping methodology described below that mouse ID 53 and 62 did not have an rd mutation (data not shown) (35). The PDE β-subunit mutation (rd) was detected by amplifying exon 7 of the PDE β-subunit with primer pairs named W149 and W150 and digested with the restriction enzyme DdeI as detailed previously (35).
Genotyping
Genomic DNA was isolated from mouse tails by using a Wizard genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's instructions with the following modifications. All procedures were done with half the recommended volumes, and DNA was precipitated with nucleic acid carrier (Pellet Paint Co-Precipitant, Novagen). PCR was performed with 20-μl reaction mixtures containing the 0.15 μm primer pair, 0.5 mm MgCl2, 0.2 mm dNTP, 0.4 units TaqDNA polymerase (Roche Applied Science), and 50–200 ng of genomic DNA. PCR amplifications were done with an initial 5 min of denaturation at 94 °C, and 35 cycles at 94 °C (30 s), 60 °C (30 s), and 72 °C (1 min) with final elongation at 72 °C (7 min). For hrhoG genotyping, PCR was accomplished with an annealing at 70 °C. Primer sequences and PCR product sizes were as follows: P23H, GenoRhoL 2163, 5′-TGGAAGGTCAATGAGGCTCT-3′, and GenoRhoR 2561, 5′-GACCCCACAGAGACAAGCTC-3′, 399 bp from WT and 573 bp from P23H (Figs. 2B, bottom, and D); P23H neo, Ch6 bef1stloxP, 5′-GGAGCAGGCAAAAATCACAT-3′, and aft1stLoxP, 5′-ACGAGATCAGCAGCCTCTGT-3′, 499 bp; Cre transgene, GenoCreL 473, 5′-GCACTGATTTCGACCAGGTT-3′, and GenoCreR 672, 5′-GCTAACCAGCGTTTTCGTTC-3′, 199 bp; opsin knock-out loci, K183-F, 5′-CTTGCCGAATATCATGGTGG-3′, and opsin exon1-R, 5′-TCTGCTCATACCTCCAAGTG-3′, ∼650 bp; opsin WT loci, opsin exon 1F, 5′-CAGTTACCTGGAGTTGCGCT-3′, and opsin exon1-R, 439 bp. The RPE65 L450M variant was identified by amplifying the RPE65 genome, including the L450M variant coding region with a primer pair as described previously (36); this was digested with the restriction enzyme MwoI that recognizes the Leu450 allele (CTG = Leu) but not the Met450 allele (ATG = Met). Lecithin-retinol acyltransferase knock-out loci and WT loci were genotyped with primer pairs named Neo1 and LRAT1S for knock-out loci and LRATWT1 and LRAT1S for WT loci as described previously (31). Human opsin-GFP fusion (hrhoG) knock-in loci and mouse opsin loci were genotyped as reported previously (33). The PDE β-subunit mutation (rd) was detected by amplifying exon 7 of the PDE β-subunit with primer pairs named W149 and W150 and digested with the restriction enzyme DdeI as detailed previously (35).
Opsin cDNA Cloning
Neural retinas from P23H/+ mice at PND 26 (mouse ID 129, 131, 132, and 141) were dissolved in 800 μl of TRIzol (Invitrogen) containing 200 μg of glycogen (Roche Applied Science). Total RNA was extracted according to the manufacturer's instructions and then precipitated by nucleic acid carrier (Pellet Paint Co-Precipitant, Novagen). Precipitates were dissolved in 88 μl of RNase-free water containing 40 units of RNase inhibitor (Roche Applied Science) and then treated with 1 unit of RNase-free DNase (RQ1, Promega) at 37 °C for 55 min. After RNA clean-up with spin columns (RNeasy MiniElute Cleanup kit, Qiagen, Valencia, CA), cDNA was synthesized with Sensiscript reverse transcriptase (Qiagen) using oligo(dT)12–18 primers. For a negative control, the same amount of total RNA was processed without reverse transcriptase. PCR was performed with 20-μl mixtures containing the following: 0.16 μm mRhol21, 5′-AAGCAGCCTTGGTCTCTGTC-3′; 0.26 μm mRhoR1291, 5′-AGTGGGGAGCCTCATTTTG-3′; a 1-unit mixture of Taq and proofreading polymerase (Expand High Fidelity Enzyme Mix, Roche Applied Science), 0.2 mm dNTP, and 4 mm MgCl2. PCR amplifications were done with an initial 2-min denaturation at 94 °C, 10 cycles at 94 °C (15 s), 57 °C (30 s), and 72 °C (55 s), 25 cycles at 94 °C (15 s), 57 °C (30 s), and 72 °C (55 s) with 5 s elongation for each successive cycle and a final 7-min elongation at 72 °C. Predicted sizes of PCR products (∼1.2 kb) were not generated by negative controls involving either nonreverse-transcribed total RNA or water as templates (data not shown). After purification by spin column chromatography (Microspin S-400, GE Healthcare), A-tailing was performed with 10-μl mixtures containing 5 units of TaqDNA polymerase (Roche Applied Science) and 0.2 mm dATP at 67 °C for 35 min. A-tailed PCR products were ligated into pGEM-T Easy vector (Promega) and transfected into JM109 cells (Promega). The presence of WT and P23H opsin was determined by sequencing. Examples of sequence chromatograms from mouse ID 129 (P23H/+) are shown in Fig. 2C. To assess the ratio between P23H and WT opsin mRNA, we sequenced 139 cDNA clones from four heterozygous mice and calculated the WT to P23H opsin cDNA ratios (Fig. 2E).
Immunohistochemistry
Eye cups were fixed with 4% paraformaldehyde in PBS (136 mm NaCl, 2 mm KCl, 8 mm Na2HPO4, 1 mm KH2PO4, pH 7.4) containing 5% sucrose for 5 h at 4 °C. Fixed tissues were dehydrated and then frozen as described previously (37). Cryo-sections 5–7 μm thick were made with a cryostat-microtome (CM1850, Leica, Bannockburn, IL) and incubated with antibodies as reported previously (38). Samples were incubated with Alexa 488 PNA (Invitrogen) at room temperature for 2.5 h. Final concentrations of antibodies were as follows: mouse 1D4 (5 μg/ml) and rabbit anti-calreticulin (2 μg/ml, Sigma); rat 13A4 anti-prominin-1 (0.5 μg/ml, from Dr. Corbeil); Cy3-conjugated goat anti-mouse IgG (3 μg/ml, Jackson ImmunoResearch, West Grove, PA); Alexa 488 goat anti-rabbit IgG (4 μg/ml, Molecular Probes); Alexa 488 goat anti-rat IgG (4 μg/ml, Molecular Probes); and Alexa 488 PNA (5 μg/ml, Invitrogen). Fluorescence was detected with a fluorescence microscope (CTR 6000, Leica). Images were captured by a CCD camera (QImaging Retiga Exi Fast 1394, QImaging, Surrey, British Columbia, Canada) and analyzed with Adobe Photoshop software. Nuclei in columns of photoreceptor cells were manually counted with “cell counter” plug-in Image J software (National Institutes of Health) in retinal sections from at least three mice stained with 12.3 μg/ml Hoechst 33342 (Invitrogen).
Light and Electron Microscopy
Eye cups were processed as described previously (39) with the modification that initial fixation with aldehyde/DMSO was done at 37 °C for 2–4 h and then eye cups were cut in half on their dorsal-ventral axis and fixed again for several minutes. To track the dorsal-ventral and nasal-temporal axis, the right and left eyes were processed in individual vials. Thick sections (300–900 nm) were stained with 0.5% toluidine blue O (Sigma) to observe morphology and with 1% toluidine blue O and 1% borax, pH 9.2, to count nuclei. Images were collected with a microscope (CTR 6000, Leica), equipped with a CCD camera (QImaging Micro Publisher 5.0 RTV QImaging), and analyzed with Adobe Photoshop software. Nuclei were counted as described previously (40, 41) with the following modifications: five continuous segments of retina, each 200 μm long, were analyzed with a cell counter plugged-in Image J software (National Institute of Health) starting 400 μm from the optic nerve and extending to the dorsal or ventral ciliary marginal zone. Average numbers of nuclei in a 200-μm length of retina were calculated from three P23H/+ mouse retinas (PND 63) and one WT mouse retina (PND 63). Our data from the PND 63 WT mouse (about 273 photoreceptor cell nuclei in a 200 μm length of retina containing ∼3% cone photoreceptor cell nuclei) were similar to those reported previously (41).
Immunoblotting Analyses
Neural retina obtained from mouse eyes was immediately frozen in liquid nitrogen. Frozen retinas then were processed in homogenization buffer (2% SDS in 12.5 mm Tris-HCl, pH 6.8). Protein concentrations were measured with a two-dimensional Quant kit (GE Healthcare). Proteins separated by SDS-PAGE in 10 or 12% acrylamide gels were transferred to PVDF membranes by a semi-dry electrophoretic transfer apparatus (Bio-Rad). Transfer was performed with 10% methanol in 10 mm Tris and 192 mm glycine at 15 V for 1 h. Resulting membranes were blocked with 0.3% nonfat dry milk (Nestle, Carnation Nestle, Vevery, Switzerland) in TBST buffer (50 mm Tris-HCl, pH 7.5, 15 mm NaCl, and 0.1% Tween 20) for 20 min, incubated with primary antibody overnight at 4 °C, washed with TBST, incubated with peroxidase-conjugated secondary antibody for 1–2 h at room temperature, washed again with TBST, and finally incubated with chemiluminescent substrate (SuperSignal West Pico Substrate, Thermo Scientific, Rockford, IL). Signals were detected by exposure to x-ray film (CL-XPosure, Thermo Scientific) and developed with an x-ray film processor (Mini-medical 90, ImageWorks, Elmsford, NY). Images captured by a CCD camera (Alpha Imager HP, Cell Biosciences, Santa Clara, CA) were analyzed with Adobe Photoshop software. Final antibody concentrations were as follows: mouse monoclonal 1D4 (42), 0.5 μg/ml; mouse monoclonal B6–30N (43), 0.26 μg/ml; rabbit polyclonal anti-GFP (Novus Biologicals Littleton, CO), 0.11 μg/ml; peroxidase-conjugated goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA), 0.02 μg/ml; and peroxidase-conjugated donkey anti-rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA), 0.13 μg/ml. For glycosylation analyses, 30 μg of protein was treated with 1,100 units of Endo H (New England Labs, Ipswich, MA) or 2,200 units of PNGase F (New England Labs) in 22 μl of reaction solution that included 44 ng of leupeptin and 1.75 μg of PMSF according to the manufacturer's protocol.
Mouse ERGs
All mouse ERG procedures were performed by published methods (44, 45). For single-flash recording, the duration of white light flash stimuli from 20 μs to 1 ms was adjusted to provide a range of illumination intensities from −3.7 to 1.6 log cd·s/m2. Three to eight recordings were made at sufficient intervals between flash stimuli (10 s to 10 min) to allow recovery from photo-bleaching effects.
Statistical Analyses of ERGs
Data representing the means ± S.D. or the means ± S.E. for the results of at least three independent experiments were compared by the one-way analysis of variance test.
RESULTS
P23H Mutation in the Opsin Gene Causing adRP, the Human Disease to Model
Typical clinical visual fields of individuals with P23H opsin mutation (Fig. 1A) and the rates of their progressive constriction with age (Fig. 1B) are illustrated. These light-adapted kinetic visual fields (mainly a cone-mediated visual task) measured with large and small stimuli can be within normal limits in a minority of patients into the 3rd decade of life (Fig. 1A, white symbols; see inset P1). Others can retain a normal field extent with a large stimulus but reduced extent to a smaller target (Fig. 1A, light gray symbols) even into the 8th decade of life. Most individuals displayed reduced visual fields to both stimulus sizes (Fig. 1A, dark gray), and the common pattern of visual loss is altitudinal, meaning greater dysfunction in the superior visual field (Fig. 1A, see inset, P2) representing the inferior retina. To determine the progression of visual field loss in individuals with the P23H opsin mutation, we considered a subset of six patients with available longitudinal data (intervals of 3–18 years; median = 6 years), and we assumed a model wherein the disease progresses exponentially after its onset (46–51). Estimated rates of visual field constriction were 7%/year for the large target and 18%/year for the small target (52, 53). Assuming an invariant exponential rate of progression, individual ages of disease onset (estimated from the intercepts) ranged from 17 to 72 years. Replotting of the longitudinal visual field data as a function of time after the age of onset showed a substantially reduced scatter (Fig. 1B). Rod ERGs in all patients in the cohort were abnormally reduced in amplitude, or signals were not detectable; cone ERGs in two of the patients were within normal limits, but all others had either reduced amplitude or no detectable cone signal.
Rod- and cone-mediated vision and the regional variations in each were also measured across the visual field in individuals with P23H opsin mutation and are represented as three stages of increasing severity (Fig. 1C). In mild disease there is some rod visual dysfunction in the superior field extending into the temporal peripheral field. At this stage of rod disease, there is less cone dysfunction, but later cone defects are also first noted in superior field loci. Intermediate disease is manifested by superior and temporal field rod scotomas (blindspots) and relative preservation of inferior and inferior-nasal loci (corresponding to the superior and superior-temporal retina). A similar pattern but with a less pronounced deficit occurs with measured cone sensitivity loss. In severe disease, there is no detectable rod function and only a small central island of reduced cone function.
In vivo histopathology with optical coherence tomography was quantitatively assessed in three patients (P4, P3, and P2) at three parafoveal loci (1.7, 2.3, and 2.9 mm away from the central fovea) in the superior retina. This region was selected because it normally contains rod and cone photoreceptors (rod:cone ratio ∼10–22) (54), and in some patients it could show remaining quantifiable retinal architecture (Fig. 1D, cross-sectional scans on left). P4, a patient with relatively mild disease, showed normal thicknesses of the outer photoreceptor nuclear layer (ONL), ROS, and COS at all three loci (Fig. 1D, bar graphs). P3, with more advanced disease, showed normal thickness of ONL, ROS, and COS at 1.7 mm transitioning to abnormal ONL and ROS but normal COS at 2.3 mm; at 2.9 mm, all three measures were abnormal. P2 showed normal thickness at 1.7 mm but abnormal thickness parameters at the more eccentric loci. Interestingly, the thickness of the inner segment layer was normal at all three parafoveal loci examined in each of these patients. Also notable was the outer retinal laminopathy in P4 at eccentricities beyond 4.5 mm (Fig. 1D, arrow). Here, there was an abrupt loss of the signal normally originating from the connecting cilium region between inner and outer segments even though distinct signals associated with the external limiting membrane and the RPE were still detectable.
The above retinal structural observations support the conjecture that one of the earliest expressions of the retinal disease in adRP patients with the P23H opsin mutation involves abnormal thinning of the ONL and shortening of the ROS; spatially lagging behind the encroaching wave of primary rod disease is shortening of the COS. With more extensive disease, there is further abnormality of inner and outer segments followed by loss of all remaining photoreceptors. Retinal function measurements (by psychophysics and electroretinography) support the structural findings by demonstrating that loss of rod function precedes loss of cone function. The spatial topography of degeneration across the rod-dominant human retina displays strong intraretinal variation; on average, the superior temporal retinal region is the last to degenerate completely. After all rods are lost, there is often some cone function remaining at the cone-only fovea.
Generation and Retinal Pathology of P23H/+ Mice
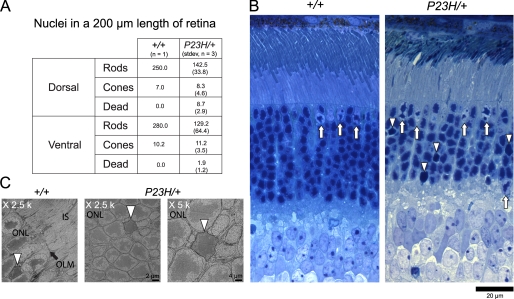
We generated P23H opsin (Fig. 2A) knock-in mice (P23H/+) by replacing the endogenous opsin gene with a mouse opsin gene carrying the P23H mutation. A neomycin cassette, first introduced into the mouse opsin gene for the purpose of clone selection, was later removed by cross-breeding with cre transgenic mice to generate P23H knock-in mice (Fig. 2B). cDNA sequencing of these heterozygotes (Fig. 2C) showed that both the WT and P23H mRNA were transcribed. To determine the ratio of WT and P23H mRNA, we sequenced more than 130 cDNA clones obtained from four mice. The number of WT and P23H clones was similar indicating that the mRNA level of the P23H opsin was comparable with that of the WT opsin (Fig. 2E). Morphological analysis of the retina revealed that at postnatal day 63 (PND 63), the ONL consisted of 9–10 rows of nuclei in WT (+/+) mice, 6–7 rows in P23H/+ mice, and 0–1 row in homozygous (P23H/P23H) mice. Moreover, the ONL had decreased to only 2–4 rows of nuclei by PND 112 in P23H/+ mice. By this age, dorsal versus ventral retinal degeneration also differed significantly. At PND 112, P23H/+ mice with four rows of nuclei in the dorsal retina had only two rows of nuclei in their ventral retina where the lengths of ROS also were shortened (Fig. 3A). In homozygote P23H/P23H mice, photoreceptor cells were almost gone by PND 63, and the inner nuclear layer (INL) was apposed to the RPE layer. Some rod photoreceptor cells survived between the INL and RPE (Fig. 3A, arrows). Quantitation of the numbers of nuclei per column of ONL for mice of different genetic backgrounds and ages is shown in Fig. 3B.
FIGURE 3.
Progressive retinal degeneration in P23H/+ mice. A, plastic sections of retinas from WT (+/+), heterozygous (P23H/+), and homozygous (P23H/P23H) mice of differing PND ages. Sections were stained with toluidine blue. OS, outer segment; IS, inner segment; ONL, outer nuclear layer; INL, inner nuclear layer; and GC/NFL, ganglion cell/nerve fiber layer. Arrows represent surviving rod nuclei in a homozygous P23H/P23H mouse. B, data showing the number of nuclei per column in genetically different mice. Cryosections from WT, P23H/+, and P23H/P23H mice of differing PND ages were used for quantification. Data were derived from three eyes of three or five eyes of five mice. P23H/+ mice show age-dependent progressive retina degeneration, which is more severe in the ventral retina compared with the dorsal retina. P23H/P23H mice lost most of their photoreceptor cells by PND 63.
To investigate the survival of cone photoreceptor cells in our animal model, we analyzed cone cells after labeling with PNA that selectively binds to cone-specific domains of the interphotoreceptor matrix (cone matrix sheath) and COS (55). By PND 35, ROS of P23H/+ mice were significantly shorter than those of WT mice (Fig. 4). However, the abundance and length of PNA-labeled cone inner and outer segments closely mimicked their counterparts in the retina of WT animals (Fig. 4). Because the distance between the RPE and OLM was significantly shorter than that in the retina of WT mice, PNA-labeled inner and OS of P23H/+ mouse retinas were tilted. We also determined the number of cone photoreceptor cells by counting their nuclei after toluidine blue staining of plastic sections; nuclei of cones and rods were readily distinguishable (Fig. 5) (40). Although rod nuclei numbers in P23H/+ mice were only about half of those of WT animals (Fig. 5A), cone nuclei numbers were close to those found in WT mice. In P23H/+ mice, some cells had lost clumps of heterochromatin and displayed a characteristic broad staining pattern of the entire nucleus (Fig. 5B, arrowheads). We considered these to be dying cells, but their nuclear appearance differed from that of pyknotic photoreceptor-cell nuclei observed in light-induced photoreceptor degeneration (56). These observations indicate that cone photoreceptor cells initially survived at least until mice were 2 months old, when nearly half of their rod photoreceptor cells had disappeared. High resolution images of all dead rod cell photoreceptor nuclei from three mice at PND 63 were similar to these found in retinas from WT animals showing characteristic necrotic structures (57) and lacked apoptotic bodies (Fig. 5C). Because apoptotic bodies disappear in 24 h (57), only a small number of apoptotic cells might have escaped detection by our method. Thus, our data suggest that the observed degeneration in adult mice was mainly due to necrosis.
FIGURE 4.
Rods and cones in P23H/+ mice. Cryo-sections from PND 35 WT and P23H/+ mice from the same litter were double-stained with opsin (1D4) antibody and lectin PNA. Images labeled with same marker were captured under identical conditions. Significant shortening of ROS was observed in P23H/+ mice, but the number of cone photoreceptor cells labeled by PNA was not reduced by this age. RPE, retinal pigmented epithelium; ONL, outer nuclear layer; INL, inner nuclear layer; IPL, inner plexiform layer; GC/NFL, ganglion cells/nerve fiber layer; DIC, differential interference contrast.
FIGURE 5.
Rod and cone nuclei numbers and images in P23H/+ mice. A, average numbers of rod and cone nuclei in 200-μm lengths of WT and P23H/+ mouse retina were determined at PND 63. Compared with WT littermates, the number of rod photoreceptor nuclei in P23H/+ mouse was almost half, whereas the numbers of cone nuclei were virtually the same. B, plastic sections of retinas from PND 63 WT and P23H/+ mice were stained with toluidine blue. Rod and cone photoreceptor nuclei were distinguishable by the shape of their chromatin; cone nuclei are indicated by arrows. We also identified dead rod photoreceptor nuclei that had lost clumps of chromatin (indicated by arrowheads). C, TEM images reveal dead photoreceptor cell nuclei (arrowheads) in both WT and P23H/+ mice. IS, inner segment; OLM, outer limiting membrane; and ONL, outer nuclear layer.
Assessment of Retinal Function with ERGs
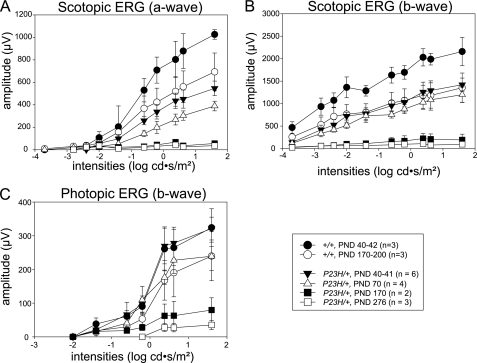
Full-field ERGs were obtained in P23H/+ mice to evaluate their age-dependent changes in retinal function. By PND 41, ERG responses in P23H/+ mice under scotopic conditions were severely reduced compared with those of control WT mice (Fig. 6, A and B). The waveforms were less than 50% of PND 40–42 WT mice at the highest stimuli (1.6 cd·s/m2) for a-waves and b-waves and less than 30% at the lowest stimulus (−3.7 cd·s/m2) for b-waves. These responses decreased further with a 10% loss in both amplitudes until PND 70, whereas no significant changes were observed in control WT mice (data not shown). By PND 170, ERG responses under scotopic conditions were nearly undetectable (Fig. 6, A and B), whereas responses observed in control WT mice by PND 170–200 of age were 65% of PND 40–42 WT mice at the highest stimuli (1.6 cd·s/m2) for a-waves and b-waves. ERG responses in P23H/+ mice under photopic conditions were maintained at comparable levels with those of WT mice at PND 40, whereas mildly reduced responses (20% loss) were observed at PND 70, and severely depressed responses similar to those under scotopic conditions were noted by PND 170. There was a continuous decline in responding until PND 276, whereas only a mild reduction (26% loss) occurred in control WT mice at this age (Fig. 6C). These observations indicate that substantial rod cell dysfunction had occurred by PND 41, well before modest cone dysfunction was evident at PND 70, which slowly progressed. Consistent with the data from human P23H adRP patient ERG and psychophysical data, rod photoreceptor function was affected from the youngest age in mice that we analyzed (PND 40), whereas cone function was not initially compromised. However, both rod and cone photoreceptor cells progressively degenerated with time.
FIGURE 6.
ERG responses in P23H/+ mice. Full field ERG responses of P23H/+ and WT (C57BL/6) mice were recorded under scotopic (A and B) and photopic (C) conditions. Compared with WT control mice, rod photoreceptor cell-evoked a- and b-wave amplitudes under scotopic conditions were reduced in P23H/+ mice at all analyzed ages, and the extent of this abnormality increased with age (A and B). Cone photoreceptor cell-dependent b-wave amplitudes under photopic conditions in P23H/+ mice were same as in WT mice at PND 40–41 but then decreased with age (C). Rod photoreceptor cell function was reduced to about half that of WT at PND 40–41 and decreased even more with age. Cone function was close to WT at PND 40–41, but reduced to about 80% of WT at PND 70, and then it decreased with age as well.
Glycosylation and Degradation of P23H Opsin
To investigate the glycosylation status of P23H opsin, we analyzed retinal homogenates by immunoblotting. WT and P23H opsin migrate similarly in SDS-PAGE so they could not be differentiated from each other. However, no detectable band was observed in P23H/P23H mice at PND 23 because severe photoreceptor degeneration had taken place by this age (Fig. 7A). To distinguish WT opsin from P23H opsin, we generated heterozygote mice with GFP-tagged WT human opsin (33) and P23H human opsin (hrhoG/P23H). In these mice, WT opsin and P23H opsin were distinguishable by their molecular weights. The molecular mass of GFP-tagged WT opsin was about 70 kDa. A GFP-truncated form of human opsin was also observed (Fig. 7B, left panel, ∼40 kDa, narrow arrow), which was also distinguishable from P23H opsin (Fig. 7B, left panel, ∼35 kDa, white arrow). Because the P23H opsin band was much less intense than that of GFP-tagged WT opsin, we next investigated the ratio of WT to P23H opsin protein levels in heterozygous mice. Whereas a weak GFP-tagged WT opsin signal was visualized (Fig. 7B, thick black arrow) in the ×1 lane, a stronger P23H opsin band was seen only in the ×100 lane (white arrow) loaded with 100 times the amount of retinal homogenate as the ×1 lane. The latter band did not stain as strongly as the comparable control band in the ×10 lane (Fig. 7B) indicating that the level of P23H opsin was between l and 10% of GFP-tagged WT opsin. To investigate post-translational glycosylation of P23H opsin, retinal homogenates from P23H/P23H mice at PND 10 were treated with different deglycosylation enzymes. Endo H is a specific endoglycosidase that cleaves asparagine-linked mannose-rich oligosaccharides of proteins undergoing post-translational modification in the ER and Golgi lumen. Unlike WT opsin, the P23H opsin was Endo H-sensitive (Fig. 7C, right panel, open arrows). In control experiments of deglycosylation, the less specific peptide:N-glycosidase F removed all carbohydrates from both proteins (Fig. 7C). These data indicate that P23H opsin undergoes incomplete glycosylation and is retained in the ER and/or Golgi where most of it is degraded.
FIGURE 7.
P23H opsin, glycosylation and ratio to WT opsin protein. A, retinal homogenates from PND 23 WT, P23H/+, and P23H/P23H mice were labeled with N-terminal (B6–30) and C-terminal (1D4) opsin antibodies. All lanes were loaded with 10 μg of homogenate. Opsin monomers (∼35 kDa) produced double bands due to differences in glycosylation. The molecular mass of opsin monomers shifted down to ∼30 kDa after treatment with PNGase F. Double bands were due to incomplete deglycosylation. No glycosylation difference was observed between WT and P23H/+ opsin. No detectable signal was observed in retinal homogenates from P23H/P23H mice. B, retinal homogenates from PND 28 mice were labeled with B6-30 and anti-GFP antibodies. Left panel, in homozygous GFP-tagged human opsin knock-in mice (hrhoG/hrhoG), strong bands were observed for the monomer of GFP-tagged human opsin (∼70 kDa) and its dimer (∼160 kDa). A band of human opsin with a truncated GFP tag (∼ 40 kDa) was also seen that was distinguishable from the P23H opsin band (∼35 kDa) observed in lane of P23H/hrhoG; a heterozygous hrhoG and P23H knock-in mouse. Two μg of protein were loaded. Middle panel, different amounts of retinal homogenate from P23H/hrhoG mice were loaded, from left to right: ×1, 0.175 μg; ×10, 1.75 μg; and ×100, 17.5 μg. A faint GFP-tagged human opsin monomer band was detected in lane ×1 (∼70 kDa, thick black arrow) and very faint P23H opsin monomer band was detected in lane ×10 (∼35 kDa, open arrow). The P23H band was obvious in lane ×100 (open arrow) where a fainter truncated GFP tagged human opsin monomer band was also seen (∼40 kDa, narrow arrow). Right panel, 2 μg of retinal homogenate from a P23H/hrhoG mouse labeled with anti-GFP antibody produced a single broad band at ∼70 kDa confirming the identity of GFP-tagged human opsin. C, immunoblots of 2 μg of protein samples treated with Endo H and PNGase F were resolved on 12% SDS-polyacrylamide gels (upper panels) and 10% gels (lower panels). Left panel, retinal homogenate from a WT mouse (C57BL/6J, 5-month-old) labeled with 1D4 antibody. Both WT opsin dimer and monomer were shifted to lower molecular weights after either Endo H- or PNGase F treatment. Middle panel, retinal homogenate from a P23H homozygous mouse (PND 10) labeled with 1D4 antibody evidenced a similar shift. Right panel, retinal homogenate from a P23H homozygous mouse (PND 10) labeled with B6-30 antibody. Arrowheads in upper panels indicate P23H opsin dimers resolved in 12% gels. Arrowheads in lower panels show P23H opsin dimers resolved in 10% gels. Open arrow, P23H opsin monomer. Differing from WT opsin (left), PNGase F-treated P23H opsin shifted only ∼4 kDa relative to Endo H-treated retinal homogenate. This ∼4-kDa difference arose from two N-acetylglucosamines left at the glycosylation sites. Thus, glycosylation of P23H opsin is Endo H-sensitive.
Subcellular Localization of WT and P23H Opsin
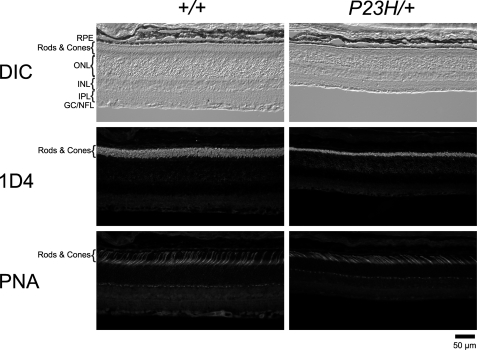
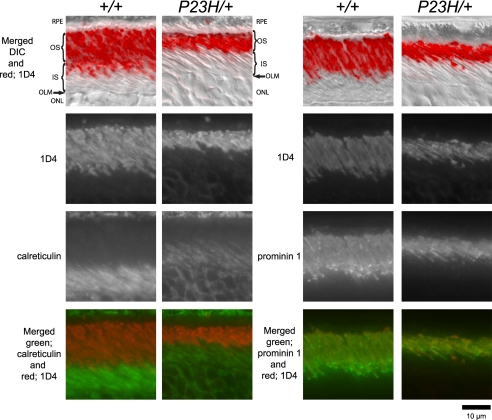
To investigate the subcellular localization of P23H and WT opsin in P23H/+ mice, we used an immunohistochemical procedure to stain retinal cryosections from WT and P23H/+ mice. The C-terminal opsin antibody, 1D4, recognized both the P23H and WT opsin. In WT mice, a strong signal was observed for the ROS, whereas only a weak signal was visible for the inner segments (IS) and ONL (Fig. 8). The signal pattern of 1D4 localization in P23H/+ mice was indistinguishable from that in WT mice, but the ROS in P23H/+ mice were comparatively angular and markedly shortened. The ROS-specific localization was confirmed by double staining with both 1D4 and anti-calreticulin (calreticulin is a chaperone protein that primarily localizes in the ER (58)). In both WT and P23H/+ mice, calreticulin recognized the IS of rod photoreceptor cells where the ER is localized but it did not colocalize with the 1D4 signal. Prominin 1, a membrane protein localized in the OS of rod and cone photoreceptor cells (59), accumulated at the base of ROS where it produced a punctate signal. Moreover, the prominin signal completely overlapped with the strong 1D4 signal in both WT and P23H/+ mice (Fig. 8), indicating that subcellular localization of both P23H and WT opsin was confined to the same compartments.
FIGURE 8.
Subcellular localization of opsin in P23H/+ mice. Retinal cryosections from a PND 35 WT mouse (+/+) and P23H/+ mouse of the same litter were double-stained with antibodies against opsin (1D4) and calreticulin or opsin (1D4) and prominin 1. The 1D4 antibody reacted with both WT and P23H opsin, localizing in the ROS of both WT and P23H/+ mice (1st and 2nd row) and illustrating the markedly shortened angular ROS in P23H/+ animals. The ER marker calreticulin localized to the RPE, inner segments, and ONL (left panel, 3rd row) and did not co-localize with the 1D4 signal (left panel, merged). Prominin 1 localized to both rod and cone OS (right panel, 3rd row). In ROS, prominin 1 accumulated at the base of OS as suggested by the punctuate signals in both WT and P23H/+ mice. The 1D4 signal did colocalize with the prominin 1 signal (right panel, merged). Thus, even though the ROS in P23H/+ mice were shorter, their subcellular localization of opsin was same as in WT mouse. RPE, retinal pigmented epithelium; OS, outer segment; IS, inner segment; OLM, outer limiting membrane; ONL, outer nuclear layer; and DIC, differential interference contrast.
Outer Segment Structures of P23H/+ Mice
Because levels of P23H protein were significantly low and it did not accumulate in the ER of P23H/+ mice, rod photoreceptor cell destruction may have been triggered by something other than accumulation of P23H protein in the ER. Because the integrity of OS structures is critical for survival of photoreceptor cells, disorganization of these structures is associated with photoreceptor cell death and retinal degeneration. To test whether the structures of ROS are affected in P23H/+ mice, we analyzed their retinas by transmission electron microscopy (TEM). Retina from three PND 63 P23H/+ mice showed a characteristic OS disorganization that differed from other types of disorganization reported to date. As indicated by the arrow in Fig. 9B, P23H/+ mice had perpendicularly placed discs. These discs are surrounded by the plasma membrane. This disorganization of discs could destabilize the ROS and impede their contact with the RPE with a negative effect on proper phagocytosis. Because P23H protein is Endo H-sensitive and most of it should be retained in ER, our results suggest expression and transport of only a small fraction of the P23H mutant protein that is not properly glycosylated to the ROS compromises its structure causing photoreceptor degeneration.
FIGURE 9.
Rod outer segment structures in WT and P23H/+ mice. Transmission electron micrographs of ROS from WT (A and A′) and P23H/+ (B–F, B′ and C′) mice. Images were taken from PND 63 littermates. Disorganization of ROS was observed in all three analyzed P23H/+ mice (B, C, and D) as perpendicularly placed discs (arrows) or elongated discs (arrowheads). C, E, and F retinal images were obtained from one mouse to show disorganization at the incisure (C) and base of ROS (E) and both types of disorganization in a single OS (F). Lower magnification images of A–C are shown in A′–C′. Arrows correspond to perpendicularly placed discs. Arrowheads, perpendicularly placed elongated discs. Open arrowheads, vesicles. Open arrow in C, an incisure.
Photoreceptor Cell Death after Genetic Ablation of 11-cis-Retinal
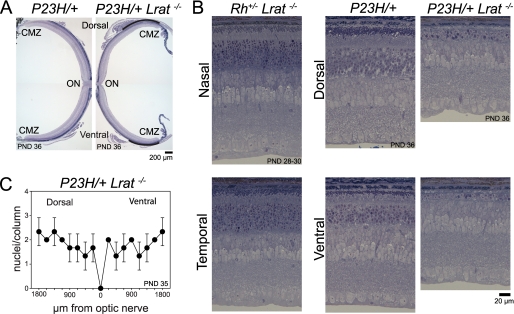
To investigate if 11-cis-retinal generated by the visual cycle affects retinal degeneration in P23H mice, we crossed P23H mice with lecithin-retinol acyltransferase knock-out (Lrat−/−) animals. Lack of lecithin-retinol acyltransferase, one of the key enzymes involved in the retinoid cycle, causes 11-cis-retinal depletion and eventual blindness (60, 61). Compared with P23H/+ mice, lack of 11-cis-retinal in P23H/+Lrat−/− mice caused more severe photoreceptor degeneration. By PND 36, the number of photoreceptor nuclei in these double mutant mice was dramatically reduced in most parts of the retina. Near the ciliary marginal zone reduction was milder (indicated by the curved line in Fig. 10A) but more severe than that seen in P23H/+ mice littermates (Fig. 10, A and B). Significant retinal degeneration in P23H/+Lrat−/− mice at PND36 (Fig. 10B) correlated with results of the TUNEL assay, which recognizes both apoptotic and necrotic cells (62). At PND 14, the ONL thickness was about equal between P23H/+ and P23H/+ Lrat−/− mice, but the TUNEL-positive photoreceptor cell nuclei number in P23H/+Lrat−/− mice was significantly higher than in P23H/+ mice. Lack of 11-cis-retinal did not cause retinal degeneration in 1-month-old mice (Rho+/+Lrat−/−) (31). However, there was a possibility that the severe retinal degeneration in P23H/+Lrat−/− mice resulted from rhodopsin haploinsufficiency. To test this possibility, we analyzed heterozygotes of the opsin knock-out mouse lacking 11-cis-retinal (Rho+/−Lrat−/−). As observed in Rho+/+Lrat−/− mice, the OS in Rho+/−Lrat−/− mice were shortened by PND 28–3, but the number of photoreceptor nuclei per column was normal. Collectively, these results indicate the lack of 11-cis-retinal production during development of ROS increased the toxicity of P23H protein.
FIGURE 10.
Effect of genetic depletion of 11-cis-retinal production on retinal degeneration. A, whole retinal images obtained from PND 36 P23H/+ and P23H/+Lrat−/− littermate mice. Plastic sections were made from dorsal-ventral axis cut and stained with toluidine blue. Both mice were heterozygous with respect to the L450M variation of RPE65 (RPE65450L/450M). The ONL, observed as a purple line in the P23H/+ mouse retina (left panel), was almost gone and only retained near the cilial marginal zone (CMZ) in the P23H/+Lrat−/− mouse (right panel; indicated by curved line). B, high magnification retinal images shown in A and Rho+/−Lrat−/−mice at PND 28–30. Images were taken from the midpoint between optic nerve (ON) and ciliary marginal zone. Genetic deletion of 11-cis-retinal production promoted retinal degeneration in the P23H/+ mouse at PND 36 but not in Rho+/−/Lrat−/− mouse at PND 28–30. In the P23H/+Lrat−/− mouse, however, the ROS were almost undetectable, and the number of photoreceptor nuclei was dramatically reduced compared with a littermate that had 11-cis-retinal (P23H/+). C, statistical data of the number of photoreceptor nuclei per ONL column in P23H/+Lrat−/−. Cryosections from PND35 P23H/+Lrat−/− RPE65450L/450L− mice were used for quantification. Data were derived from three eyes of three mice. P23H/+Lrat−/− mice showed severe retinal degeneration, and numbers of photoreceptor nuclei per column were similar to those in the P23H/P23H mouse (Fig. 3B, PND 35).
Subcellular Localization and Amount of Opsin after Genetic Ablation of 11-cis-Retinal
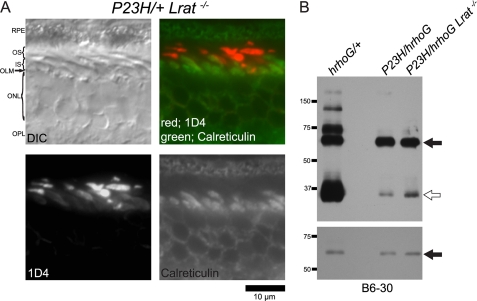
In contrast to P23H transgenic Xenopus (63, 64) and cultured cell lines (65), no opsin accumulation was observed in the rod cell ER of PND 35 P23H/+ mice that exhibited substantial loss of ROS (Fig. 8). Different from P23H/+ mice, retinal degeneration was so severe in P23H/+Lrat−/− mice that rows of rod photoreceptors were less than half those of P23H/+ mice at 1 month of age (Fig. 10C). However, few ROS did survive, and opsin was localized in shortened ROS; mislocalization to either IS or ONL was not obvious (Fig. 11A, 1D4 label). After increasing imaging sensitivity, a faint opsin signal was seen in rod IS, but this signal was significantly less than that in the OS that appeared saturated at the higher imaging sensitivity needed to detect the IS signal (Fig. 11A, 1D4 label). The opsin signal in IS of WT retina was also observed when the imaging sensitivity was increased (data not shown). Thus, photoreceptor degeneration accelerated by the lack of 11-cis-retinal did not result from opsin accumulation in the ER. Because 11-cis-retinals are molecular chaperons of P23H opsin in cultured cells (10, 12), our result suggests that lack of 11-cis-retinal increases P23H toxicity by increasing the P23H protein level. To test this, we analyzed the P23H protein ratio between GFP-tagged human opsin and P23H opsin by immunoblotting at an early stage of photoreceptor degeneration at PND 14. At this age, mature ROS were just formed (66), and the majority of photoreceptor cells were still present, and TUNEL-positive photoreceptor cell nuclei were abundant in the P23H/+Lrat−/− mice (data not shown). The signal intensity of the P23H protein (Fig. 11B, lane P23H/hrhoG Lrat−/−, open arrow) was significantly lower than GFP-tagged human opsin (Fig. 11B, lane P23H/hrhoG Lrat−/−, arrow) even though lecithin-retinol acyltransferase was absent. The signal intensity of P23H opsin (Fig. 11B, lane P23H/hrhoG Lrat−/−, open arrow) in P23H/hrhoG Lrat−/− was higher than that of P23H/hrhoG (Fig. 11B, lane of P23H/hrhoG, open arrow), but signal intensity of GFP-tagged human opsin (∼70 kDa) in P23H/hrhoG Lrat−/− was also higher than that of P23H/hrhoG (lower panel, indicated by arrow). Thus, as in P23H/hrhoG mice, the P23H protein was significantly lower than that of GFP-tagged human opsin in the absence of 11-cis-retinal. Different from the P23H protein, the signal intensity of WT opsin (Fig. 11B, ∼35 kDa, lane hrhoG/+, open arrow) was higher than that of the GFP-tagged human opsin (Fig. 11B, lane hrhoG/+, arrow). Taken together, these findings indicate that the increased toxicity of P23H protein lacking 11-cis-retinal did not result from either an increased level of P23H protein in the ROS or its ER accumulation.
FIGURE 11.
Subcellular localization and protein levels of opsin after genetic deletion of 11-cis-retinal production in Lrat−/− mice. A, cryosections of retina from P23H/+Lrat−/− mice were subjected to immunohistochemical analyses with antibodies against opsin (1D4) and calreticulin at PND 35. Even though massive photoreceptor degeneration is seen, subcellular localization of opsin was normal. B, retinal homogenates from mice at PND 14 were labeled with N-terminal (B6-30) opsin antibodies. All lanes were loaded with 2 μg of homogenate. hrhoG/+ denotes the sample from a heterozygous GFP-tagged human knock-in opsin (hrhoG) and endogenous WT opsin mouse. P23H/hrhoG, heterozygous hrhoG and P23H mouse. P23H/hrhoG Lrat−/−; a heterozygous hrhoG and P23H knock-in mouse lacking lecithin-retinol acyltransferase. GFP-tagged human opsin (∼70 kDa) is indicated by an arrow. Opsin monomers (∼35 kDa) are indicated by an open arrow. Top panel, 4-min exposure was used to analyze the ratio between P23H opsin or WT opsin and GFP-tagged human opsin. Lower panel, 1-s exposure was used to analyze the signal intensity of GFP-tagged human opsin in each lane. The P23H protein level was significantly lower than GFP-tagged human opsin even though 11-cis-retinal production was genetically depleted. DIC, differential interference contrast. RPE, retinal pigmented epithelium; OS, outer segment; IS, inner segment; OLM, outer limiting membrane; ONL, outer nuclear layer; OPL, outer plexiform layer.
DISCUSSION
Human P23H Mutant adRP, Photoreceptor Structure and Natural History
Mutations in the opsin gene constitute one of the most common molecular causes of human RP accounting for nearly 10% of all cases in North America (67). Moreover, P23H is, by far, the most common opsin mutation (8, 68). We investigated the P23H-induced disease with modern techniques to improve understanding of its expression, retinal distribution, and rate of progression. These analyses in patients also provide a standard for comparison with the phenotype of the P23H mutant mouse model being developed. We found a spectrum of disease severity (Fig. 1A) as had previously been well documented (25, 52, 69–72). Longitudinal data (some obtained over decades) were consistent with the hypothesis that patients exhibited variable ages of disease onset followed by invariant rates of disease progression (Fig. 1B). Although versions of this hypothesis have previously been tested in other hereditary retinal degenerations (46–51), this is the first such approach to the disease caused by P23H mutations. Intermediate stages of this retinopathy showed an intraretinal gradient of disease with an altitudinal spatial pattern preferentially affecting the inferior retinal region (25, 70–73).
The human P23H disease showed greater functional deficits of the rod than the cone system (Fig. 1C) (52, 72) as can be expected from the selective expression of rhodopsin in rod photoreceptors. Importantly, we demonstrated that some patients could have normal rod function, at least in some regions of the retina and at some early stages of disease (Fig. 1C). The latter finding implied the existence of a pre-degenerative state of the mutant retina with normal numbers of rod photoreceptors and normal rod outer segments. To test this hypothesis, we examined in vivo the human retinal architecture of representative mutant retinas with high resolution methods (27) and with special emphasis on the OS layer, extending previous work in normal subjects (30). Rod and cone photoreceptor structure could be normal, as determined by the thickness of the ONL and ROS and COS layers (Fig. 1D). The limited post-mortem retinal histopathology from patients with adRP due to P23H opsin mutations (74–76) has largely been from severely affected retinas. Consistent with our noninvasive results, the donor eye with the mildest disease expression had retained rod and cone nuclei, as well as retained ROS and COS (76).
Modeling the Human Disease in Experimental Cell Lines, Transgenic Animal Models, and the P23H Rhodopsin Knock-in Mouse
For this study, we generated novel P23H rhodopsin knock-in mice to model the disease in human patients (52, 72). These mice recapitulated the human retinopathy and showed greater functional deficits of the rod photoreceptor system as compared with the cone system (Fig. 6). There was a strong intraretinal gradient of degeneration in P23H knock-in mice with greater involvement of the inferior retina (Figs. 3, 5, and 10), as noted in the human phenotype (Fig. 1) (25, 27, 70, 71, 77).
There is a long history of attempting to model P23H opsin human adRP. Transgenic rodents (17, 18, 78) have differed in retinal disease distribution. Results in the P23H transgenic mice that carried additional two point mutations have shown greater involvement of the inferior retina (79), whereas P23H transgenic rats showed greater involvement of superior retina (78). Models have also included in vitro P23H opsin-transfected cultured cells (10, 12, 80, 81) and transgenic Xenopus laevis (63). In P23H opsin-transfected cultured cells, P23H opsin accumulated and formed aggregates in the ER (10, 12, 80, 81), and in transgenic X. laevis, this mutant opsin accumulated in the ER of rod photoreceptor cell IS as well (63). But in two transgenic rodent models, there was no evidence of abnormal localization of the mutant opsin (82, 83), although in another transgenic mouse there was evidence for rhodopsin missorting (20). Some investigators considered that abnormal accumulation of P23H opsin initiated ER-induced cellular apoptosis through the unfolded protein response (21). Whether tissue culture experiments accurately mimic human retinal conditions and how the relative dosage of the WT and mutant opsin affects ROS integrity and retinal pathology in transgenic animal models have been topics of debate. Uncertainty about conclusions derived from the previous models fueled our interest in examining the mechanistic details of this mutation in the P23H opsin knock-in mice.
Pathology and Cell Biological Dysfunction in P23H Opsin Knock-in Mice
We found that the mouse P23H protein was inadequately glycosylated with levels between 1 and 10% that of WT opsin. Because the consensus sequence for glycosylation at Asn15 is at a distance from P23H, and because Pro23 is at the center of a chromophore cap formed by five distorted β-strands (strands three to five are formed by residues from Phe13 to Pro34 and strand 4 connects Ser14–Asn15 in the N-terminal region of the molecule with Pro23), it seems reasonable to speculate that a significant change in the structure of the extracellular (intradiscal) cap for P23H opsin fundamentally affects glycosylation of Asn15 and not recognition of a consensus sequence for glycosylation. Moreover, the P23H protein did not accumulate in the endoplasmic reticulum, as described in some transgenic rodent opsin-mutant models, but instead it disrupted the rod photoreceptor disks. The other noteworthy characteristic of this structural disorganization was the perpendicularly oriented elongated discs (Fig. 9, arrowheads). This structural anomaly is similar to the described disorganization caused by defects in the OS structural microtubular protein (84), prominin 1 (59) and GARP (85). Such mice also featured perpendicularly oriented elongated discs not surrounded by the plasma membrane, considered to be caused by incomplete membrane fusion of disc precursors during disc morphogenesis (59, 85). Different from those mice, however, the elongated discs in P23H/+ mice were surrounded by plasma membrane. So how might the P23H/+ disorganization have arisen? As indicated by in Fig. 9 (open arrowheads), vesicles were frequently observed next to the disorganized discs. This type of vesicle had previously been noted at the incisure of a disc in horizontal sections (86). In fact, one of the elongated discs in a P23H/+ mouse retina (Fig. 9C, arrowheads) was also located next to an incisure (Fig. 9C, open arrow). These data indicate that photoreceptors may have degenerated due to disorganization of OS at the incisure. It is intriguing to speculate that some of the abnormal cross-sectional retinal images from human P23H disease (for example, the abrupt loss of the signal from outer segments in Fig. 1D) may be due to change in back-scatter characteristics of abnormally oriented photoreceptor discs.
New Therapeutic Approaches to Combat P23H Retinopathy
Generated P23H opsin knock-in mice appear to be the most suitable model to date for further study of human adRP retinal pathology and disease mechanisms and for proof-of-concept studies of potential therapies. We found that most of P23H protein was degraded, and its retinal cytotoxicity was enhanced by lack of the 11-cis-retinal chromophore. Ideally, stabilization of the mutant opsin with chromophore, chromophore analogs, and nonretinoid compounds appears to be the most straightforward preventative strategy. In cultured cells, the P23H opsin partially regenerates in the presence of 11-cis-retinal (11, 87), and treatment with 9-cis-retinal or 11-cis-7-ring retinal promotes appropriate trafficking of this mutant opsin to the plasma membrane (10, 12). Although it is not known if the P23H opsin actually regenerates normally with 11-cis-retinal in the retina, the relationship between light exposure and retinal degeneration in humans (70), animal models (88, 89), and vitamin A-depleted transgenic Xenopus (90) indicates that 11-cis-retinal might protect against the P23H mutant-induced retinal degeneration (91).
Acknowledgments
We thank Dr. Hisashi Fujioka (Case Western University) for helpful instruction and suggestions for sample preparation and TEM imaging, and Kiet Luc and Hitomi Midori (Case Western University) for their technical assistance with TEM sample preparation and printing of negatives. We also thank Dr. John H. Wilson (Baylor College of Medicine) for providing us with human opsin-GFP fusion knock-in mice, Dr. Janis Lem (Tufts University) for opsin knock-out mice, Cristina Mullins and Melani Oliveras (Scheie Eye Institute) for data analyses, Denis Corbeil (Max-Planck-Institute, Dresden, Germany) for anti-prominin 1 antibodies, and David Salom (Polgenix, Cleveland, OH) for 1D4 and B6-30 antibodies. We also thank Dr. L. T. Webster, Jr. (Case Western Reserve University) for comments and technical support.
This work was supported, in whole or in part, by National Institutes of Health Grants K08EY019880, K08EY019880, EY019031, EY009339, R24 EY021126, and P30 EY11373. This work was also supported by the Foundation Fighting Blindness.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and data 1 and 2 and additional references.
- RP
- retinitis pigmentosa
- adRP
- autosomal dominant RP
- COS
- cone outer segment
- ERG
- electroretinogram
- PND
- postnatal day ages
- ROS
- rod outer segment
- OS
- outer segment
- RPE
- retinal pigmented epithelium
- RPE65
- retinal pigmented epithelium-specific protein 65 kDa (retinoid isomerase)
- TEM
- transmission electron microscopy
- Endo H
- endoglycosidase H
- PNGase F
- peptide:N-glycosidase F
- ER
- endoplasmic reticulum
- cd
- candela
- LRP
- longitudinal reflectivity profile
- ONL
- outer nuclear layer
- IS
- inner segment
- INL
- inner nuclear layer
- PNA
- peanut agglutinin lectin.
REFERENCES
- 1. Farrar G. J., Kenna P. F., Humphries P. (2002) EMBO J 21, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palczewski K. (2006) Annu. Rev. Biochem. 75, 743–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rattner A., Sun H., Nathans J. (1999) Annu. Rev. Genet. 33, 89–131 [DOI] [PubMed] [Google Scholar]
- 4. Dryja T. P., Li T. (1995) Hum. Mol. Genet. 4, 1739–1743 [DOI] [PubMed] [Google Scholar]
- 5. Hartong D. T., Berson E. L., Dryja T. P. (2006) Lancet 368, 1795–1809 [DOI] [PubMed] [Google Scholar]
- 6. Dryja T. P., McGee T. L., Reichel E., Hahn L. B., Cowley G. S., Yandell D. W., Sandberg M. A., Berson E. L. (1990) Nature 343, 364–366 [DOI] [PubMed] [Google Scholar]
- 7. Chapple J. P., Cheetham M. E. (2003) J. Biol. Chem. 278, 19087–19094 [DOI] [PubMed] [Google Scholar]
- 8. Sohocki M. M., Daiger S. P., Bowne S. J., Rodriquez J. A., Northrup H., Heckenlively J. R., Birch D. G., Mintz-Hittner H., Ruiz R. S., Lewis R. A., Saperstein D. A., Sullivan L. S. (2001) Hum. Mutat. 17, 42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noorwez S. M., Malhotra R., McDowell J. H., Smith K. A., Krebs M. P., Kaushal S. (2004) J. Biol. Chem. 279, 16278–16284 [DOI] [PubMed] [Google Scholar]
- 10. Noorwez S. M., Kuksa V., Imanishi Y., Zhu L., Filipek S., Palczewski K., Kaushal S. (2003) J. Biol. Chem. 278, 14442–14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaushal S., Khorana H. G. (1994) Biochemistry 33, 6121–6128 [DOI] [PubMed] [Google Scholar]
- 12. Saliba R. S., Munro P. M., Luthert P. J., Cheetham M. E. (2002) J. Cell Sci. 115, 2907–2918 [DOI] [PubMed] [Google Scholar]
- 13. Illing M. E., Rajan R. S., Bence N. F., Kopito R. R. (2002) J. Biol. Chem. 277, 34150–34160 [DOI] [PubMed] [Google Scholar]
- 14. Rajan R. S., Kopito R. R. (2005) J. Biol. Chem. 280, 1284–1291 [DOI] [PubMed] [Google Scholar]
- 15. Lewin A. S., Drenser K. A., Hauswirth W. W., Nishikawa S., Yasumura D., Flannery J. G., LaVail M. M. (1998) Nat. Med. 4, 967–971 [DOI] [PubMed] [Google Scholar]
- 16. Ranchon I., LaVail M. M., Kotake Y., Anderson R. E. (2003) J. Neurosci. 23, 6050–6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Olsson J. E., Gordon J. W., Pawlyk B. S., Roof D., Hayes A., Molday R. S., Mukai S., Cowley G. S., Berson E. L., Dryja T. P. (1992) Neuron 9, 815–830 [DOI] [PubMed] [Google Scholar]
- 18. Naash M. I., Hollyfield J. G., al-Ubaidi M. R., Baehr W. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5499–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White D. A., Fritz J. J., Hauswirth W. W., Kaushal S., Lewin A. S. (2007) Invest. Ophthalmol. Vis. Sci. 48, 1942–1951 [DOI] [PubMed] [Google Scholar]
- 20. Roof D. J., Adamian M., Hayes A. (1994) Invest. Ophthalmol. Vis. Sci. 35, 4049–4062 [PubMed] [Google Scholar]
- 21. Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., Walter P. (2007) Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacobson S. G., Yagasaki K., Feuer W. J., Román A. J. (1989) Exp. Eye Res. 48, 679–691 [DOI] [PubMed] [Google Scholar]
- 23. Jacobson S. G., Roman A. J., Aleman T. S., Sumaroka A., Herrera W., Windsor E. A., Atkinson L. A., Schwartz S. B., Steinberg J. D., Cideciyan A. V. (2010) Invest. Ophthalmol. Vis. Sci. 51, 1079–1085 [DOI] [PubMed] [Google Scholar]
- 24. Roman A. J., Schwartz S. B., Aleman T. S., Cideciyan A. V., Chico J. D., Windsor E. A., Gardner L. M., Ying G. S., Smilko E. E., Maguire M. G., Jacobson S. G. (2005) Exp. Eye Res. 80, 259–272 [DOI] [PubMed] [Google Scholar]
- 25. Cideciyan A. V., Hood D. C., Huang Y., Banin E., Li Z. Y., Stone E. M., Milam A. H., Jacobson S. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7103–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jacobson S. G., Cideciyan A. V., Iannaccone A., Weleber R. G., Fishman G. A., Maguire A. M., Affatigato L. M., Bennett J., Pierce E. A., Danciger M., Farber D. B., Stone E. M. (2000) Invest. Ophthalmol. Vis. Sci. 41, 1898–1908 [PubMed] [Google Scholar]
- 27. Aleman T. S., Cideciyan A. V., Sumaroka A., Windsor E. A., Herrera W., White D. A., Kaushal S., Naidu A., Roman A. J., Schwartz S. B., Stone E. M., Jacobson S. G. (2008) Invest. Ophthalmol. Vis. Sci. 49, 1580–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jacobson S. G., Aleman T. S., Sumaroka A., Cideciyan A. V., Roman A. J., Windsor E. A., Schwartz S. B., Rehm H. L., Kimberling W. J. (2009) Invest. Ophthalmol. Vis. Sci. 50, 1886–1894 [DOI] [PubMed] [Google Scholar]
- 29. Maeda T., Cideciyan A. V., Maeda A., Golczak M., Aleman T. S., Jacobson S. G., Palczewski K. (2009) Hum. Mol. Genet. 18, 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Srinivasan V. J., Monson B. K., Wojtkowski M., Bilonick R. A., Gorczynska I., Chen R., Duker J. S., Schuman J. S., Fujimoto J. G. (2008) Invest. Ophthalmol. Vis. Sci. 49, 1571–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Batten M. L., Imanishi Y., Maeda T., Tu D. C., Moise A. R., Bronson D., Possin D., Van Gelder R. N., Baehr W., Palczewski K. (2004) J. Biol. Chem. 279, 10422–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Batten M. L., Imanishi Y., Tu D. C., Doan T., Zhu L., Pang J., Glushakova L., Moise A. R., Baehr W., Van Gelder R. N., Hauswirth W. W., Rieke F., Palczewski K. (2005) PLoS Med. 2, e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chan F., Bradley A., Wensel T. G., Wilson J. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9109–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lem J., Krasnoperova N. V., Calvert P. D., Kosaras B., Cameron D. A., Nicolò M., Makino C. L., Sidman R. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 736–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pittler S. J., Baehr W. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8322–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim S. R., Fishkin N., Kong J., Nakanishi K., Allikmets R., Sparrow J. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11668–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barthel L. K., Raymond P. A. (1990) J. Histochem. Cytochem. 38, 1383–1388 [DOI] [PubMed] [Google Scholar]
- 38. Sakami S., Etter P., Reh T. A. (2008) Mech. Dev. 125, 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Imanishi Y., Sun W., Maeda T., Maeda A., Palczewski K. (2008) J. Biol. Chem. 283, 25091–25102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carter-Dawson L. D., LaVail M. M. (1979) J. Comp. Neurol. 188, 263–272 [DOI] [PubMed] [Google Scholar]
- 41. Carter-Dawson L. D., LaVail M. M. (1979) J. Comp. Neurol. 188, 245–262 [DOI] [PubMed] [Google Scholar]
- 42. Hodges R. S., Heaton R. J., Parker J. M., Molday L., Molday R. S. (1988) J. Biol. Chem. 263, 11768–11775 [PubMed] [Google Scholar]
- 43. Adamus G., Zam Z. S., Arendt A., Palczewski K., McDowell J. H., Hargrave P. A. (1991) Vision Res. 31, 17–31 [DOI] [PubMed] [Google Scholar]
- 44. Maeda A., Maeda T., Imanishi Y., Kuksa V., Alekseev A., Bronson J. D., Zhang H., Zhu L., Sun W., Saperstein D. A., Rieke F., Baehr W., Palczewski K. (2005) J. Biol. Chem. 280, 18822–18832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maeda T., Lem J., Palczewski K., Haeseleer F. (2005) Invest. Ophthalmol. Vis. Sci. 46, 4320–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Berson E. L., Sandberg M. A., Rosner B., Birch D. G., Hanson A. H. (1985) Am. J. Ophthalmol. 99, 240–251 [DOI] [PubMed] [Google Scholar]
- 47. Birch D. G., Anderson J. L., Fish G. E. (1999) Ophthalmology 106, 258–268 [DOI] [PubMed] [Google Scholar]
- 48. Iannaccone A., Kritchevsky S. B., Ciccarelli M. L., Tedesco S. A., Macaluso C., Kimberling W. J., Somes G. W. (2004) Invest. Ophthalmol. Vis. Sci. 45, 784–792 [DOI] [PubMed] [Google Scholar]
- 49. Herrera W., Aleman T. S., Cideciyan A. V., Roman A. J., Banin E., Ben-Yosef T., Gardner L. M., Sumaroka A., Windsor E. A., Schwartz S. B., Stone E. M., Liu X. Z., Kimberling W. J., Jacobson S. G. (2008) Invest. Ophthalmol. Vis. Sci. 49, 2651–2660 [DOI] [PubMed] [Google Scholar]
- 50. Cideciyan A. V., Swider M., Aleman T. S., Tsybovsky Y., Schwartz S. B., Windsor E. A., Roman A. J., Sumaroka A., Steinberg J. D., Jacobson S. G., Stone E. M., Palczewski K. (2009) Hum. Mol. Genet. 18, 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Massof R., Finkelstein D. (1987) in A Two-stage Hypothesis for the Natural Course of Retinitis Pigmentosa (Zrenner E., Krastel H., Goebel H.-H., eds) Vol. 62, pp. 29–58, Pergamon Press Ltd., Oxford [Google Scholar]
- 52. Berson E. L., Rosner B., Weigel-DiFranco C., Dryja T. P., Sandberg M. A. (2002) Invest. Ophthalmol. Vis. Sci. 43, 3027–3036 [PubMed] [Google Scholar]
- 53. Oh K. T., Longmuir R., Oh D. M., Stone E. M., Kopp K., Brown J., Fishman G. A., Sonkin P., Gehrs K. M., Weleber R. G. (2003) Am. J. Ophthalmol. 136, 306–313 [DOI] [PubMed] [Google Scholar]
- 54. Curcio C. A., Sloan K. R., Kalina R. E., Hendrickson A. E. (1990) J. Comp. Neurol. 292, 497–523 [DOI] [PubMed] [Google Scholar]
- 55. Szél A., von Schantz M., Röhlich P., Farber D. B., van Veen T. (1993) Invest. Ophthalmol. Vis. Sci. 34, 3641–3645 [PubMed] [Google Scholar]
- 56. Hao W., Wenzel A., Obin M. S., Chen C. K., Brill E., Krasnoperova N. V., Eversole-Cire P., Kleyner Y., Taylor A., Simon M. I., Grimm C., Remé C. E., Lem J. (2002) Nat. Genet. 32, 254–260 [DOI] [PubMed] [Google Scholar]
- 57. Kerr J. F., Wyllie A. H., Currie A. R. (1972) Br. J. Cancer 26, 239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Clementi E., Racchetti G., Zacchetti D., Panzeri M. C., Meldolesi J. (1992) Eur. J. Neurosci. 4, 944–953 [DOI] [PubMed] [Google Scholar]
- 59. Yang Z., Chen Y., Lillo C., Chien J., Yu Z., Michaelides M., Klein M., Howes K. A., Li Y., Kaminoh Y., Chen H., Zhao C., Chen Y., Al-Sheikh Y. T., Karan G., Corbeil D., Escher P., Kamaya S., Li C., Johnson S., Frederick J. M., Zhao Y., Wang C., Cameron D. J., Huttner W. B., Schorderet D. F., Munier F. L., Moore A. T., Birch D. G., Baehr W., Hunt D. M., Williams D. S., Zhang K. (2008) J. Clin. Invest. 118, 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McBee J. K., Palczewski K., Baehr W., Pepperberg D. R. (2001) Prog. Retin Eye Res. 20, 469–529 [DOI] [PubMed] [Google Scholar]
- 61. von Lintig J., Kiser P. D., Golczak M., Palczewski K. (2010) Trends Biochem. Sci. 35, 400–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. de Torres C., Munell F., Ferrer I., Reventós J., Macaya A. (1997) Neurosci. Lett. 230, 1–4 [DOI] [PubMed] [Google Scholar]
- 63. Tam B. M., Moritz O. L. (2006) Invest. Ophthalmol. Vis. Sci. 47, 3234–3241 [DOI] [PubMed] [Google Scholar]
- 64. Tam B. M., Moritz O. L. (2007) J. Neurosci. 27, 9043–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sung C. H., Schneider B. G., Agarwal N., Papermaster D. S., Nathans J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8840–8844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Obata S., Usukura J. (1992) Cell Tissue Res. 269, 39–48 [DOI] [PubMed] [Google Scholar]
- 67. Seyedahmadi B. J., Rivolta C., Keene J. A., Berson E. L., Dryja T. P. (2004) Exp. Eye Res. 79, 167–173 [DOI] [PubMed] [Google Scholar]
- 68. Dryja T. P., McEvoy J. A., McGee T. L., Berson E. L. (2000) Invest. Ophthalmol. Vis. Sci. 41, 3124–3127 [PubMed] [Google Scholar]
- 69. Berson E. L., Rosner B., Sandberg M. A., Dryja T. P. (1991) Arch. Ophthalmol. 109, 92–101 [DOI] [PubMed] [Google Scholar]
- 70. Heckenlively J. R., Rodriguez J. A., Daiger S. P. (1991) Arch. Ophthalmol. 109, 84–91 [DOI] [PubMed] [Google Scholar]
- 71. Stone E. M., Kimura A. E., Nichols B. E., Khadivi P., Fishman G. A., Sheffield V. C. (1991) Ophthalmology 98, 1806–1813 [DOI] [PubMed] [Google Scholar]
- 72. Kemp C. M., Jacobson S. G., Roman A. J., Sung C. H., Nathans J. (1992) Am. J. Ophthalmol. 113, 165–174 [DOI] [PubMed] [Google Scholar]
- 73. Cideciyan A. V., Jacobson S. G., Aleman T. S., Gu D., Pearce-Kelling S. E., Sumaroka A., Acland G. M., Aguirre G. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5233–5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kolb H., Gouras P. (1974) Invest. Ophthalmol. 13, 487–498 [PubMed] [Google Scholar]
- 75. John S. K., Smith J. E., Aguirre G. D., Milam A. H. (2000) Mol. Vis. 6, 204–215 [PubMed] [Google Scholar]
- 76. To K., Adamian M., Dryja T. P., Berson E. L. (2002) Am. J. Ophthalmol. 134, 290–293 [DOI] [PubMed] [Google Scholar]
- 77. Jacobson S. G., Kemp C. M., Sung C. H., Nathans J. (1991) Am. J. Ophthalmol. 112, 256–271 [DOI] [PubMed] [Google Scholar]
- 78. LaVail M. M., Yasumura D., Matthes M. T., Drenser K. A., Flannery J. G., Lewin A. S., Hauswirth W. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11488–11493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Naash M. L., Peachey N. S., Li Z. Y., Gryczan C. C., Goto Y., Blanks J., Milam A. H., Ripps H. (1996) Invest. Ophthalmol. Vis. Sci. 37, 775–782 [PubMed] [Google Scholar]
- 80. Sung C. H., Davenport C. M., Hennessey J. C., Maumenee I. H., Jacobson S. G., Heckenlively J. R., Nowakowski R., Fishman G., Gouras P., Nathans J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 6481–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rajan R. S., Illing M. E., Bence N. F., Kopito R. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13060–13065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu X., Wu T. H., Stowe S., Matsushita A., Arikawa K., Naash M. I., Williams D. S. (1997) J. Cell Sci. 110, 2589–2597 [DOI] [PubMed] [Google Scholar]
- 83. Green E. S., Menz M. D., LaVail M. M., Flannery J. G. (2000) Invest. Ophthalmol. Vis. Sci. 41, 1546–1553 [PubMed] [Google Scholar]
- 84. Vaughan D. K., Fisher S. K. (1989) Invest. Ophthalmol. Vis. Sci. 30, 339–342 [PubMed] [Google Scholar]
- 85. Zhang Y., Molday L. L., Molday R. S., Sarfare S. S., Woodruff M. L., Fain G. L., Kraft T. W., Pittler S. J. (2009) J. Cell Sci. 122, 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cohen A. I. (1960) Am. J. Anat. 107, 23–48 [DOI] [PubMed] [Google Scholar]
- 87. Liu X., Garriga P., Khorana H. G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4554–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Vaughan D. K., Coulibaly S. F., Darrow R. M., Organisciak D. T. (2003) Invest. Ophthalmol. Vis. Sci. 44, 848–855 [DOI] [PubMed] [Google Scholar]
- 89. Walsh N., van Driel D., Lee D., Stone J. (2004) Brain Res. 1013, 194–203 [DOI] [PubMed] [Google Scholar]
- 90. Tam B. M., Qazalbash A., Lee H. C., Moritz O. L. (2010) Invest. Ophthalmol. Vis. Sci. 51, 1327–1334 [DOI] [PubMed] [Google Scholar]
- 91. Palczewski K. (2010) Trends Pharmacol. Sci. 31, 284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Science 289, 739–745 [DOI] [PubMed] [Google Scholar]